Abstract
The Cardiotoxicity of Chemotherapy Knowledgebase (CATCH-KB) contains information extracted from articles investigating an association between germline genetic polymorphisms and the development of chemotherapy-induced cardiotoxicity (CIC) in cancer patients receiving antineoplastic treatments. CATCH-KB also contains integrated gene and drug information from open biomedical resources such as PharmGKB1 and SIDER2. Furthermore, the genetic polymorphisms, drugs, and cancer types detailed in CATCH-KB are standardized according to appropriate biomedical ontologies, such as SNOMED-CT3 and RxNorm4. CATCH-KB currently contains information on 49 research papers published between 2004 and 2017 investigating a total of 2,210 variants in over 280 genes for an association with CIC. By centralizing this information and linking it to external open-access biomedical resources, CATCH-KB will facilitate hypothesis generation and meta-analysis efforts and will ultimately accelerate the use of genetic screening in preventing CIC. CATCH-KB is publicly accessible via http://catchkb.org.
Introduction
Chemotherapy-induced cardiotoxicity (CIC) is a broad term encompassing cardiac complications such as ischemia, arrhythmia, hypertension, and heart failure5. Cardiotoxicity has been primarily observed as a side effect of anthracyclines, but is also implicated in treatment with targeted cancer therapies such as tyrosine kinase inhibitors (e.g., imatinib) and antibody therapies (e.g., trastuzumab). These are very commonly used cancer therapies, but cardiac complications cause the cessation of these otherwise effective and life-saving treatments. Estimates of the prevalence of cardiotoxicity vary widely, but a meta-analysis of anthracycline-related cardiotoxicity showed that 6.3% of patients developed clinically overt heart failure and 17.9% developed subclinical cardiotoxicity5. Reports7, 8 have also shown that the incidence of cardiotoxicity is increased when trastuzumab is used in combination with anthracycline-based chemotherapy for the treatment of HER2-positive breast cancer. Although this regimen has reduced the risk of all-cause death by 33%9-11, up to 4%10-12 of patients will develop severe congestive heart failure and up to 23% of patients will develop a significant decline in left ventricular ejection fraction (LVEF). CIC is of great interest as it not only interferes with the delivery of curative cancer therapy, but also results in cardiovascular-related morbidity and mortality, yet its mechanisms are still not completely understood. While acute cardiotoxicity presents immediately or within a year of treatment, late cardiotoxicity does not manifest until a year or more after treatment, in some instances as much as a decade later13. Thus, with increased cancer survival rates, the number of cancer survivors affected by cardiotoxicity is also expected to rise14.
Current monitoring techniques rely mainly on echocardiogram measurements like LVEF to detect a decrease in heart function. As it takes a significant amount of cardiomyocyte damage to induce an observable decrease in LVEF, there is a delay in treating cardiotoxicity, which often leads to suboptimal recovery. With only 42% of anthracycline-induced cardiotoxic patients achieving complete cardiac recovery6, there is a clear need to develop alternative screening techniques to identify at-risk patients, and, where feasible, establish early initiation of potentially cardio-protective therapy. Some risk factors, such as age>65, pre-existing cardiac conditions, and cumulative anthracycline dose have been identified15. However, these risk factors alone cannot accurately identify which patients will develop CIC14. This high degree of unexplained variability in treatment response and side effects indicates that genetics likely play an important role and that genetic testing could provide vital insights into identifying patients at risk for CIC5.
To date, candidate gene association studies attempting to identify genes of interest to CIC a) have examined genes based on our current limited knowledge of the mechanism of CIC, and b) have produced inconsistent results across papers examining the same variant. Thus, potentially cardiotoxic cancer therapies are administered across entire populations which are then monitored for side effects instead of identifying susceptible patients beforehand and tailoring their treatment accordingly.
While many genes have been examined for an association with CIC, to the best of our knowledge, none are currently utilized in patient care. Consolidating existing study data in one centralized knowledge repository will help identify promising variants and accelerate the translation from research to clinical practice. Furthermore, curating details of the design and implementation of the analyses will facilitate comparisons and pattern recognition across studies to further guide research efforts. Finally, through integration with other existing resources, such as the Pharmacogenomics Knowledgebase1 (PharmGKB) and Kyoto Encyclopedia of Genes and Genomes16 (KEGG), a centralized CIC knowledge resource will enable novel hypothesis generation. With these goals in mind, we have created the Cardiotoxicity of Chemotherapy Knowledgebase (CATCH-KB; http://catchkb.org).
Materials and Methods
Development of CATCH-KB:
We performed a literature review of existing papers via PubMed (as of January 2018) that have explored associations between genetic variants and CIC. Our search criteria in PubMed was (“cardiotoxicity”[MeSH Terms] OR “cardiotoxicity”[All Fields]) AND (“genetic therapy”[MeSH Terms] OR (“genetic”[All Fields] AND “therapy”[All Fields]) OR “genetic therapy”[All Fields] OR “genetic”[All Fields] OR “gene”[All Fields] OR “variant”[All Fields]). While we tried other searches, such as “chemotherapy-induced cardiotoxicity” and “anthracycline cardiotoxicity,” this query allowed us to more broadly search for all genes associated with cardiotoxicity. We included only primary research papers investigating how germline variants affected the development of cardiotoxicity related to antineoplastic drugs. This search was supplemented with a manual search of cited references from retrieved articles. Of the 738 results returned, we identified 49 primary research papers of interest done in humans and 50 done in other model organisms that are included in CATCH-KB (Figure 1). All of the human-based papers referenced in recent reviews17, 18 are included in CATCH-KB, leading us to believe our search was exhaustive for analyses done in humans, the main focus of CATCH-KB. The existing data in CATCH-KB for papers on model organisms is preliminary.
Figure 1.
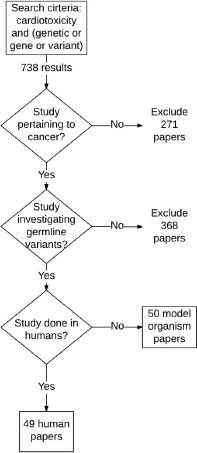
Inclusion criteria for CATCH-KB
In CATCH-KB, information can be queried by PMID, gene ID, or variant rsID for human papers and by gene ID or PMID for model organism papers. Information was manually extracted from papers and, for papers investigating variants in humans, includes details on the genes and variants examined as well as paper details, such as the date of the paper, candidate gene or GWAS approach, any association found, the paper-defined threshold for significance, the drug exposure, the cardiotoxicity definition, the length of follow up, the sequencing platform used, the cohort size, and population details including age, race or ethnicity, and cancer type. For model organism-based papers, the type of animal used, the gene studied, how cardiotoxicity was measured, the drug exposed to, and a description of the association is provided.
The gene name, drug name, and cancer type defined in CATCH-KB are all mapped to standardized biomedical ontologies using application programming interfaces (APIs) from BioPortal19 for Human Genome Organization Gene Nomenclature Committee20 (HGNC),RxNorm,4 and Systematized Nomenclature of Medicine – Clinical Terminology3 (SNOMED-CT). Additionally, the grade of cardiotoxicity is defined according to the Common Terminology Criteria for Adverse Events21 (CTCAE). Gene family information is also taken from HGNC20. We used the BioPortal API to perform an automated pull of the results from the BioPortal class search. Specifically, for gene names we pulled the gene symbol and for the drugs and cancer types, we pulled the “notation” element which corresponds with a single unique RxNorm Concept Unique Identifier (RxCUI) and SNOMED-CT Identifier (ID), respectively. If no value was returned, we manually verified the results by performing a search through BioPortal’s website. For example, when “5-fluorouracil” returned no results, we performed a manual search for “fluorouracil” and retrieved the RxCUI, which we confirmed through a search in RxNav22. 100% of genes mapped to an HGNC-approved gene symbol, 100% of cancer types matched to SNOMED-CT IDs, and 21 of 23 or 91% of drugs mapped to RxNorm – although the other two were mapped via manual review as described above.
This level of data standardization within CATCH-KB helps with data integration by addressing ambiguities that arise when papers use different terms to represent the same concept. Further, data standardization will help draw relevant comparisons between papers and allow their data to be better integrated with existing standardized resources.
Variant information is shown as an rsID. For studies that did not list the rsID, we used tools like PharmGKB1, SNPedia23, and dbSNP24 to identify the corresponding rsIDs. Our analysis indicates that occasionally papers do not report the risk allele for their analysis. CATCH-KB provides risk allele information along with a general description of the association analysis in the “association detail” section. For papers that do not report this information, the risk allele is not available in CATCH-KB. The lack of information from specific studies is in part accommodated in CATCH-KB by providing the user with links to external resources, such as PharmGKB and the National Human Genome Research Institute (NHGRI) and European Bioinformatics Institute (EBI) GWAS Catalog25.
CATCH-KB integrates information from multiple open-access biomedical resources by providing URL links to relevant pages. CATCH-KB links users the relevant PubMed entry for each paper. For each chemotherapy drug examined in the papers included in CATCH-KB, there are links to drug–gene information in PharmGKB1 and to drug warning labels in Side Effect Resource2 (SIDER). Links to association studies summarized in the NHGRI-EBI GWAS Catalog25 and gene pathway information from KEGG16 are also provided for genes described in CATCH-KB. Linking these distributed, heterogeneous resources helps make CATCH-KB users aware of existing information available and provides context on the drugs and genes being detailed. We anticipate that this will facilitate hypothesis generation by making connections to other phenotypes, genes, and drugs that CIC-associated genes interact with.
In order to make CATCH-KB easily accessible and usable for other studies, we developed a web-based platform using the Ruby on Rails Web application framework. Rails interfaces with a relational database by default, enabling us to easily link the MySQL database housing CATCH-KB to the web platform. CATCH-KB uses a simple data model (Figure 2). This facilitates the description of a single paper reporting multiple association experiments with several genes. CATCH-KB’s web interface provides a search functionality with which a user can query the knowledgebase.
Figure 2.
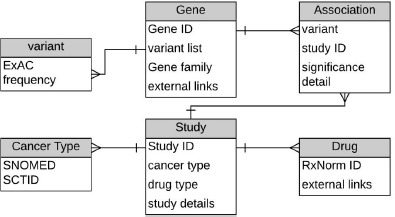
Model of CATCH-KB data structure
Database maintenance:
In order to maintain the quality of information stored in CATCH-KB, we perform automated and manual quality checks. Automated checks determine the completeness of updates to the knowledgebase, requiring certain fields be provided in an update, whether it is a manual input or an automated input from a linked external resource. We are also working to enable a flagging feature for users to notify us of information they believe to be inaccurate.
In order to keep CATCH-KB up to date, we receive a notification each time a new paper that meets our search criteria is released via PubMed. We implemented an administrator portal in CATCH-KB that allows us to manually update the knowledgebase. We continue to add new relevant papers as they are published. As our current listing of model organism-based papers investigating CIC is preliminary, we are expanding it to include more papers and additional information on each paper. In future, we plan to automate the paper curation process using text mining tools to screen through abstracts, identify papers of interest based on natural language processing parameters, and retrieve the desired information for updating CATCH-KB.
Further updates include developing an API to allow users to programmatically retrieve information from CATCH-KB. This will allow information from CATCH-KB to be incorporated into other resources and biomedical applications. For example, at Weill Cornell Medicine, we anticipate that cardiotoxicity information from CATCH-KB can be incorporated into the Precision Medicine Knowledgebase (PMKB; https://pmkb.weill.cornell.edu/)52. Further, our collaborators at the Electronic Medical Records and Genomics (eMERGE) Network53 and the All of Us Research Program54 can use data from CATCH-KB in their on-going efforts to study and implement genomic medicine.
We also plan to expand CATCH-KB into a fully defined service-oriented architecture (SOA) that integrates information from external resources, namely PharmGKB, SIDER, NHGRI-EBI GWAS Catalog, and KEGG, via dynamic APIs as opposed to static linkages. While KEGG has an API that we can use, the remaining resources do not currently have open-access APIs. Thus, implementing the SOA will require more programmatically extensive steps, such as usage of a web crawler to scrape the data. Presenting this information all in one place will allow users of CATCH-KB to more easily draw out patterns by quickly searching for variants and studies associated with the same pathway, drug, or phenotype of interest. Once these API-based linkages are made, we will add additional quality checks to notify us when changes have been made to the externally linked resources. In addition, we plan to incorporate standardized phenotype definitions, such as the ones available from Phenotype Knowledge Base (PheKB)55, in future releases of CATCH-KB.
An example search scenario using CATCH-KB:
We discuss below how a user might interact with CATCH-KB. For example, a search for the gene ID “CELF4” in CATCH-KB would return a list of the reported association information on variants within the CELF4 gene – namely rs1786814 – and the analyses describing its association with CIC (Figure 3). CATCH-KB will report the association statistics for any discovery, replication, and combined (discovery + replication) analyses reported in the respective papers. Links at the bottom of this page allow the user to navigate to pages with details on the relevant gene and paper(s). The gene detail tab shows the HGNC gene family and provides URL links that take the user to the specific page of KEGG, GWAS Catalog, or PharmGKB that displays the results of a search for the gene of interest, in this case CELF4, on the respective site. The paper detail tab (Figure 4) provides information on the paper that tested CELF4 for an association with CIC. The date, cancer type, and cardiotoxicity definition with the corresponding CTCAE level is provided. Additional details about the cohort and any replication populations are also provided, including the length of follow up, sex, age, race, and ethnicity. Further details on SNOMED-CT concepts corresponding to the cancer types are provided by clicking on the “cancer detail” link, and RxCUI, drug class, and additional information pulled from the RxNav22 API is available on the drugs investigated in the paper via the “drug detail” link.
Figure 3.
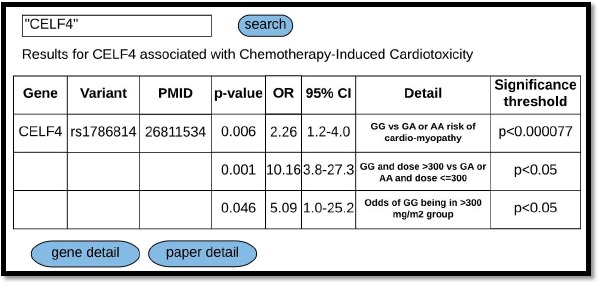
CATCH-KB search results for “CELF4”
Figure 4.
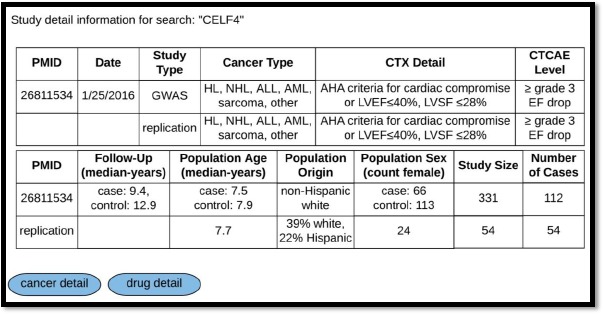
Study detail for “CELF4.” Abbreviations: CTX cardiotoxicity, HL Hodgkin’s lymphoma, NHL non-Hodgkin’s lymphoma, ALL acute lymphoblastic leukemia, AML acute myelogenous leukemia, AHA American Heart Association, LVEF left ventricular ejection fraction, LVSF left ventricular shortening fraction
Results
Statistics:
As of January 2018, CATCH-KB details the findings of 49 papers studying CIC in humans that examine 2,210 SNPs in 281 genes. The vast majority of papers (43 of 49, or 88%) have evaluated anthracycline-related CIC. 30 papers have reported a significant association between a variant and CIC with a total of 80 significant associations reported. The significance of the associations was determined by comparing the reported p-value of the association to the p-value threshold set by the paper. Both p-values along with other reported statistics, such as odds ratio and confidence interval, are presented in CATCH-KB. These 80 associations involve 53 distinct variants in 43 distinct genes. Of the 53 variants, 37 have been reported to be associated with CIC by a single study, 9 have been reported by two separate studies, and 7 have been reported by three or more separate studies. For these purposes, “study” is defined as a unique study population. Thus, the association findings reported for a replication cohort are counted separately from those reported for the discovery cohort, even though they are described within the same paper. 6 of the 7 variants that have been reported to be associated with CIC by 3 or more separate studies – rs1136201, rs1695, rs1883112, rs13058338, rs4673, rs7853758 – have been reported by at least 2 other studies as not being significantly assodated with CIC (Table 1.)
Table 1.
Variants associated with CIC by 3 or more studies
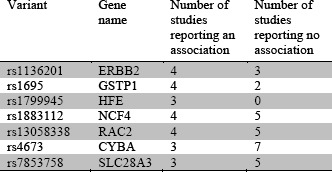
This lack of consistency is likely due to Variant Gene Number of Number of name studies studies small cohort sizes, as most studies are underpowered, especially to detect variants with small effect sizes. The average study cohort size (N) taken across all papers in CATCH-KB is 182.80 (standard deviation: 171.71). The average study cohort size (N) taken across only papers reporting a significant finding is not statistically different – 212.72 (standard deviation: 158.28). This shows us that studies finding significant associations are not simply larger and more powered, but rather studies of similar sizes are yielding inconsistent results. For example, in Roca et al,26 ERBB2 rs1136201 is positively associated with CIC, but Stanton et al27 reports a negative finding. Both papers, however, had similar cohort sizes (132 and 140 subjects, respectively) and were performed in adult females with HER2 positive breast cancer and trastuzumab exposure. This indicates that larger cohort sizes are needed, not only to ensure that studies are adequately powered to find associations, but also to ensure that studies do not detect spurious associations. Larger studies or metaanalyses, such as the recent analysis from Leong et al28, are needed to conclude if associations previously found are valid or not. Of the seven variants listed in Table 1, Leong et al examined rs4673 in CYBA, rs13058338 in RAC2, rs7853758 in SLC28A3, and rs1883112 in NCF4, and found rs4673 and rs13058338 to be significantly associated with CIC, although their study was limited to anthracycline-induced cardiotoxicity.
Some other factors leading to inconsistencies across the studies potentially include lack of replication cohorts within individual papers and differences in study population characteristics. Only seven 13,29-34 of the 49 papers included a replication cohort in which to verify their findings – and only one replication cohort included non-European individuals. Of the 49 studies, only 16 provided the race and ethnicity background of their cohorts while an additional 12 reported the nationality (e.g. German, Italian) of their cohort. CATCH-KB reports this information when such data is provided by the manuscripts. Studies also varied in the age and cancer type of their cohorts as well as the homogeneity of their populations. Some studies26, 34-41 tested a single treatment regimen in a single cancer type within a narrow age range while others had less stringent inclusion criteria. The difference in criteria and in clinical endpoints could also affect the inconsistencies in findings. Finally, since cardiotoxicity can take a decade after treatment to present, studies42-50 performed within shorter windows could be misclassifying cases and controls and diluting potentially significant associations.
Additional tools and resources:
In addition to summarizing the existing research in the field, CATCH-KB is linked to multiple external open-access biomedical resources to facilitate integration of information across different domains and drive hypothesis generation.
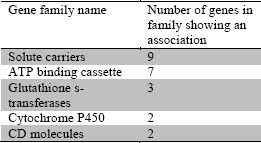
For example, CATCH-KB provides gene family information derived from HGNC. Of the 80 associations reported, a number of them involved genes within the same gene family, as described in Table 2. This may help provide mechanistic explanations for CIC as well as potential candidate genes to investigate as other genes in these families may be good candidates for future examination. With a similar focus in mind, a recent paper by Wells et al51 performed a pathway-based association analysis via KEGG when examining genetic predispositions to CIC. As they point out, a typical single SNP analysis will overlook the compound effect of several variants with small effect sizes that can be highlighted by identifying entire pathways associated with CIC. As it provides links to KEGG as well as other resources, CATCH-KB will facilitate such examinations.
Table 2.
Gene families associated with CIC
Comparison to similar resources:
Our work in establishing CATCH-KB is motivated by the success of existing resources, such as PharmGKB, which provides similar information on the relationship between drugs and variants of interest, and the GWAS Catalog, which lists genes identified from association studies. The specificity of CATCH-KB, however, allows it to include candidate gene studies and provide more detailed study-specific information, such as the cardiotoxicity definition, CTCAE level, and length of follow-up. These and other additional data elements combined with reports from animal papers will better aid researchers in designing both replication and novel CIC gene association studies. Furthermore, CATCH-KB is standardized according to appropriate biomedical ontologies and data standards, which aids in comprehension and integration of studies across different research groups. Additionally, while still preliminary, CATCH-KB includes associations seen in model organisms. The papers described were published between 1997 and 2017. All but one of the 50 papers examined the effects of doxorubicin with doses ranging from 15 to 32 mg/kg. The majority of papers (39 of 50, or 78%) used mice, with others using rats and cell lines. Of the papers reporting the gender of the animals, 65% were done in males only, 13% were restricted to females, and the rest used both genders. Only two of the papers studied animals with cancer; the remainder examined the effects of chemotherapy in animals without cancer. Common metrics of cardiotoxicity were survival, echocardiogram measurements, and morphological cardiac changes assessed via microscopy. Including information from animal papers will help researchers design novel experiments by identifying genes that have been implicated in CIC but not yet studied in humans. Intriguingly, 38 genes studied in model organisms have not yet, to the best of our knowledge, been evaluated in humans. Within CATCH-KB, we also incorporate data from other resources, including PharmGKB and the GWAS Catalog, to provide continuity across the resources.
Discussion
Impact:
By providing specific study details, CATCH-KB will help highlight any underlying patterns, such as variants that seem to be associated with CIC only in certain ages, ethnicities, or in relation to certain drugs or cancer types. This level of detail will also facilitate meta-analyses. CATCH-KB will bring to light promising variants that need further validation as well as variants that have not shown any association with CIC even after several investigations. In this way, CATCH-KB will help to drive research in the field. By providing information from external resources as mentioned above, CATCH-KB will provide context for the genes and drugs found to be associated with CIC, which will promote hypothesis generation. Additional genes in gene pathways and families with members associated with CIC could be candidates for future association studies. Information on a drug or gene being associated with other conditions could similarly provide insight into the mechanism of CIC and potential gene candidates for future examination. Including animal papers in particular will help researchers identify novel candidates for examination in humans.
CATCH-KB user groups
We anticipate that CATCH-KB will be used primarily by pharmacogenomic researchers. It is likely many users will visit CATCH-KB when designing new association studies between polymorphisms and CIC. The association information available in CATCH-KB will help these users identify variants suitable for replication, and the model organism information will help them identify genes to investigate for the first time in humans. Users may be interested in particular genes or drugs, which is why we provide details like the information retrieved from HGNC and RxNorm. Users will likely also utilize CATCH-KB to compare and identify studies for inclusion in meta-analyses. The study-specific details in CATCH-KB will help users understand the context in which the association analyses were done, such as the drugs the subjects were exposed to, how their cardiotoxicity was measured, and how long they were followed for.
Limitations to the field:
The inability of many studies to replicate previously found associations between variants and CIC prohibits the implementation of genetic screening to identify patients at risk of developing CIC. While this failure to replicate associations could be because there is no underlying association, it could also be due to the late onset of CIC or variation in associations across populations of different ages, races, ethnicities, and cancer types. The biggest hindrance to consistent results, however, is the relatively small sample size of studies. As CIC is a complex trait demonstrating many associations with a small effect size, larger studies are required to adequately identify variants of interest. These larger cohorts are hard to create, however. Small sample size is a problem across pharmacogenomics research, as discussed in a recent review56. Sample sizes are limited because only a portion of individuals with a specific condition are treated with the drug of interest, and only a fraction of those cases have the baseline and on-treatment measurements necessary to see the effect of the drug.
While it would be ideal to perform large scale prospective analyses through consortium efforts to address these shortcomings, projects of this scale take large investments of time and resources. Alternatively, efforts such as the NIH-funded Electronic Medical Records and Genomics (eMERGE) Network53, 57 are aimed at the use of electronic health record (EHR)-derived phenotypes in genetic association studies. We anticipate that a knowledge resource, such as CATCH-KB, will be of immense utility for investigators in the eMERGE Network, and other large-scale national consortia conducting research using EHR-linked biobanks. With the recent launch of the All of Us Research Program,54 these EHR-linked biobanks are becoming more prevalent and present a valuable research opportunity.
Finally, while identifying variants that are consistently associated with CIC will be a vital step towards clinical implementation of genetic screening to prevent CIC, candidate genes should be validated in mechanistic experiments. For such purposes, experimental approaches using induced pluripotent stem cells (iPSCs) or CRISPR58 modified lines may prove most useful, especially as an iPSC line was recently established as a reliable model of cardiotoxicity59. Additionally, the vast majority of papers are focused on anthracycline-induced cardiotoxicity. While this is the most prevalent form of cardiotoxicity to date, other therapies, including newer targeted therapies, have also been linked with cardiotoxicity. This cardiotoxicity likely operates through different mechanisms and presents with different clinical endpoints, necessitating further research.
Conclusion
CATCH-KB is an open-source, publicly available knowledgebase that summarizes existing research investigating an association between genetic variants and the development of chemotherapy-induced cardiotoxicity (CIC). It also provides links to information about relevant gene pathways, gene associations with other diseases, and gene–drug associations. Additionally, the information in CATCH-KB is standardized according to appropriate biomedical ontologies to make information in CATCH-KB readily linkable to other knowledgebases using web APIs. CATCH-KB can be accessed via http://catchkb.org.
Acknowledgement
This research was funded in part by R01 GM105688, R01 GM103859, and the Englander Institute for Precision Medicine at Weill Cornell Medicine.
References
- 1.Whirl-Carrillo M, McDonagh E, Hebert J, et al. Pharmacogenomics knowledge for personalized medicine. Clinical Pharmacology & Therapeutics. 2012;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn M, Letunic I, Jensen L, Bork P. The SIDER database of drugs and side effects. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.SNOMED CT & Other Terminologies, Classifications & Code Systems. [Accessed October 31, 2016].
- 4.Nelson S, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. Journal of the American Medical Informatics Association. 2011;18(4):441–448. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown S, Sandhu N, Herrmann J. Systems biology approaches to adverse drug effects: the example of cardio-oncology. Nature Reviews Clinical Oncology. 2015;12(12):718–731. doi: 10.1038/nrclinonc.2015.168. [DOI] [PubMed] [Google Scholar]
- 6.Lotrionte M, Biondi-Zoccai G, Abbate A, et al. Review and meta-analysis of incidence and clinical predictors of anthracycline cardiotoxicity. American Journal of Cardiology. 2013;112(12):1980–1984. doi: 10.1016/j.amjcard.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 7.van Hasselt J, Boekhout A, Beijnen J, Schellens J, Huitema A. Population pharmacokinetic-pharmacodynamic analysis of trastuzumab-associated cardiotoxicity. Clin Pharmacol Ther. 2011;90(1):126–132. doi: 10.1038/clpt.2011.74. [DOI] [PubMed] [Google Scholar]
- 8.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER 2. New England Journal of Medicine. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 9.Piccart-Gebhart M, Procter M, Leyland-Jones B, et al. Trastuzumab after Adjuvant Chemotherapy in HER2-Positive Breast Cancer. New England Journal of Medicine. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 10.Tan-Chiu E, Yothers G, Romond E, et al. Assessment of Cardiac Dysfunction in a Randomized Trial Comparing Doxorubicin and Cyclophosphamide Followed by Paclitaxel, With or Without Trastuzumab As Adjuvant Therapy in Node-Positive, Human Epidermal Growth Factor Receptor 2-Overexpressing Breast Cancer: NSABP B-31. Journal of Clinical Oncology. 2005;23(31):7811–7819. doi: 10.1200/JCO.2005.02.4091. [DOI] [PubMed] [Google Scholar]
- 11.Piotrowski G, Gawor R, Stasiak A, Gawor Z, Potemski P, Banach M. Cardiac complications associated with trastuzumab in the setting of adjuvant chemotherapy for breast cancer overexpressing human epidermal growth factor receptor type 2 – a prospective study. Arch Med Sci. 2012;8(2):227–235. doi: 10.5114/aoms.2012.28549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarantini L, Cioffi G, Gori S, et al. Trastuzumab adjuvant chemotherapy and cardiotoxicity in real-world women with breast cancer. J Card Fail. 2012;18(2):113–119. doi: 10.1016/j.cardfail.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Sun C, Quinones-Lombrana A, et al. CELF4 Variant and Anthracycline-Related Cardiomyopathy: A Children’s Oncology Group Genome-Wide Association Study. J Clin Oncol. 2016;34(8):863–870. doi: 10.1200/JCO.2015.63.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen B, McLeod H. Pharmacogenomics as a risk mitigation strategy for chemotherapeutic cardiotoxicity. Pharmacogenomics. 2013;14(2):205–213. doi: 10.2217/pgs.12.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemieux J, Diorio C, Cote M, et al. Alcohol and HER2 polymorphisms as risk factor for cardiotoxicity in breast cancer treated with trastuzumab. Anticancer Res. 2013;33(6):2569–2576. [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aminkeng F, Ross C, Rassekh S, et al. Recommendations for genetic testing to reduce the incidence of anthracycline-induced cardiotoxicity. Br J Clin Pharmacol. 2016;82(3):683–695. doi: 10.1111/bcp.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Westerop L, Arts-de Jong M, Hoogerbrugge N, de Hullu J, Maas A. Cardiovascular risk of BRCA1/2 mutation carriers: A review. Maturitas. 2016;91:135–139. doi: 10.1016/j.maturitas.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Noy N, Shah N, Whetzel P, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009;37(Web Server Issue):W170–W173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray K, Yates B, Seal R, Wright M, Bruford E. genenames.org: the HGNC resources in 2015. Nucleic Acids Research. 2015;43:D1079–1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CTCAE v4.0 2016. 2016. [Accessed November 21, 2016]. https://ctep.cancer.gov/protocolDevelopment/electronic applications/ctc.htm.
- 22.Zeng K, Bodenreider O, Kilbourne J, Nelson S. RxNav: a web service for standard drug information; AMIA Annual Symposium Proceedings; 2006. [PMC free article] [PubMed] [Google Scholar]
- 23.Cariaso M, Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherry S, Ward M, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Research. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research. 2014;42(Database Issue):D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roca L, Dieras V, Roche H, et al. Correlation of HER2, FCGR2A, and FCGR3A gene polymorphisms with trastuzumab related cardiac toxicity and efficacy in a subgroup of patients from UNICANCER-PACS 04 trial. Breast Cancer Research and Treatment. 2013;139(3):789–800. doi: 10.1007/s10549-013-2587-x. [DOI] [PubMed] [Google Scholar]
- 27.Stanton S, Ward M, Christos P, et al. Pro1170 Ala polymorphism in HER2-neu is associated with risk of trastuzumab cardiotoxicity. BMC Cancer. 2015;15:267. doi: 10.1186/s12885-015-1298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leong S, Chaiyakunapruk N, Lee S. Candidate gene association studies of anthracycline-induced cardiotoxicity: a systematic review and meta-analysis. Scientific Reports. 2017;7(1):39. doi: 10.1038/s41598-017-00075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visscher H, Rassekh S, Sandor G, Caron H, van Dalen E, LC K. Genetic variants in SLC22A17 and SLC22A7 are associated with anthracycline-induced cardiotoxicity in children. Pharmacogenomics. 2015;16(10):1065–1076. doi: 10.2217/pgs.15.61. [DOI] [PubMed] [Google Scholar]
- 30.Visscher H, Ross C, Rassekh S, et al. Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline-induced cardiotoxicity in children. Pediatr Blood Cancer. 2013;60(8):1375–1381. doi: 10.1002/pbc.24505. [DOI] [PubMed] [Google Scholar]
- 31.Visscher H, Ross C, Rassekh S, et al. Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J Clin Oncol. 2012;30(13):1422–1428. doi: 10.1200/JCO.2010.34.3467. [DOI] [PubMed] [Google Scholar]
- 32.Aminkeng F, Bhavsar A, Visscher H, et al. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nature Genetics. 2015;47(9):1079–1084. doi: 10.1038/ng.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Liu W, Sun C, et al. Hyaluronan synthase 3 variant and anthracycline-related cardiomyopathy: a report from the children’s oncology group. J Clin Oncol. 2014;32(7):647–653. doi: 10.1200/JCO.2013.50.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krajinovic M, Elbared J, Drouin S, et al. Polymorphisms of ABCC5 and NOS3 genes influence doxorubicin cardiotoxicity in survivors of childhood acute lymphoblastic leukemia. Pharmacogenomics. 2016;16(6):530–535. doi: 10.1038/tpj.2015.63. [DOI] [PubMed] [Google Scholar]
- 35.Rossi D, Rasi S, Franceschetti S, et al. Analysis of the host pharmacogenetic background for prediction of outcome and toxicity in diffuse large B-cell lymphoma treated with R-CHOP 21. Leukemia. 2009;23(6):1118–1126. doi: 10.1038/leu.2008.398. [DOI] [PubMed] [Google Scholar]
- 36.Wojnowski L, Kulle B, Schirmer M, et al. NAD(P)H oxidase and multidrug resistance protein genetic polymorphisms are associated with doxorubicin-induced cardiotoxicity. Circulation. 2005;112(24):3754–3762. doi: 10.1161/CIRCULATIONAHA.105.576850. [DOI] [PubMed] [Google Scholar]
- 37.Vulsteke C, Pfeil A, Maggen C, et al. Clinical and genetic risk factors for epirubicin-induced cardiac toxicity in early breast cancer patients. Breast Cancer Res Treat. 2015;152(1):67–76. doi: 10.1007/s10549-015-3437-9. [DOI] [PubMed] [Google Scholar]
- 38.Lubieniecka J, Liu J, Heffner D, et al. Single-nucleotide polymorphisms in aldo-keto and carbonyl reductase genes are not associated with acute cardiotoxicity after daunorubicin chemotherapy. Cancer Epidemiol Biomarkers Prev. 2012;21(11):2118–2120. doi: 10.1158/1055-9965.EPI-12-1037. [DOI] [PubMed] [Google Scholar]
- 39.Reichwagen A, Ziepert M, Kreuz M, et al. Association of NADPH oxidase polymorphisms with anthracycline-induced cardiotoxicity in the RICOVER-60 trial of patients with aggressive CD20(+) B-cell lymphoma. Pharmacogenomics. 2015;16(4):361–372. doi: 10.2217/pgs.14.179. [DOI] [PubMed] [Google Scholar]
- 40.Windsor R, Strauss S, Kallis C, Wood N, Whelan J. Germline genetic polymorphisms may influence chemotherapy response and disease outcome in osteosarcoma: a pilot study. Cancer. 2012;118(7):1856–1867. doi: 10.1002/cncr.26472. [DOI] [PubMed] [Google Scholar]
- 41.Kitagawa K, Kawada K, Morita S, et al. Prospective evaluation of corrected QT intervals and arrhythmias after exposure to epirubicin, cyclophosphamide, and 5-fluorouracil in women with breast cancer. Annals of Oncology. 2012;23(3):743–747. doi: 10.1093/annonc/mdr296. [DOI] [PubMed] [Google Scholar]
- 42.Lubieniecka J, Graham J, Heffner D, et al. A discovery study of daunorubicin induced cardiotoxicity in a sample of acute myeloid leukemia patients prioritizes P450 oxidoreductase polymorphisms as a potential risk factor. Front Genet. 2013;4:231. doi: 10.3389/fgene.2013.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCaffrey T, Tziros C, Lewis J, et al. Genomic profiling reveals the potential role of TCL1A and MDR1 deficiency in chemotherapy-induced cardiotoxicity; Int J Biol Sci; 2013. pp. 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wells Q, Mosley J, Van Driest S, et al. Abstract 15509: Genomwide-Association Identifies a Novel Locus for Anthracycline Cardiotoxicity. Circulation. 2013;128(22) [Google Scholar]
- 45.Cascales A, Pastor-Quirante F, Sanchez-Vega B, et al. Association of anthracycline-related cardiac histological lesions with NADPH oxidase functional polymorphisms. Oncologist. 2013;18(4):446–453. doi: 10.1634/theoncologist.2012-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boekhout A, Gietema J, Milojkovic Kerklaan B, et al. Angiotensin II-Receptor Inhibition With Candesartan to Prevent Trastuzumab-Related Cardiotoxic Effects in Patients With Early Breast Cancer: A Randomized Clinical Trial. JAMA Oncology. 2016;2(8):1030–1037. doi: 10.1001/jamaoncol.2016.1726. [DOI] [PubMed] [Google Scholar]
- 47.Gomez Pena C, Davila-Fajardo C, Martinez-Gonzalez L, et al. Influence of the HER2 Ile655Val polymorphism on trastuzumab-induced cardiotoxicity in HER2-positive breast cancer patients: a metaanalysis. Pharmacogenetics and Genomics. 2015;25(8):388–393. doi: 10.1097/FPC.0000000000000149. [DOI] [PubMed] [Google Scholar]
- 48.Beauclair S, Formento P, Fischel J, et al. Role of the HER2 [Ile655Val] genetic polymorphism in tumorogenesis and in the risk of trastuzumab-related cardiotoxicity. Ann Oncol. 2007;18(8):1335–1341. doi: 10.1093/annonc/mdm181. [DOI] [PubMed] [Google Scholar]
- 49.Armenian S, Ding Y, Mills G, et al. Genetic susceptibility to anthracycline-related congestive heart failure in survivors of haematopoietic cell transplantation. Br J Haematol. 2013;163(2):205–213. doi: 10.1111/bjh.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volkan-Salanci B, Aksoy H, Kiratli P, et al. The relationship between changes in functional cardiac parameters following anthracycline therapy and carbonyl reductase 3 and glutathione S transferase Pi polymorphisms. J Chmother. 2012;24(5):285–291. doi: 10.1179/1973947812Y.0000000037. [DOI] [PubMed] [Google Scholar]
- 51.Wells Q, Veatch O, Fessel J, et al. Genome-wide association and pathway analysis of left ventricular function after anthracycline exposure in adults. Pharmacogenet Genomics. 2017;27(7):247–254. doi: 10.1097/FPC.0000000000000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang L, Fernandes H, Zia H, et al. The cancer precision medicine knowledge base for structured clinical-grade mutations and interpretations. J Am Med Inform Assoc. 2016;24(3):513–519. doi: 10.1093/jamia/ocw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCarty C, Chisholm R, Chute C, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The future of health begins with All of Us. 2017; 2017. [Accessed June 2, 2017]. https://allofus.nih.gov/
- 55.Kirby J, Speltz P, Rasmussen L, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. JAMIA. 2016;23(6):1046–1052. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giacomini K, Yee S, Mushiroda T, Weinshilboum R, Ratain M, Kubo M. Genome-wide association studies of drug response and toxicity: an opportunity for genome medicine. Nature Reviews Drug Discovery. 2017;16(1):1. doi: 10.1038/nrd.2016.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rasmussen-Torvik L, Stallings S, Gordon A, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther. 2014;96(4):482–489. doi: 10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bauer D, Canver M, Orkin S. Generation of Genomic Deletions in Mammalian Cell Lines via CRISPR/Cas9. J Vis Exp. 2015;(95):e52118. doi: 10.3791/52118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burridge P, Li Y, Matsa E, et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nature Medicine. 2016;22:547-556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]


