Abstract
Integration of genetic information is becoming increasingly important in clinical practice. However, genetic information is often ambiguous and difficult to understand, and clinicians have reported low-self-efficacy in integrating genetics into their care routine. The Health Level Seven (HL7) Infobutton standard helps to integrate online knowledge resources within Electronic Health Records (EHRs) and is required for EHR certification in the US. We implemented a prototype of a standards-based genetic reporting application coupled with infobuttons leveraging the Infobutton and Fast Healthcare Interoperability Resources (FHIR) Standards. Infobutton capabilities were provided by Open Infobutton, an open source package compliant with the HL7 Infobutton Standard. The resulting prototype demonstrates how standards-based reporting of genetic results, coupled with curated knowledge resources, can provide dynamic access to clinical knowledge on demand at the point of care. The proposed functionality can be enabled within any EHR system that has been certified through the US Meaningful Use program.
Introduction
Access to genetic information in medical practice is becoming increasingly prevalent1. Since the end of the Human Genome Project in 2003, over 1000 genetic tests have become available2, allowing clinicians to more easily obtain an individual’s genetic information3. Methods in informatics are permitting these vast amounts of data to be integrated into clinically relevant systems4. Once discovered and translated into clinical practice, biomarkers in the genome, such as an existing gene or variant, can provide clinical help in diagnoses, predictive medicine, and drug treatment (i.e. pharmacogenomics)5. Pharmacogenomics research, as one example, has been found in some cases to reduce healthcare cost6 and improve patient care7. Pharmacogenomics knowledge is rapidly increasing, but the assimilation of this and other kinds of genetic information into clinical practice has been slow8.
In medical practice, questions that clinicians pose often remain unanswered due to lack of time to search available knowledge resources and lack of self-efficacy in choosing and searching multiple potentially relevant resources9. Unanswered questions associated with knowledge gaps can lead to errors and lower quality care10. Clinically actionable genetic information, in particular, can be difficult to obtain. Genetic knowledge resources are especially needed because clinicians report poor knowledge in genetics and low self-efficacy integrating genetics into their decision-making process8. Therefore, care providers may benefit from access to clinically actionable recommendations to guide them in the integration of genetics into routine patient care decisions. Several online resources exist to provide healthcare professionals and patients genetic information, such as Clin Var, Genetic Testing Resources (GTR), and Gene Reviews11,12. However, these resources were designed to serve multiple purposes and audiences, with clinically actionable genetic information being dispersed within and across resources8.
The ClinGen (Clinical Genome Resource) project aims to enable “an authoritative central resource that defines the clinical relevance of genes and variants for use in precision medicine and research”13. ClinGen seeks to produce open resources by means of community collaboration to continuously improve understanding of genetic variation in a standardized fashion14. However, even with genetic standards, genetic information needs to be easily accessible within a clinical setting. Therefore, the ClinGen Electronic Health Record Working Group (EHR WG) was tasked to integrate available genomic resources (including ClinGen resources) into electronic health record (EHR) systems via clinical decision support (CDS) tools such as “infobuttons”15,16. Based on the context of the interaction between a clinician and the EHR, infobuttons anticipate clinicians’ information needs and provide automated links to relevant resources. Infobuttons are typically placed on the EHR screen next to data such as problems, medications, and laboratory tests17. A recent systematic review has reported the effect of infobuttons on clinicians, particularly on increased search efficiency, decision enhancement, and learning17. Infobuttons can be implemented in a standards-based fashion through the Health Level Seven (HL7) Infobutton Standard, which is required for EHR certification in the US Meaningful Use Program18. In a previous study, Heale et al. enabled infobutton access to several genetic information resources through Open Infobutton, an open source, HL7-compliant software suite designed to help healthcare organizations enable infobuttons within their EHR systems19. In the present study, we aimed to implement HL7- compliant infobuttons within a prototype EHR coupled with a standards-based genetic reporting application to demonstrate how this kind of CDS tool could be used to help provide clinicians with seamless access to genetic knowledge resources.
Methods
To accomplish the goals of this study, we leveraged an information display of a multi-gene sequencing panel report that was designed and evaluated in a previous study20. We developed a user interface to represent a prototypical EHR coupled with the multi-gene sequencing panel report and incorporated infobutton features to help clinicians understand other genetic-related information, such as diseases and medications (i.e., pharmacogenomics). The prototype was developed in three steps: (1) configuration of access to genetic resources within Open Infobutton; (2) development of the prototype EHR user interface with the gene sequencing panel reports coupled with infobutton functionality; and (3) population of the panel prototype with data from sample laboratory tests and the EHR prototype with mock patient data, respectively. The patient data were represented using HL7 Fast Healthcare Interoperability Resources (FHIR). Infobutton functionality, compliant with the HL7 Infobutton Standard, was implemented with Open Infobutton. Brief summaries of the multi-gene sequencing panel report, the HL7 Infobutton Standard, OpenInfobutton, and FHIR are provided below.
Multi-gene sequencing panel report design. In a study done by Cutting et al., a multi-gene sequencing panel report was iteratively designed and evaluated through clinician feedback on suitable genetic reporting20. The report, known as Genomic Medicine Assistant (GMA), featured the clinically significant gene and variant names discovered in the panel with infobuttons next to those names. The report also featured the clinical significance of the findings, recommended actions, and other relevant features. Clinicians in the study rated the usability of the GMA and the results were favorable.
HL7 Infobutton Standard. EHR certification under the US “Meaningful Use” program requires the implementation of infobutton functionality compliant with the HL7 Context-Aware Knowledge Retrieval (“Infobutton”) Standard21. The infobutton standard defines a means to integrate EHR systems with health knowledge resources22. Infobuttons utilize the context of the EHR to anticipate clinicians’ and patients’ information needs; select relevant Web resources; and automatically retrieve relevant information from the selected resources to address those needs. The Infobutton Standard allows EHRs to capture and send context in a standard context information model according to four dimensions: the patient (e.g., age, gender, conditions), the infobutton user (e.g., clinician, patient), the care setting (e.g., inpatient, outpatient), and the EHR task (e.g., order entry, medication list review, laboratory test result review, problem list entry)19. In addition, the standard allows capturing one or more concepts that the user might be interested in, such as a specific medication, gene, or laboratory test.
OpenInfobutton (www.OpenInfobutton.org). Infobutton functionality can be implemented through a Web service called an “infobutton manager”, which serves as a broker between EHR systems and multiple knowledge resources. All configuration of infobutton behavior is done within the infobutton manager’s knowledge base. OpenInfobutton is an HL7 compliant, open source software suite that includes an infobutton manager and a Web-based infobutton configuration application called LITE (Librarian Infobutton Tailoring Environment). LITE allows knowledge resource experts, such as medical librarians, to collaboratively configure access to knowledge resources within the infobutton manager’s knowledge base23,24. Knowledge resource configuration is stored in XML files called knowledge resource profiles, which include the resource’s base URL, the contexts in which the resource is relevant, and the search parameters and terminologies that the resource’s Web service application program interface (API) supports. For resources that are not HL7-compliant, the resource profile also includes mappings between search parameters in the resource’s API and the HL7 Infobutton Standard. Once a resource profile is configured in LITE, it can be made available for download to all institutions using OpenInfobutton via LITE’s resource profile store.
FHIR (www.hl7.org/fhir/). HL7 FHIR is an emerging standard that is rapidly gaining attention and adoption by the health IT industry and health care organizations25. The basic building block of FHIR is a resource, which consists of a set of data attributes that are defined based on FHIR data types and bound to standard terminologies whenever applicable26. A few examples of FHIR resources include Patient, Condition, and Observation 27. FHIR profiles are available to define the semantic structure of resources for specific use cases through constraints and extensions28.
Configuration of ClinGen and other genetic resources in OpenInfobutton
A set of clinically useful genetic resources was chosen by consensus among ClinGen EHR WG members. Also with input from the ClinGen EHR WG, resource profiles were selected and fine-tuned from those initially developed by Heale et al19. In addition, new profiles were created in LITE for APIs that became available after the work done by Heale et al. All knowledge resource profiles were validated using LITE’s testing tool.
Development of a prototype EHR with gene sequencing panel reports coupled with infobuttons
The goal of our infobutton-enabled prototypical EHR was to demonstrate how genetic knowledge resources can be integrated within EHR systems to help clinicians’ meet their genetic information needs. The prototype user interface was developed in Javascript, HTML, and CSS. Using the same approach, gene sequencing panel reports based on the design by Cutting et al.20 were included in the prototype EHR. Infobutton “i” icons were placed next to each clinical data element in both the EHR and embedded panel reports. Infobutton icons contained hyperlinks with HL7-compliant OpenInfobutton requests.
Population of the prototypes with data from sample laboratory tests and FHIR compliant JSON files
The gene panel prototype was populated with data from three case vignettes developed by Cutting et al.20 and sample laboratory test reports from Integrated Genetics29. To demonstrate the use of the FHIR standard, data in the EHR prototype were represented according to FHIR resources. Example FHIR resources were selected from two openly available FHIR servers: HAPI (hapifhir.io) and Health Services Platform Consortium (HSPC) Sandbox (sandbox.hspconsortium.org)30. Using these sample resources as guidelines, FHIR compliant JSON files with FHIR resources were created for three sample patients. These FHIR compliant JSON files were utilized to populate the prototype EHR.
Results
Configuration of ClinGen and other genetic resources in OpenInfobutton
Eight genetic knowledge resources selected by members of the ClinGen EHR Work Group were configured for EHR integration using LITE (Table 1). Although only one (PharmGKB) out of the eight genetic knowledge resources was compliant with the HL7 Infobutton Standard, we were able to successfully connect with all eight resources. Profiles were created for each resource by setting the base URLs, EHR contexts, support for standard terminologies, and mappings between resource-proprietary parameters and the HL7 Infobutton Standard (for resources that were not HL7-compliant). The infobutton user was selected as “Provider” for each resource.
Table 1.
Summary of the knowledge resources connected through the HL7 Infobutton Standard via LITE.
| Infobutton Resource | Information Available | EHR Context |
|---|---|---|
| ClinGen Allele Registry | Identifiers for genetic variants | Genetic Lab Review/Entry |
| ClinGen Curated Summaries | Gene-drug interactions and genetically caused diseases | Problem List Review/Entry |
| ClinVar | Clinical significance of knowngenetic variants | Genetic Lab Review/Entry |
| CPIC | Clinical guidelineson genetic testing and treatment related to pharmacogenomics | Medication List Review/Entry |
| GeneReviews | Genetic disease knowledge management | Problem List Review/Entry |
| Genetics Home Reference | Consumer health information | Genetic Lab Review/Entry |
| Genetic Practice Guidelines | Geneticdisease guidelines | Problem and Medication List Review/Entry |
| Genetic Testing Registry | Genetic testsand laboratories | Problem List Review/Entry |
| PharmGKB | Curated pharmacogenomics knowledge base | Medication List Review/Entry |
Development of a prototype EHR with gene sequencing panel reports coupled with infobuttons
The EHR prototype is available at http://lite.bmi.utah.edu/ClinGenDemo/Interface Project/main.html. Four sections were created in the prototype EHR user interface (Figure 1): Patient Profile, Problems, Medications, and Results. The Patient Profile tab contains patient demographics, allergies, future appointments, and a history of past appointments. These data were populated from the following FHIR resources respectively: Patient, Allergy Intolerance, Appointment, and Encounter. The Problems, Medications, and Results tabs were populated by Condition, Medication Statement, and Observation FHIR resources respectively. The Problems and Medications tabs are formatted as lists of buttons that, when clicked, present relevant metadata (e.g., onset date, “last updated”, prescription date). The Results tab is also presented as a list of buttons that, when clicked, show other genetic information such as the clinical significance and a single nucleotide polymorphisms (SNP) identifier (if available), as well as a link to the referenced multi-gene sequencing panel report (Figure 2). Each panel report contains infobuttons next to the gene and variant names. Infobuttons were also integrated into the EHR next to the conditions, medications, and genes/variants. A dropdown list on the top right of the EHR prototype contains the patient names. When a name is selected, the information from the corresponding patient’s JSON file is extracted and presented via the EHR interface.
Figure 1.

Main screen of the EHR prototype showing the Profile, Problems, Medications, and Results tabs which can be accessed by clicking on the respective tab headings.
Figure 2.

An example of genetic laboratory results as shown in the Results tab. A clinician can click on individual results to view the Clinical Significance, SNP, and a link to the full laboratory report from which each result is retrieved. The infobutton “i” icons can be clicked to send HL7-compiant requests to OpenInfobutton to retrieve information on the clinical interpretation of each result.
Infobutton links included a URL-based request to OpenInfobutton including seven parameters, four of which represent the context of the EHR (Table 2). Figures 3 and 4 show examples of the user interface returned by OpenInfobutton after an infobutton is clicked.
Table 2.
List of parameters included in the HL7-compliant infobutton request sent by the prototype EHR to OpenInfobutton.
| Infobutton Request URL Segment | Use | Example values |
|---|---|---|
| http://service.oib.utah.edu:8080/infobutton-service/infoRequest | Base URL of the knowledge resource | --- |
| represented Organization.id.root | Domain name of requesting organization | ClinicalGenome.org |
| taskContext.c.c | Context of EHR task | PROBLISREV, MLREV, GENEREV, VARREV |
| mainSearchCriteria.v.c | Search term code | LOINC, or SNOMED-CTcodes |
| mainSearchCriteria.v.cs | Search term code system | Code system identifier |
| mainSearchCriteria.v.dn | Search term | Display name |
| informationRecipient.languageCode.c | Language | English, Spanish |
| performer | Role of person wanting results | PROV (provider) |
Figure 3.

User interface returned from OpenInfobutton in response to a request for information on the gene “HNF1A” when a provider is reviewing the patient’s lab result list. This example shows a resource that could be used for both clinicians and patients.
Figure 4.

User interface returned from OpenInfobutton in response to a request for information on the variant “c.681G>A” when a provider is reviewing the patient’s lab result list. This shows an example of a clinician resource known as “ClinVar.”
Population of the EHR prototype with data from sample laboratory tests and FHIR compliant JSON files
The gene panel prototypes were populated with information about the variant results of the test, the clinical significance, a brief interpretation summary, clinical recommendations, and information about the test itself (Figure 5). References sited within the test are also provided, allowing clinicians to check the sources.
Figure 5.
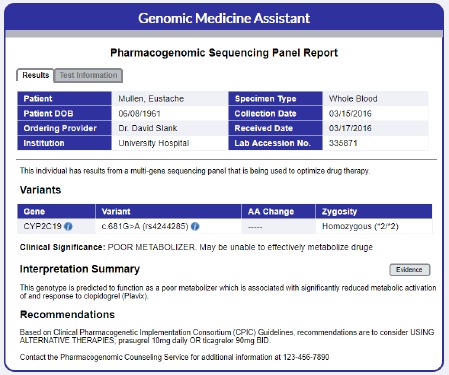
Multi-gene paneling report, which can be accessed from the Results tab. The infobutton links request provider reference information related to genes or variants in the context of genetic test results review.
A Bundle FHIR resource, which holds an embedded list of other FHIR resources, was created for three sample patients, each containing all the resources necessary to populate the EHR prototype (see table 3 for all resources implemented). Table 4 displays the genetic FHIR extensions required to bind each genetic data type, allowing for additional genetic information to be supported outside of the Observation resource. Standard terminologies were used within each FHIR resource as follows: SNOMED-CT for problem list items, medications, and allergies; HGNC for gene names; and LOINC for laboratory tests. The resources for each patient were validated as FHIR compliant by uploading them to the HAPI server, which passes all data through a FHIR-JSON validator before storing to the backend30.
Table 3.
Links to example JSON FHIR resources.
Table 4.
FHIR Extension references for genetic data types.
| Genetic Resource Type | Extension URL |
|---|---|
| Gene | http://hl7.org/fhir/StructureDefinition/observation-geneticsGene |
| DNA Sequence Variant Name | http://hl7.org/fhir/StructureDefinition/observation-geneticsDNASequenceVariantName |
| Allelic state (Zygosity) | https://www.hl7.org/fhir/extension-observation-geneticsallelicstate.html |
| Amino Acid Change Name | http://hl7.org/fhir/extension-observation-geneticsaminoacidchangename.html |
| DNA Variant Id (SNP) | http://hl7.org/fhir/StructureDefinition/observation-geneticsDNAVariantId |
Discussion
Clinical Significance
We have developed a prototype EHR with a multi-gene sequencing panel report to test and demonstrate the feasibility of infobuttons to provide clinicians with access to relevant genetic knowledge resources at the point of care. This project demonstrated that several knowledge resources could be integrated with a prototypical EHR through a standards-based approach. It also shows how genetic data and infobutton capabilities can be integrated into an EHR user interface. Health care organizations can follow similar steps to implement this functionality within their EHRs in real health care settings.
We were able to configure eight genetic knowledge resources using LITE, with no need for programming or changes to the underlying OpenInfobutton software. Our experience supports the findings by a usability study conducted by Jing et al., which found that medical librarians were able to configure resource profiles in LITE without any formal training31. URL-based, HL7-compliant infobutton requests were also easy to implement into the prototype EHR. Similar experiences have been reported in interviews with implementers of the HL7 Infobutton Standard18. Meaningful Use certified EHRs must be able to connect to knowledge resources using the HL7 Infobutton Standard18. Therefore, the functionality demonstrated in our prototype EHR can also be implemented within any certified EHR that is configured to use OpenInfobutton. Detailed documentation on how to deploy and use OpenInfobutton is available at http://www.openinfobutton.org/documentation. Three deployment alternatives are available: (1) access a cloud-based instance, such as the one hosted at the University of Utah (not production-level, available only for development and demonstration purposes); (2) download and install a virtual machine that includes all the OpenInfobutton software; and (3) pull the source code from OpenInfobutton’s GitHub repository and build it within the target deployment environment.
A usability issue to consider when implementing infobuttons in an EHR system is how the infobutton interface is presented to the user. The prototype in this study is coded in such a way to have the interface appear in a new window. However, the infobutton interface can be presented in any way the EHR system developer sees fit, whether in a new window or as a “popup” in the same window. An advantage of showing the interface through a “popup” window is to minimize workflow interruptions. However, showing the interface in a new window allows for multiple infobutton interfaces to be open at once. EHR system implementers will need to consider which option best suits their system when integrating infobuttons.
Challenges and Limitations
The vast majority of genetic knowledge resources were not designed for use by clinicians at the point of care and are not yet compliant with the HL7 Infobutton Standard. Therefore, most of the resources required manual mapping of the resources’ API parameters to the Infobutton standard. Those mappings were stored within the resources’ profiles in the infobutton manager and are available for use by any healthcare organization that uses OpenInfobutton.
We faced several interoperability challenges related to the lack of support for standard terminologies in the genetics domain. Knowledge resource APIs and EHR systems do not support gene standard terminologies, such as HGNC.19 To address this limitation, we created infobutton requests only with gene names. A similar problem was faced with allele variants. Representing genetic panel reports within FHIR requires the use of a standard terminology to uniquely identify the different kinds of panels. EHR Meaningful Use criteria requires adoption of LOINC to encode laboratory test panels and results. However, LOINC does not currently provide a comprehensive set of concepts for the growing number of available the genetic sequencing panels. Therefore, in the Observation resources created, a general LOINC code, corresponding with the term “Genetic analysis report”, was used for the terminology code in the Observation resources.
Genetic labs still provide their reports primarily in unstructured, PDF format, which imposes a challenge to infobutton integration since discrete genetic results are required for auto-generated infobutton requests32. The sample laboratory reports leveraged to populate the prototype multi-gene sequencing panels were all in PDF format, thus the data had to be loaded into the prototype EHR manually. Similar difficulties will be found in clinical practice until structured, standards-compliant genomic testing reports are more widely implemented.
Future Work
Future studies include investigating the usefulness of infobutton responses to clinical genomics inquiries in clinical practice. Ongoing efforts are focusing on continuously expanding, updating and improving OpenInfobutton resource profiles for clinical genomics. Future plans also include deploying access to clinical genomics resources via infobuttons in patient care settings. As mentioned previously, several options are available to facilitate deployment of OpenInfobutton at different healthcare organizations.
Conclusion
There is a critical need to provide clinicians with efficient access to clinically actionable genetic information at the point of care. We developed a prototype that demonstrates an approach to help address this need using standards-based functionality that is required for EHR certification in the Meaningful Use program.
Acknowledgements
This project was funded by grant number 1U01HG007437-01 Grant name: Clinically Relevant Genetic Variants Resource: A Unified Approach for Identifying Genetic Variants for Clinical Use
References
- 1.Knoppers BM, Zawati MH, Sénécal K. Return of genetic testing results in the era of whole-genome sequencing. Nat Med [Internet] 2015;16(9):553–9. doi: 10.1038/nrg3960. Available from: http://www.nature.com/doifinder/10.1038/nrg3960%5Cnpapers3://publication/doi/10.1038/nrg3960. [DOI] [PubMed] [Google Scholar]
- 2.Group U. Personalized Medicine: Trends and Prospects for the New Science of Genetic Testing and Molecular Diagnostics - Working Paper 7. 2012. [cited 2017 Aug 17]. Available from: http://www.unitedhealthgroup.eom/~/media/uhg/pdf/2012/unh-working-paper-7.ashx.
- 3.The Cost of Sequencing a Human Genome - National Human Genome Research Institute (NHGRI) [Internet] [cited 2017 Aug 17]. Available from: https://www.genome.gov/27565109/the-cost-of-sequencing-a-human-genome/
- 4.Hudson KL. Genomics, health care, and society. N Engl J Med. 2011;365:1033–41. doi: 10.1056/NEJMra1010517. [DOI] [PubMed] [Google Scholar]
- 5.Roychowdhury S, Chinnaiyan AM. Translating genomics for precision cancer medicine. Annu Rev Genomics Hum Genet [Internet] 2014;15:395–415. doi: 10.1146/annurev-genom-090413-025552. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25184532. [DOI] [PubMed] [Google Scholar]
- 6.Stallings SC, Huse D, Finkelstein SN, Crown WH, Witt WP, Maguire J, et al. A framework to evaluate the economic impact of pharmacogenomics. Pharmacogenomics. 2006;7(6):853–62. doi: 10.2217/14622416.7.6.853. [DOI] [PubMed] [Google Scholar]
- 7.Kazani S, Wechsler ME, Israel E. The role of pharmacogenomics in improving the management of asthma. Journal of Allergy and Clinical Immunology. 2010;125:295–302. doi: 10.1016/j.jaci.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heale BSE, Khalifa A, Stone BL, Nelson S, Del Fiol G. Physicians’ pharmacogenomics information needs and seeking behavior: a study with case vignettes. [cited 2017 Aug 17];BMC Med Inform Decis Mak [Internet] 2017 Dec 1;17(1):113. doi: 10.1186/s12911-017-0510-9. Available from: http://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-017-0510-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ely JW, Osheroff JA, Chambliss ML, Ebell MH, Rosenbaum ME. Answering physicians’ clinical questions: Obstacles and potential solutions. J Am Med Informatics Assoc. 2005;12(2):217–24. doi: 10.1197/jamia.M1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leape LL, Bates DW, Cullen DJ, Cooper J, Demonaco HJ, Gallivan T, et al. Systems analysis of adverse drug events. ADE Prevention Study Group. [Internet] Journal of the American Medical Association. 1995;274:35–43. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7791256. [PubMed] [Google Scholar]
- 11.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. [cited 2017 Aug 17];Nucleic Acids Res [Internet] 2013 Jan 1;41(D1):D8–20. doi: 10.1093/nar/gkx1095. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23193264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pagon RA, Adam MP, Ardinger HH. GeneReviews [Internet] 2017 Available from: https://www.ncbi.nlm.nih.gov/books/NBK5191/ [Google Scholar]
- 13.About ClinGen - ClinGen | Clinical Genome Resource [Internet] [cited 2017 Aug 17]; Available from: https://www.clinicalgenome.org/about/ [Google Scholar]
- 14.Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, et al. ClinGen - The Clinical Genome Resource. N Engl J Med [Internet] 2015;372(23):2235–42. doi: 10.1056/NEJMsr1406261. Available from: http://www.nejm.org/doi/abs/10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guilherme Del Fiol RARPDC. Infobuttons at Intermountain Healthcare: Utilization and Infrastructure. AMIA Annu Symp Proc [Internet] 2006;2006:180. Available from: /pmc/articles/PMC1839474/?report=abstract%5Cnpapers2://publication/uuid/EE2995D4-AC79-4B07-BDE6-06511A4EA554. [PMC free article] [PubMed] [Google Scholar]
- 16.Cimino JJ, Elhanan G, Zeng Q. Supporting infobuttons with terminological knowledge. Proc AMIA Annu Fall Symp [Internet] 1997:528–32. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=9357682&retmode=ref&cmd=prlinks%5Cnpapers2://publication/uuid/7E8A906E-347E-4C1A-A15E-6A1D8BF95846. [PMC free article] [PubMed] [Google Scholar]
- 17.Cook DA, Teixeira MT, Heale BSE, Cimino JJ, Del Fiol G. Context-sensitive decision support (infobuttons) in electronic health records: A systematic review. Journal of the American Medical Informatics Association. 2017;24:460–8. doi: 10.1093/jamia/ocw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Fiol G, Huser V, Strasberg HR, Maviglia SM, Curtis C, Cimino JJ. Implementations of the HL7 Context-Aware Knowledge Retrieval (“Infobutton”) Standard: Challenges, strengths, limitations, and uptake. J Biomed Inform. 2012;45(4):726–35. doi: 10.1016/j.jbi.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heale BSE, Overby CL, Del Fiol G, Rubinstein WS, Maglott DR, Nelson TH, et al. Integrating Genomic Resources with Electronic Health Records using the HL7 Infobutton Standard. Appl Clin Inform [Internet] 2016;7(3):817–31. doi: 10.4338/ACI-2016-04-RA-0058. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27579472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cutting E, Banchero M, Beitelshees AL, Cimino JJ, Del Fiol G, Gurses AP, et al. User-centered design of multi-gene sequencing panel reports for clinicians. J Biomed Inform. 2016;63:1–10. doi: 10.1016/j.jbi.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Federal Register Volume 77, Number 45. 2012. Mar 7, pp. 13832–13885.pp. 13847
- 22.Strasberg HR, Del Fiol G, Cimino JJ. Terminology challenges implementing the HL7 context-aware knowledge retrieval (’Infobutton’) standard. J Am Med Inf Assoc [Internet] 2013;20(2):218–23. doi: 10.1136/amiajnl-2012-001251. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23077131%5Cnhttp://jamia.bmj.com/content/20/2/218.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cimino JJ, Jing X, Del Fiol G. Meeting the electronic health record “meaningful use” criterion for the HL7 infobutton standard using OpenInfobutton and the Librarian Infobutton Tailoring Environment (LITE) AMIA Annu Symp Proc [Internet] 2012;2012:112–20. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3540503&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 24.Del Fiol G, Curtis C, Cimino JJ, Iskander A, Kalluri ASD, Jing X, et al. Disseminating context-specific access to online knowledge resources within electronic health record systems. In: Studies in Health Technology and Informatics. 2013:672–6. [PMC free article] [PubMed] [Google Scholar]
- 25.Lamprinakos GC, Mousas AS, Kapsalis AP, Kaklamani DI, Venieris IS, Boufis AD, et al. Using FHIR to develop a healthcare mobile application. In: Proceedings of the 2014 4th International Conference on Wireless Mobile Communication and Healthcare - “Transforming Healthcare Through Innovations in Mobile and Wireless Technologies”; 2014; MOBIHEALTH. 2015. pp. 132–5. [Google Scholar]
- 26.Bender D, Sartipi K. HL7 FHIR: An agile and RESTful approach to healthcare information exchange. In: Proceedings of CBMS 2013 - 26th IEEE International Symposium on Computer-Based Medical Systems. 2013:326–31. [Google Scholar]
- 27.Resourcelist - FHIR v3.0.1 [Internet] [cited 2017 Aug 17]. Available from: http://www.hl7.org/implement/standards/fhir/resourcelist.html.
- 28.Mandel JC, Kreda DA, Mandl KD, Kohane IS, Ramoni RB. SMART on FHIR: A standards-based, interoperable apps platform for electronic health records. J Am Med Informatics Assoc. 2016;23(5):899–908. doi: 10.1093/jamia/ocv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sample Reports | Integrated Genetics [Internet] [cited 2017 Aug 17]. Available from: https://www.integratedgenetics.com/clients/sample-result-reports.
- 30.FHIR Connectathon 16 - HL7Wiki [Internet] [cited 2017 Aug 17]. Available from: http://wiki.hl7.org/index.php?title=FHIR_Connectathon_16.
- 31.Jing X, Cimino JJ, Del Fiol G. Usability and acceptance of the librarian infobutton tailoring environment: An open access online knowledge capture, management, and configuration tool for openinfobutton. J Med Internet Res. 2015;17(11) doi: 10.2196/jmir.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarczy-Hornoch P, Amendola L, Aronson SJ, Garraway L, Gray S, Grundmeier RW, et al. A survey of informatics approaches to whole-exome and whole-genome clinical reporting in the electronic health record. Genet Med. 2013;15(10):824–32. doi: 10.1038/gim.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]


