Abstract
Drug repositioning for available medications can be preferred over traditional drug development, which requires substantially more effort to uncover new insights into medications and diseases. Genome-Wide Association Studies (GWAS) and Phenome-Wide Association Studies (PheWAS) are two complimentary methods for finding novel associations between genes and diseases. We hypothesize that discoveries from these studies could be leveraged to find new targets for existing drugs. Thus, we propose a framework to learn opportunities for inferring such relationships via overlapped genes between disease-associated genes (e.g. GWAS and PheWAS findings) and drugtargeted genes. We use drug indications found in Medication Indication Resource (MEDI) as a gold standard to evaluate if drug indications learned from GWAS and PheWAS findings have clinical indications. We examined 151,011 <drug, GWAS phenotype> pairs from 987 drugs across 153 diseases and 763 pairs were statistically significant. Out of these 763 pairs, 16 of them were found to have clinical indications.
Introduction
The importance of drug therapy, or the employment of medication to treat diseases, can be observed in the expansive application of pharmacology in the clinical sphere of medicine1, 2. New drug therapies are established through two major methods: i) drug discovery: bringing a new medication onto the market3; or ii) drug repositioning: reapplying existing medications to new medical conditions4, 5. The disadvantages of drug discovery include its long and expensive process and the repeated failures of newly-developed drugs to pass clinical trials2, 3, 6, 7. Applications of drug repositioning have led to potential and successful remedies for various medical conditions, including but not limited to cancer8, cardiovascular disease9, and Parkinson’s disease10. Notably, drug repositioning can be looked towards for a more efficient and effective alternative for identifying novel pharmaceutical treatments4, 5, 11.
Drug repositioning is commonly approached from identification of new targets for a particular medication. A well-known successful example of drug reposition is thalidomide, which was originally used as a sedative and has been found to possess anticancer activity12. More recently, metformin, a medication originally used to treat type 2 diabetes, has been recently reported and validated with evidence from electronic health records (EHRs) to have drug repositioning association with cancer survival13. There are two typical approaches when conducting drug repositioning research: i) designing biochemical experiments to find uncovered genetic targets10, 14; and ii) conducting clinical studies to discover disease targets4, 5, 13. The first type aims to investigate relationships between drugs and targeted genes, which require gathering genetic evidence to authenticate the possibility of newly found targeted genes. The second type aims to identify relationships between drugs and diseases from clinical information. While the former requires a substantial number of biochemical experiments, each of which will take time to validate the results and expend resource costs, the second type of repositioning is limited when finding the hidden reasons (e.g. genetic variations) that a drug has positive indications for a disease. Thus, we propose a data-driven framework, which only relies on the publically available genetic and clinical resources, to learn opportunities for discovering novel drug targeted diseases. Notably, for each targeted disease, we are able to identify their associated genetic variations.
For this study, genetic evidence comes in the form of data found from two comparable analytic tools: genome-wide association studies (GWAS) and phenome-wide association studies (PheWAS). While both operate in a similar fashion, the former samples a large number of genetic variants for association with a single phenotype whereas the latter does the same procedure with many phenotypes to one gene. We will leverage findings of the associations between genotypes and phenotypes from these studies and drug targets in DrugBank, a comprehensive online database detailing over 9,000 medications and their pathways and targets, to infer drug targeted diseases. Medication Indication Resource (MEDI), a comprehensive online database documenting drugs and its clinical disease targets15, is then leveraged to assess the plausibility of these inferred drug targeted diseases.
Study Materials
This study leverages findings learned from GWAS and PheWAS and drug-targeted gene-sets from DrugBank to infer novel indications for drugs and then uses MEDI to assess the plausibility of learned drug indications. To orient the audience, we introduce four major data resources involved in this study: i) DrugBank on drugs and their protein targets14; ii) GWAS findings on the associations between genes and GWAS phenotypes (diseases/traits)16; iii) PheWAS findings on clinical phenotypes and their relationships with genes17; and iv) clinically implemented drug-indications managed in MEDI15. Because this investigation utilizes analytic tools and databases that have been recently pioneered, we take a moment here to explain the developments and applications of each resource.
DrugBank: Associations between drugs and their protein targets
DrugBank is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information14. It is widely used by the drug industry, medicinal chemists, pharmacists, physicians, and the general public. Its extensive drug and drug-target data have enabled the discovery and repurposing of a number of existing drugs to treat rare and newly identified illnesses18. In this study, we will leverage drug targets (genes) recorded in DrugBank and relationships between genes and phenotypes in GWAS and PheWAS findings to build connections between drugs and phenotypes.
GWAS: Associations between genes and GWAS phenotypes
The GWAS catalog (https://www.ebi.ac.uk/gwas/) manages the majority of all associations between single nucleotide polymorphisms (SNPs) and GWAS phenotypes learned from GWAS over the past decade16. Genomic regions can be defined for each SNP by calculating linkage disequilibrium with other SNPs in the region (r2 > 0.6) to define genomic boundaries. Genes will be included if they fall at least partially within that genomic region. Interval-based Enrichment Analysis (INRICH) can then be applied to test for enrichment of genes from a set (e.g. drug targets) within the genomic regions compared to randomly generated sets of genomic regions accounting for number of genes18, 19. This approach has been previously demonstrated in schizophrenia20. An example of broad graphical depiction of enriched gene-sets associated with distinct chromosomal regions is provided in Figure 1. We will transform associations between a GWAS trait and SNPs to relationships between the trait and a set of genes via INRICH.
Figure 1.
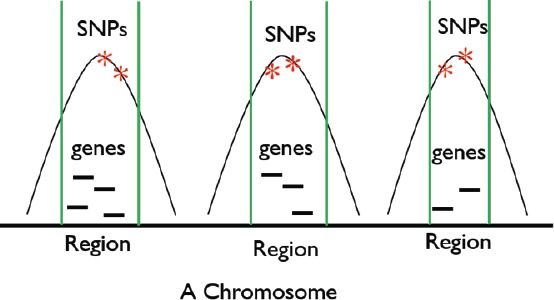
Enriched gene-sets for three different regions, each of which contain SNPs that are confirmed to be associated with a specific GWAS phenotype.
PheWAS: Associations between clinical phenotypes and GWAS phenotypes
PheWAS codes, which is broadly utilized to characterize clinical phenotypes17, was developed to accompany PheWAS. Denny et.al17, 21 built mapping associations between PheWAS codes and International Classification of Diseases, 9th Edition (ICD-9) codes because GWAS phenotypes cannot be directly compared to clinical conditions. Additionally, they build connections between GWAS phenotypes and PheWAS codes, and such connections have been established based on both GWAS (relationship between a GWAS phenotype and genes) and PheWAS findings (relationship between a gene and PheWAS codes). Algorithms for such connection establishments are managed in Phenotype KnowledgeBase (PheKB, https://phekb.org)22.
MEDI: Associations between drugs and clinical phenotypes
MEDication Indication resource (MEDI) was created with the intention of supporting EHRs through its aggregation of clinical treatment data into a comprehensive dataset, which may provide assistance with EHR applications and research15. To date, MEDI contains 3,112 medications and 63,343 medication indications in the form of <drug, clinical phenotype> pairs, where medications are represented by RxNorm concept unique identifiers (RxCUI) and clinical phenotypes are identified by ICD-9 codes. <drug, clinical phenotype> pairs were extracted from four different medication resources: 1) RxNorm – a National Library of Medicine (NLM)-organized drug knowledge platform; 2) SIDER 2 – a drug database handling FDA-retrieved adverse drug reactions; 3) Medline Plus – a website that provides health information for patients and healthcare providers; and 4) Wikipedia – a collaborative online encyclopedia. MEDI also employs formal and prevalent identifications for the information it contains, allowing more productive and straightforward mapping and merging of data. We utilize <drug, clinical phenotype> pairs managed in MEDI to assess plausibility of <drug, clinical phenotype> pairs we learned from GWAS and PheWAS findings.
Methods
Our investigation begins with learning associations between drugs and clinical phenotypes (drug indications) via DrugBank and GWAS and PheWAS findings. We then assess the plausibility of learned drug indications via MEDI and utilize statistical models to assess the significances of the learned drug indications. The pipeline to learn drug indications and their evaluations are depicted in Figure 2.
Figure 2.
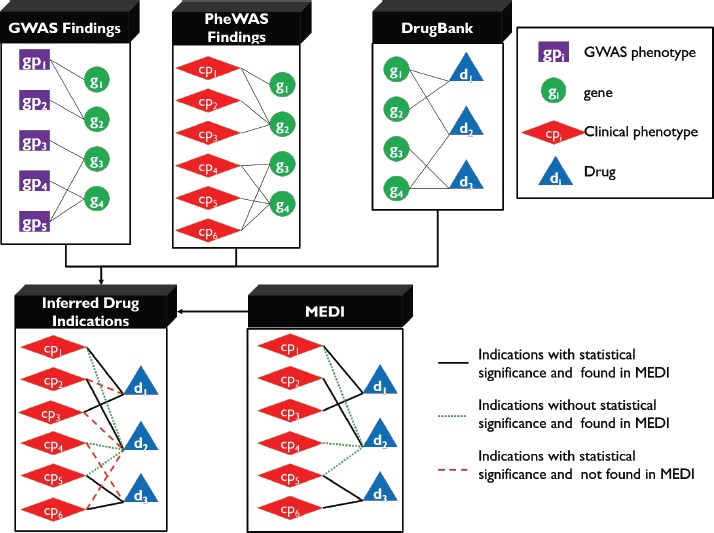
Methods pipeline. Top left: <GWAS phenotype, gene> relationships are inferred from information in GWAS catalogs and INRICH analysis. Top center: <clinical phenotype, gene> relationships are provided by already conducted PheWAS studies. Top right: <drug, gene> relationships are found in DrugBank. These three forms of data are mapped to produce learned drug indications (bottom left). The learned drug indications are then checked for documentation in MEDI.
First, we infer associations between clinical phenotypes and genes via GWAS and PheWAS findings. Specially, we align <GWAS phenotype, genes> to <PheWAS code, genes>.
Second, we leverage overlapped genes between GWAS traits associated genes derived from GWAS findings and drug-targeted genes to learn drug indications, or <drug, GWAS phenotype> pairs, and test their statistical significances.
Third, we will align the learned <drug, GWAS phenotype> pairs to <drug, PheWAS code> pairs and then leverage MEDI as a gold standard to evaluate the plausibility of the pairs. Through this process, we can categorize learned indications into three groups: i) indications with significant genetic relations (e.g. overlapped genes) between the drugs and clinical phenotypes and are also confirmed by MEDI; ii) indications without significant genetic relations and are confirmed by MEDI; and iii) indications with significant genetic relations and not confirmed by MEDI.
Finding potential drug indications and test their statistical significance
For this investigation, we determine the degree of association (in the form of P values) between drug and GWAS phenotype for 151,011 pairs (987 drugs crossed with 153 GWAS phenotypes). These P values are calculated by comparing the observed number of drug targets within associated regions of a particular GWAS trait to randomly permuted regions of the exact length and number, which ensures that the total number of genes is the same. For instance, in Figure 3, the degree of association between a GWAS phenotype and a drug is dependent on the number of overlapping genes between their associated gene-sets.
Figure 3.
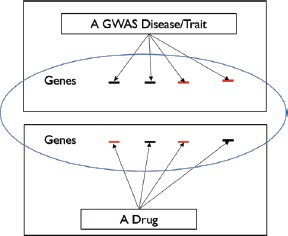
An example to measure association between a GWAS phenotype and a drug via their overlapped genes, which are shown in red.
For each <drug, GWAS phenotype> pair, we infer: i) the number of gene targets for the medication, which is obtained from DrugBank; ii) number of genes overlapped between drug-targeted genes and GWAS phenotype associated genes; and iii) the P value indicating the significance of the association between the drug and GWAS phenotype.
Thereafter, we transform <drug, GWAS phenotype> pairs into <drug, clinical phenotype> pairs using mapping information between PheWAS Codes and GWAS traits/diseases learned from both GWAS and PheWAS findings. We represent clinical phenotypes in the form of PheWAS codes as opposed to GWAS phenotypes because GWAS phenotypes are not completely analogous to clinical conditions and transitioning to clinical phenotypes allows for precise confirmation of learned indications.
Assessing plausibility of learned <drug, clinical phenotype> pairs
We use MEDI as a gold standard to confirm learned drug indications. In order to perform confirmation, we must first resolve the gap regarding identification of drug and phenotype between learned indications and those in MEDI. As shown in Table 1, a drug indication in MEDI is represented as a RxNorm ID (RxCUI) and ICD-9 code pair whereas a learned drug indication is represented as a DrugBank ID and PheWAS code pair.
Table 1.
Differences in representation of data in MEDI and of learned indications.
| Drug Indications | Drug | Clinical Phenotype |
|---|---|---|
| Inferred | Drug in DrugBank | PheWAS code |
| MEDI | Drug in RxNorm | ICD-9 code |
For the inconsistency of clinical phenotypes between inferred indications and those in MEDI, we use mappings between ICD-9 codes and PheWAS codes to replace ICD-9 codes in MEDI with PheWAS codes. This is because PheWAS codes have been validated to be more accurate to represent clinical phenotypes than ICD-9 codes by various PheWAS studies21, 23. For the inconsistency of drugs between DrugBank and RxNorm, we map DrugBank drug IDs to RxNorm IDs using the FDA’s UNII system and INCHI keys. Such mapping information has been widely used under the Observational Health Data Sciences and Informatics (OHDSI) framework24. After drugs and clinical phenotypes alignments between different systems (e.g., PheWAS, DrugBank, RxNorm, and MEDI), we use indications in MEDI to confirm if the learned indications exist in clinical setting.
One of our ultimate goals is to utilize GWAS and PheWAS findings to find novel drug indications and then recommend these indications to be further investigated to reveal more clinical treatment options. To achieve such a goal, we first need to demonstrate that the occurrence of learned significant drug indications found in MEDI is not random. Our hypothesis was designed as follows: the learned significant <drug, clinical phenotype> pairs confirmed in MEDI did not occur by chance. To test this hypothesis, we use two types of statistical tests: i) random permutation; and ii) chi-square.
For the random permutation, we generate random pairs based on the learned significant drug indications. Specifically, we randomly shuffle between drugs and clinical phenotypes as shown in the bottom of Figure 4. To perform an empirical permutation test, we repeat the random process 100 times. Within each permutation, we count the number of drug indications confirmed by MEDI. The P value to test that the learned drug indications confirmed by MEDI are not randomly inferred is calculated as: , where Num(permutations) is defined as the number of permutations whose number of indications confirmed by MEDI is equal or greater than the number of learned indications confirmed by MEDI.
Figure 4.
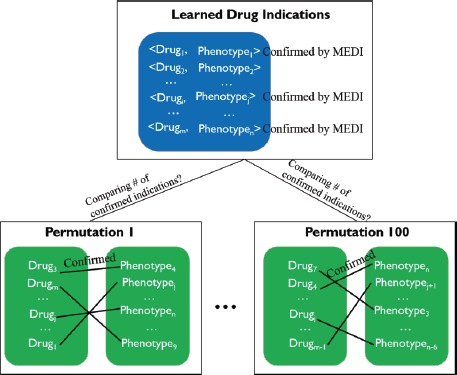
Process to count the number of permutations whose confirmed number of drug indications are equal or larger than the number of learned drug indications confirmed in MEDI.
For the chi-square test, we also run 100 random permutations and conduct a chi-square test for each permutation. Our null hypothesis is that the learned significant <drug, clinical phenotype> pairs confirmed by MEDI occurred by chance. To calculate the chi-square and p-values, we build a 2x2 contingency table with rows represented by learned/random pairs and columns represented by in/not in MEDI. The details are shown in Table 2. Because we perform 100 trials, we will compute and average the chi-square and P values for all trials.
Table 2.
Information used by Chi-square test to determine if learned indications confirmed by MEDI occur by chance.
| Number of drug indications | ||
|---|---|---|
| In MEDI | Not in MEDI | |
| Learned drug indications | a | b |
| Permutated drug indications | c | d |
Results
Learned <drug, clinicalphenotype> pairs
151,011 <drug, GWAS phenotype> pairs were transformed into 156,933 <drug, clinical phenotype> pairs in order to confirm each pair in MEDI. After checking for documentation for these 156,933 pairs, we then condensed the results back down to 151,011 <drug, GWAS phenotype> pairs. To do so, we gathered all <drug, clinical phenotype> pairs that map to a single <drug, GWAS phenotype> pair and sum the number of pairs that were documented in MEDI. Out of the 151,011 pairs, 1,431 were confirmed in MEDI and 763 were statistically significant with a P value less than 0.05. The results are shown in Figure 5.
Figure 5.
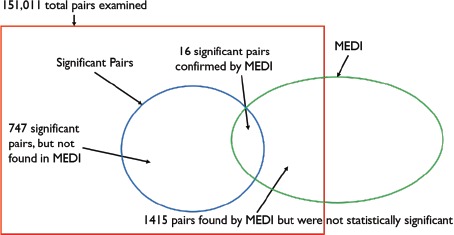
A Venn diagram depicting the learned drug indications. There were 1,431 learned indications (<drug, GWAS phenotype> pair) confirmed in MEDI and 763 pairs that were tested to be significant. Out of all total pairs examined, 16 pairs were both significant and confirmed in MEDI.
From the figure, it can be seen that out of 763 significant pairs, 16 were confirmed to be in MEDI as shown in Table 3, which indicates these drug indications are genetically supported. In other words, the drug and the clinical phenotype in these pairings have overlapping genes and their genetic relation is statistically significant. At the same time, the drug used to treat the clinical phenotype was also found to be used in clinical practice (confirmed by MEDI).
Table 3.
The 16 significant pairs that were found in MEDI.
| RxCUI IDs | Drug | GWAS Phenotype | PheCode | P value |
|---|---|---|---|---|
| 5691 | Imipramine | Bipolar Disorder | 296.1 | 0.002598 |
| 5691 | Imipramine | Schizophrenia | 295.1 | 0.039195 |
| 89013 | Aripiprazole | Schizophrenia | 295.1 | 0.049467 |
| 6779 | Mesoridazine | Schizophrenia | 295.1 | 0.044916 |
| 8331 | Pimozide | Schizophrenia | 295.1 | 0.003172 |
| 8704 | Prochlorperazine | Schizophrenia | 295.1 | 0.027732 |
| 115698 | Ziprasidone | Schizophrenia | 295.1 | 0.049467 |
| 1525 | Bezafibrate | Type 2 Diabetes | 250.2 | 0.024596 |
| 4821 | Glipizide | Type 2 Diabetes | 250.2 | 0.005061 |
| 274332 | Nateglinide | Type 2 Diabetes | 250.2 | 0.000051 |
| 73044 | Repaglinide | Type 2 Diabetes | 250.2 | 0.012215 |
| 72610 | Troglitazone | Type 2 Diabetes | 250.2 | 0.000069 |
| 106955 | Cortisone | Ulcerative Colitis | 555.2 | 0.003842 |
| 155323 | Methylprednisolone | Ulcerative Colitis | 555.2 | 0.001987 |
| 8640 | Prednisone | Ulcerative Colitis | 555.2 | 0.001457 |
| 142442 | Naproxen | Urate Levels | 274.1 | 0.009905 |
For each pair of the remaining 747 significant drug indications that were not confirmed in MEDI, the drug and the clinical phenotype in these pairings have significant genetic association, but the drug was not found to be used to treat the clinical phenotype in practice. This provides a great opportunity for drug companies to repositioning these drugs to further investigate their novel indications.
There are 1,415 drug indications that were not significant but found in MEDI. These indications are more complex, and could be further investigated in the following directions. First, they may be genetically associated but have notbeen investigated; if this is the case, we can conduct more GWAS and PheWAS studies to study their genetic relationship. Second, the drug and the clinical phenotype within a pair might not be genetically correlated and the drug has been employed to treat the clinical phenotype in practice; in this scenario, drug companies or clinicians can conduct studies to learn more about the pairing from the clinical perspective. Examples of both significant drug indications that were not confirmed in MEDI and not significant drug indications that were found in MEDI are shown in Table 4.
Table 4.
Left of the table provides examples of significant pairs that were not found in MEDI. Right of the table provides examples of non-significant pairs that were found in MEDI.
| Significant pairs not found in MEDI | Non-significant pairs found in MEDI | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| RxCUI IDs | Drug | GWAS Phenotype | PheCode | P value | RxCUI IDs | Drug | GWAS Phenotype | PheCode | P value |
| 51499 | Irinotecan | Asthma | 495.0 | 0.033482 | 3628 | Dopamine | Bipolar Disorder | 296.1 | 1 |
| 85248 | Alosetron | Bladder Cancer | 189.21 | 0.005432 | 328134 | Gefitinib | Lung Cancer | 165.1 | 0.209136 |
| 18747 | Balsalazide | Obesity | 278.1 | 0.000005 | 5640 | Ibuprofen | Migraine | 340.0 | 1 |
| 89784 | Isopropamide | Type 2 Diabetes | 250.2 | 0.012065 | 73044 | Repaglinide | Type 1 Diabetes | 250.1 | 1 |
| 1525 | Bezafibrate | Melanoma | 172.1 | 0.023378 | 10432 | Thalidomide | Prostate Cancer | 185.0 | 0.179429 |
Statistical test results of 16 significant drug indications confirmed by MEDI
We implemented the random permutation procedure and obtained an expected value of 0 all 100 times. Consequently, the P value provided by the empirical permutation test is 0/100. However, because P values cannot be 0 and we performed 100 trials, the best approximation for our P value is <0.01. The chi-square test corroborated these results. The average chi-square value and P value were calculated to be 16.170 and <0.0001, respectively.
According to the results from random permutation and chi-square test, we confirmed that the 16 significant learned drug indications did not occur by chance, which indicates the drug and the clinical phenotype in each of these pairs have genetic relations and the drugs have been used in the clinical practice for treatments.
In other words, it is unlikely that 16 of 763 observed significant pairs were found in MEDI by chance and there exists a correlation between genetic significance and clinical success. For that reason, there is benefit in conducting additional investigations (i.e. reviewing drug-phenotype associations, clinical trials, etc.) to interpret and discover the remaining 747 pairs with genetic significance but not found in the clinical practice as well as to investigate the MEDI pairs for which genetic and drug target information existed but no significant overlap was found.
Discussion
Our work intends to identify potential drug therapies by way of drug repositioning using GWAS and PheWAS, DrugBank, and MEDI data. Drug repositioning represents a more efficient method for acquiring undiscovered drug treatment options than the traditional drug discovery route. Implementing drug repositioning can involve gathering genetic evidence in support of possible therapies and testing for association with clinical documentation. For this study, we learned drug-GWAS phenotype correlations from GWAS and PheWAS findings and employed the MEDI database as our gold standard to assess plausibility of learned drug indications. Our drug indication learning and evaluating framework outlined above offers insight into the viability of drug repositioning given public resources (e.g. GWAS, PheWAS, DrugBank and MEDI). The contributions of this work are as follows:
-
1)
First, we proposed a framework to build connections among drugs, genes, and phenotypes via available public resources. For example, we use PheWAS findings to build connections between GWAS phenotypes and clinical phenotypes, which provides an opportunity to relate genotypes and clinical phenotypes. Additionally, we leverage overlapped genes to connect drugs with GWAS phenotypes.!
-
2)
Second, we filled the gap between drug indications managed in MEDI and those learned from GWAS and PheWAS findings, which provides an opportunity to perform comparisons between drug indications learned from the genetic perspective and those learned from the clinical perspective.
-
3)
Third, we deduced several interesting findings. For instance, we found 1,415 drug-phenotype pairs that are not significantly correlated but are documented in MEDI. Explanations for this phenomenon include the increasing need for conducting more GWAS and PheWAS studies as well as the understanding that diseases/traits can be environmentally influenced.
-
4)
Fourth, we deduced several interesting findings. For instance, we found 747 drug-phenotype pairs that have significant overlap between genetic findings and drug targets but are not documented in MEDI. While many of these will be false positives, we expect some proportion of these pairs could represent drug repositioning opportunities. For the five of the 747 significant pairs not located in MEDI listed in Table 4, the drugs mostly look to be unrelated to the clinical phenotypes. For example, Alosetron is used to treat diarrhea symptoms for irritable bowel syndrome (IBS) but was discovered to have genetic correlation with bladder cancer. Another example is Irinotecan, which is more generally employed to treat colon cancer but was discovered to be genetically related with a lung condition like asthma. However, further research is needed to determine whether these genetic associations have clinical significance.
We note that there are several limitations regarding data representation and alignments, which could have specifically lowered the number of confirmed significant pairs learned from GWAS and PheWAS findings. For instance, the alignments between drugs in DrugBank and those in RxNorm; connections between GWAS phenotypes and clinical phenotypes; and mappings between ICD-9 codes and PheWAS codes. Also, we realize that not all significant <drug, GWAS phenotype> pairs can result in successful drug repositioning outcomes. Ultimately, however, based on the statistical analysis performed, we have found promising developments in terms of drug repositioning with regards to the resources utilized in this study.
Conclusion
Drug repositioning introduces a productive approach to identifying novel drug therapies. The process involves first pinpointing new disease targets for existing medications on the market and evaluating whether these newly formed drug indications are applicable from both of genetic and clinical perspectives. In this study, we proposed a framework to build connections between drugs, genes and phenotypes via available public resources and then to deduce novel drug indications and evaluate their plausibility via MEDI. We investigated 151,011 <drug, GWAS phenotype> pairs and found that: i) 1,431 pairs were confirmed by MEDI; and ii) 763 pairs with significant genetic relations and 16 of them were confirmed by MEDI. Furthermore, we conducted statistical analysis (e.g. empirical permutation and chi-square test) to illustrate that the learned significant pairs (drug indications) confirmed by MEDI did not occur by chance.
Future data-driven drug repositioning investigations can be conducted by altering our methods in several ways, including but not limited to utilizing other genetic information outside of GWAS and PheWAS databases to link medications with indications, employing different drug indication databases for determining validity for learned pairs, and focusing drug reposition for one particular drug for potentially better accuracy. While there were limitations concerning data alignment (e.g. mapping between GWAS phenotypes and PheWAS codes, mapping between DrugBank IDs and RxNorm IDs, etc.), these outcomes depict optimistic developments for pharmaceutical therapy discovery by way of drug repositioning.
Acknowledgements
This research was supported, in part, by grants T15LM00740, and R00LM011933 of the National Library of Medicine, National Institutes of Health.
References
- 1.Gambardella A, Labate A, Mumoli L, Lopes-Cendes I, Cendes F. Role of pharmacogenomics in antiepileptic drug therapy: current status and future perspectives. Curr Pharm Des. 2017 Sep 10; doi: 10.2174/1381612823666170911111536. doi: [DOI] [PubMed] [Google Scholar]
- 2.Mould DR, Hutson PR. Critical Considerations in Anticancer Drug Development and Dosing Strategies: The Past, Present, and Future. J Clin Pharmacol. 2017;57(Suppl 10):S116–S128. doi: 10.1002/jcph.983. doi: [DOI] [PubMed] [Google Scholar]
- 3.Tripathi R, Lee-Verges E, Higashi M, et al. New drug discovery approaches targeting recurrent mutations in chronic lymphocytic leukemia. Expert Opin Drug Discov. 2017;12(10):1041–52. doi: 10.1080/17460441.2017.1362387. [DOI] [PubMed] [Google Scholar]
- 4.Luo Y, Zhao X, Zhou J, et al. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat Commun. 2017;8(1):573. doi: 10.1038/s41467-017-00680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khaladkar M, Koscielny G, Hasan S, et al. Uncovering novel repositioning opportunities using the Open Targets platform. Drug Discov Today. 2017;S1359-6446(17):30189–7. doi: 10.1016/j.drudis.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Tsubamoto H, Ueda T, Inoue K, Sakata K, Shibahara H, Sonoda T. Repurposing itraconazole as an anticancer agent. Oncology letters. 2017;14(2):1240–46. doi: 10.3892/ol.2017.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbett A, Pickett J, Burns A, Corcoran J, Dunnett SB, Edison P, et al. Drug repositioning for Alzheimer’s disease. Nat Rev Drug Discov. 2012;11(11):833–46. doi: 10.1038/nrd3869. [DOI] [PubMed] [Google Scholar]
- 8.Lu ZN, Tian Bl, Guo XL. Repositioning of proton pump inhibitors in cancer therapy. Cancer Chemother Pharmacol. 2017 doi: 10.1007/s00280-017-3426-2. doi: [DOI] [PubMed] [Google Scholar]
- 9.Ishida J, Konishi M, Ebner N, Springer J. Repurposing of approved cardiovascular drugs. J Transl Med. 2016;14:269. doi: 10.1186/s12967-016-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harikrishna Reddy D, Misra S, Medhi B. Advances in drug development for Parkinson’s disease: present status. Pharmacology. 2014;93(5-6):260–71. doi: 10.1159/000362419. [DOI] [PubMed] [Google Scholar]
- 11.Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3(96):96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shim JS, Liu JO. Recent advances in drug repositioning for the discovery of new anticancer drugs. Int J Biol Sci. 2014;10(7):654–63. doi: 10.7150/ijbs.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu H, Aldrich MC, Chen Q, et al. Validating drug repurposing signals using electronic health records: a case study of metformin associated with reduced cancer mortality. J Am Med Inform Assoc. 2015;22:179–91. doi: 10.1136/amiajnl-2014-002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wishart D S, Knox C, Guo A C, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic acids research. 2006;34(suppl_1):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W, Bastarache L, Lasko TA, Xu H, Cronin RM, Denny JC. Development and evaluation of an ensemble resource linking medications to their indications. Journal of the American Medical Informatics Association: JAMIA. 2013;20(5):954–61. doi: 10.1136/amiajnl-2012-001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research. 2014;42(D1):D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26(9):1205–10. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Re M, Valentini G. Network-based drug ranking and repositioning with respect to DrugBank therapeutic categories. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2013;10(6):1359–71. doi: 10.1109/TCBB.2013.62. [DOI] [PubMed] [Google Scholar]
- 19.Purcell SM, Thomas B, O’Dushlaine C, Lee PH. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28(13):1797–99. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruderfer DM, Charney AW, Readhead B, et al. Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach. The Lancet Psychiatry. 2016;3(4):350–7. doi: 10.1016/S2215-0366(15)00553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roden DM, Bastarache L, Denny JC. Phenome-Wide Association Studies as a Tool to Advance Precision Medicine. Annu Rev Genomics Hum Genet. 2016;17:353–73. doi: 10.1146/annurev-genom-090314-024956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016;23(6):1046–52. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–10. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyce RD, Horn JR, Hassanzadeh O, de Waard A, Schneider J, Luciano JS, Rastegar-Mojarad M, Liakata M. Dynamic enhancement of drug product labels to support drugsafety, efficacy, and effectiveness. J Biomed Semantics. 2013;4(1):5. doi: 10.1186/2041-1480-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


