Abstract
Standards-based modeling of electronic health records (EHR) data holds great significance for data interoperability and large-scale usage. Integration of unstructured data into a standard data model, however, poses unique challenges partially due to heterogeneous type systems used in existing clinical NLP systems. We introduce a scalable and standards-based framework for integrating structured and unstructured EHR data leveraging the HL7 Fast Healthcare Interoperability Resources (FHIR) specification. We implemented a clinical NLP pipeline enhanced with an FHIR-based type system and performed a case study using medication data from Mayo Clinic’s EHR. Two UIMA-based NLP tools known as MedXN and MedTime were integrated in the pipeline to extract FHIR MedicationStatement resources and related attributes from unstructured medication lists. We developed a rule-based approach for assigning the NLP output types to the FHIR elements represented in the type system, whereas we investigated the FHIR elements belonging to the source of the structured EMR data. We used the FHIR resource “MedicationStatement” as an example to illustrate our integration framework and methods. For evaluation, we manually annotated FHIR elements in 166 medication statements from 14 clinical notes generated by Mayo Clinic in the course of patient care, and used standard performance measures (precision, recall and f-measure). The F-scores achieved ranged from 0.73 to 0.99 for the various FHIR element representations. The results demonstrated that our framework based on the FHIR type system is feasible for normalizing and integrating both structured and unstructured EHR data.
Introduction
With the widespread usage of electronic health records (EHRs) in healthcare organizations, there are huge requirements for semantic interoperability and computable phenotyping in clinical and translational research. The lack of EHR data interoperability between institutions, however, makes it challenging for secondary use of EHR data, especially in collaborative research across institutions. Representing EHR data using a standard data model would assist in achieving large-scale data research collaboration, and meanwhile support the rapid generation of computable phenotypes.
Several data models have been developed to provide a standardized data representation for EHR data, including, amongst others, the HL7 Consolidated Clinical Document Architecture (CDA)1, the HL7 Reference Information Model (RIM)2, the OHDSI Common Data Model (CDM)3, the National Quality Forum (NQF) Quality Data Model (QDM)4, the Informatics for Integrating Biology and the Bedside (i2b2)5, the HL7 Fast Healthcare Interoperability Resources (FHIR)6. Meanwhile, clinical Natural Language Processing (NLP) plays an important role in extracting information from clinical text to a structured representation. A number of standards-based data modeling approaches have been studied using a variety of NLP technologies, such as using NLP to automatically generate entry level CDA7, mapping textual queries to OHDSI CDM8, using NLP for clinical research on i2b2 data sets9, using NLP supported diagnostic criteria QDM representation10. Some research has discussed using NLP with output to the FHIR data model, e.g. integration of NLP data with other sources of cancer data to build FHIR-based cancer phenotypes11. These studies, however, are limited in application to a specific clinical domain. To the best of our knowledge, a generic integration approach for modeling EHR data with the FHIR data model has not yet been well studied.
As an emerging next generation standard framework, FHIR was developed to meet clinical interoperability needs, and has the benefit of being relatively easy to implement and rapid to deploy. In addition, FHIR leverages the latest web standards and places a strong focus on implementability. Meanwhile, in regards to the NLP modules and tools developed independently in the NLP research community, there are broad requirements for achieving direct interoperability through a common type system with integrating applications. To extract structured data from unstructured data so as to model EHR data into the FHIR data model, a computationally processable type system that is shareable amongst multiple NLP systems is required. The Apache-licensed project Unstructured Information
Management Architecture (UIMA) provides such a software framework for building type systems while supporting interaction between multiple NLP components.
Another significant challenge is related to the terminology binding. Many elements in the FHIR resources have a coded value; some in the form of a fixed string (a sequence of characters) assigned as one of a set of fixed values defined in the FHIR specification; some in the form defined as “concept” where external terminologies or ontologies (e.g. LOINC, RxNorm or SNOMED CT) are used. In some cases, a locally maintained dictionary and/or look up table are even used as a part of FHIR profiles. Normalizing non-standard data into the coded FHIR fields thus poses a challenge.
Therefore, the overall objective of our study is to develop and evaluate a generic framework and accompanying methods for integrating structured and unstructured EMR data to a standard and interoperable format. We implement a clinical NLP pipeline enhanced with an FHIR-based type system and perform a case study using medication data from Mayo Clinic’s EHR.
Materials and Methods
Materials
Data corpus
We used a data set that was composed of 14 clinical notes randomly selected from those generated by Mayo Clinic and contained 166 manually annotated medication statement resources and their associated elements. The annotation set was used to evaluate the performance of our methods.
UIMA and UIMA-based NLP tools
UIMA is a data-driven architecture where individual components are able to communicate with one another through a data structure called the Common Analysis System (CAS), which uses a specified hierarchical type system. A common type system was thus defined under UIMA to meet the need for interoperability between different NLP12. As there are many clinical NLP tools developed on UIMA architecture, our NLP implementation was also designed using UIMA so as to increase interoperability, Currently, UIMA based clinical NLP tools include: 1) cTAKES13- cTAKES provides the pipeline for selecting which descriptors are used together to support the clinical NLP tasks; 2) MedXN14- MedXN is a medication entity and attribute extraction and normalization NLP tool; and 3) MedTime15- MedTime extracts and normalizes TIMEX3-based temporal expressions from clinical text.
FHIR Specificaiton
FHIR is based on a notion of “resource.” The FHIR specification defines a set of core resources and an infrastructure for handling resources16. The core FHIR resources represent a wide range of healthcare related concepts, both clinical and administrative. Through aggregating granular clinical concepts, the resources can support the representation of complex clinical senarios. Currently, FHIR has 3 available exchange formats: JSON, XML and RDF17.
HAPI FHIR
HAPI FHIR is an open-source implementation of the FHIR specification in Java18. HAPI FHIR defines model classes for every resource type and data type defined by the FHIR specification. The class FhirContext acts as a factory for most other parts of the API as well as a runtime cache of information that HAPI needs to operate. As the HAPI FHIR APIs supports the data model as defined by FHIR specifications, we used the HAPI FHIR API to serialize data stored in our UIMA-based FHIR type system into standard FHIR XML and JSON representations.
Methods
For each different types of FHIR resource, corresponding EHR data may exist as either structured data, e.g. patient demographics and laboratory test observations stored within a database, or as unstructured clinical notes, e.g. medication lists and problem lists. In many cases, the unstructured data is embedded within semi-structured EHR data, which provides some information on the nature of the data’s content. According to different characteristics of the EHR data, we designed an integrated framework for modeling EHR data into the FHIR specification. The FHIR data modeling tasks are separated into two workflows, as shown in Figure 1. The first workflow is for modeling unstructured EHR data in FHIR. This requires using different clinical NLP tools to recognize elements from different clinical note sections, and to transform these clinical elements into their respective elements within the FHIR data model. The second workflow combines information from structured data with the NLP output to build a complete FHIR resource. For this workflow, the metadata mapping and data standardization are the core tasks before conducting data transformation. An FHIR-based type system is developed to integrate different FHIR elements from unstructured structured data into a generic framework that supports direct generation of the data in FHIR standard format.
Figure 1.
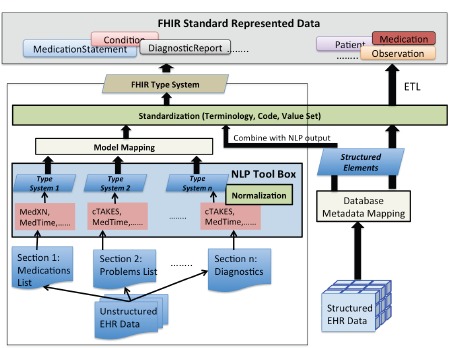
The framework for integrating structured and unstructured EHR data using FHIR
Considering medication statement data is often recorded within clinical narratives, in this paper, we discuss our methods in detail using one type of FHIR resource, “MedicationStatement”. To demonstrate our expected modeling results, a standard FHIR representation of the textual medication statement “Guaifenesin 100-mg/5 mL give 10 mL every six hours as-needed for cough.” is shown in Figure 2.
Figure 2.

An example of an FHIR MedicationStatement instance
Transforming FHIR resources into annotation schemas
An annotation schema is necessary for annotating unstructured data elements, so as to guide the extraction of entities from textual notes. The FHIR RDF specification was loaded as a schema for annotation purpose, and a Protégé plugin annotation tool Knowtator19 was used to annotate the textual clinical notes. The annotation process was to identify and categorize the annotated fields into the FHIR-based annotation schema. To focus the annotation field and create operational annotation guideline, we tailored the FHIR annotation schema for sectional annotation tasks. The clinically relevant sections defined in HL7 CDA20 are used for identifying target clinical notes sections. For example, the sections contain Allergies, Medications, Problems, immunizations, Diagnostics, etc., in this study, we used the FHIR “MedicationStatement” resource and associated resources for the annotation tasks. In a word, instances of FHIR elements belonging to the FHIR MedicationStatement were manually annotated for evaluation.
Acquiring FHIR elements from unstructured data
A number of different clinical NLP tools were integrated into the overall pipeline to perform different specialized tasks depending on the specific clinical note section. In our use case of extraction information from a medication list, we recognized 3 subtasks for which we integrated 3 different NLP tools (Figure 3). MedXN was responsible for the extraction of standardized medication concept mentions as well as its related attributes; MedTime was used to extract temporal elements defined in FHIR; and finally, other entities that could not be directly extracted by any existing NLP tools, such as Dosage.addtionalInstruction and MedicationStatement.reasonCode, were directly extracted from free-text through the development of specific NLP entity extraction modules.
Figure 3.
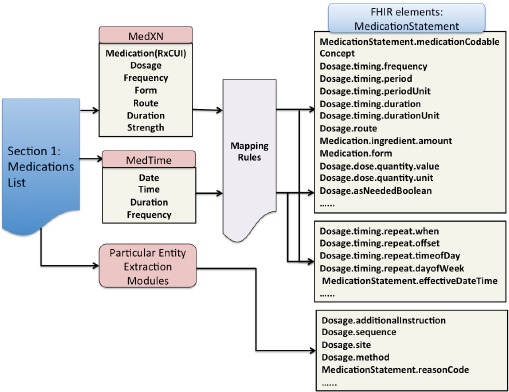
Populating the FHIR resource “MedicationStatement” from a textual medication list
Mapping the NLP output type to FHIR element type
As the output type systems of the different NLP tools all differed from our target FHIR element definition, we created a set of rules to classify the detected NLP elements. In terms of the mapping analysis of FHIR elements from the output of MedXN and MedTime, we concluded that while some of the NLP output types could be directly mapped to FHIR elements without semantic differences, e.g. MedXN:Medication.form maps directly to FHIR:Medication.form; in most situations, there were semantic gaps among different data models due to differing semantic granularity We therefore needed to do semantic converting, including semantic combination and separation, through the creation of mapping rules. After the model mapping, each entity and attribute recognized from NLP tools was classified into an FHIR element field, as shown in Figure 3.
Creating an FHIR type system to interoperate with UIMA-based NLP tools
UIMA is a NLP framework architecture commonly used in clinical NLP domain, and as such several NLP tools have been successfully implemented based on UIMA, especially under the OHNLP Consortium21. Since a significant portion of the adopted NLP tools in our integration framework were based on UIMA, we created an FHIR-based type system using the UIMA architecture to allow for rapid integration with the output from the available UIMA-based NLP tools, e.g. MedXN, MedEX, cTAKES, MedKAT, and MedTime. Using the FHIR specification schemas, we generated a corresponding FHIR type system in UIMA, which maintains consistency with the naming of elements (with the exception of certain UIMA reserved words such as begin, end, and start, for which corresponding FHIR fields were renamed from “{field}” to “fhir{field}”), structure hierarchy, and data restrictions present within the FHIR specification.
Combining structured data with NLP output
In regards to the unstructured notes within the EHR, mentions extracted from the text using NLP tools comprised the majority of the content for the corresponding FHIR resource backbone. There were still, however, several pieces of information that needed to be captured from structured EHR data and integrated with the NLP output to complete the population of the corresponding FHIR resource’s content. We take the FHIR resource “MedicationStatement” as an example here. While certain elements could be mapped from NLP output types, e.g. MedicationStatement.medicationCodableConcept, Dosage.timing. (frequency|period|duration), Dosage.route, Medication.form, Dosage.asneededBoolean, some of the other FHIR elements, such as MedicationStatement.status, MedicationStatement.subject, MedicationStatement.taken, MedicationStatement, category, could be directly mapped and imported from EHR structured tables. The key steps for combining structured data with NLP output were to: 1) Set templates for mapping the database metadata to the corresponding FHIR resource elements 2) extract the instance data; 3) Link the structured instance data with NLP output through a primary key reference or directly as an attributes defined within the FHIR resource.When populating each FHIR “MedicationStatement” resource instance, for example, we could directly get its “subject” (Who is/was taking the medication) information from structured EHR, and linked each “subject” to the specific “MedicationStatement” instance through the Reference (Patient|Group) identifier of the FHIR resource Patient or the FHIR resource Group. “Status” information (active | completed | entered-in-error | intended | stopped | on-hold), as another example, could be extracted directly from existing EHR tables but needed to be normalized using the FHIR defined value set.
Standardization and FHIR representation
FHIR names its methods of defining codes collectively as “code systems”. For example, for the MedicationStatement.route element, it specifies how a drug enters the body, the “SNOMED CT Route Codes” (http://hl7.org/fhir/ValueSet/route-codes) are used by FHIR to standardize different textual mention. For example, “by mouth”, “sprays each nostril”, and “topically” would be normalized to “Oral route|26643006”, “Nasal route|46713006”, and “topical route|6064005” with their associated SNOMED CT codes. A part of the normalization work has been achieved by NLP tools: for instance, MedXN converts medication information to the RxNorm standard and maps it to the corresponding RxCUI; cTAKES extracts medical concepts and assigns UMLS concept unique identifiers (CUIs) and SNOMED CT codes by integrating dictionary lookups for the UMLS, SNOMED CT and other dictionaries as part of its name entity recognition pipeline.
Evaluation design
We evaluated the overall performance of our rule-based classification algorithm for populating the FHIR type system. We used individual MedicationStatement resource instances (n = 166) from a collection of medication list sections from 14 clinical notes. These manually annotated MedicationStatements and their related attributes were then used as the gold standard to evaluate the performance of the rule based classification algorithm. Two authors (N.H., F.S.) annotated these medication lists, and the inter-observer agreement achieved was 0.95, which was deemed sufficiently consistent. Three standard measures were used to measure the performance of the extractors: precision (P), recall (R), and F-measure (F), where , where TP stands for True Positive, FP stands for False Positive, and FN stands for False Negative. For the capture of structured data, a review on the element definitions of the FHIR resource was used to evaluate the feasibility of gathering the FHIR elements from the structured Mayo Clinic EHR data.
Results
As an initial implementation, our annotation task focused on building the FHIR resource “MedicationStatement” and its related resources and elements. A screenshot of the FHIR annotation schema in Protégé for a MedicationStatement annotation is shown in Figure 4.
Figure 4.
The FHIR annotation schema in Protégé (MedicationStatement)
Table 1 shows our mapping relations between the MedXN and MedTime types and the FHIR elements in the “MedicationStatement”. After the model analysis, we found that the data elements mapping relations were generally 1:n maps with a few 1:1 maps, due to the FHIR model describing resources on a more refined granularity. We use abbreviation for the multi-level FHIR name system for display in Table 1. For example, the FHIR element “Dosage.timing.repeat.frequency” is displayed as “Dosage.frequency”.
Table 1.
Model Elements Mapping Between MedXN/MedTime and FHIR
| NLP tools and Output Types | FHIR Elements (MedicationStatement) | Mapping Rules and Examples |
|---|---|---|
| MedXN:Drug | MedicationStatement.medication CodableConcept | 1:1 Example: Guaifenesin [RxCUI: 5032] |
| MedXN: | Dosage.frequency | 1:n |
| Drug:attributes:type=’frequency’ | Dosage.frequencyMax | Examples: |
| Dosage.period | once daily | |
| MedTime: | Dosage.periodMax | →1[frequency], 1[period], |
| Dosage.periodUnit | d[periodUnit] | |
| MedTimex3:type=’SET’ | Dosage.asNeededBoolean | 4-6 times |
| Dosage.dayofWeek | →4[frequency], 6[frequencyMax] | |
| Dosage.when | Regular: once daily every six hours | |
| Irregular: as needed for pain | ||
| MedXN: | Dosage.duration | 1:n |
| Drug:attributes:type=’duration’ | Dosage.durationMax | Example: |
| MedTime: | Dosage.durationUnit | 3 days |
| MedTimex3:type=’DURATION’ | →3[duration], d[durationUnit] | |
| MedXN: | Dosage.route | 1:1 |
| Drug:attributes:type=’route’ | Examples: | |
| by mouth [oral route] | ||
| MedXN: | Medication.ingredient.amount. | 1:n |
| Drug:attributes:type=’strength’ | numerator.quantity.value | Examples: |
| Medication.ingredient.amount. | Regular: 500 mg /5 ml→ | |
| numerator.quantity.unit | 500[numerator.quantity.value], | |
| Medication.ingredient.amount. | mg[numerator.quantity.unit], | |
| denumerator.quantity.value | 5[denumerator.quantity.value], ml | |
| Medication.ingredient.amount. | [denumerator.quantity.unit] | |
| denumerator.quantity.unit | Irregular: 200 mg → Default assign: | |
| 1[denumerator.quantity. | ||
| MedXN: | Medication.form | 1:1 |
| Drug:attributes:type=’form’ | Examples: | |
| tab[Tablet] | ||
| MedXN: | Dosage.doseQuantity.value | 1:n |
| Drug:attributes:type=’dosage’ | Dosage.doseQuantity.unit | |
| Dosage.doseQuantity.Range.low.value | 10 ml →10[value], ml[unit] | |
| Dosage.doseQuantity.Range.low.unit | 2-3 tabs →2[range.low.value], tab[range.low.unit], | |
| Dosage.doseQuantity.Range.high.value | 3[range.high.value], tab[range.high.unit] | |
| Dosage.doseQuantity.Range.high.unit | Notes: we currently both map “tab” to dose unit and medication form | |
| MedTime: MedTimex3:type=“DATE” | MedicationStatement.effectiveD atetime | 1:1 Examples: April 16th |
| MedTime: | MedicationStatement.effectiveD atetime | 1:n Examples: |
| MedTimex3:type=“TIME” | Dosage.timeofDay | April 8, 2008 at 04:38 PM |
The FHIR Type system in XML format used for interoperability with existing NLP tools was built on the FHIR STU 3 v1.8.0 specification, which includes the full structure definition of FHIR specification, a fragment is displayed in Figure 5.
Figure 5.

The FHIR type system in UIMA
We used RxNorm to normalize medication concepts, and used the FHIR-defined value sets to normalize FHIR attribute elements. Currently, we have normalized 11 FHIR elements using locally configured data constraints, with work in progress on all FHIR elements standardization work, dependent to a large extent on the FHIR terminology services interfaces. Currently normalized elements comprise:
-
-
MedicationStatement.medicationCodeableConcept(RxNorm),
-
-
Dosage,timing.frequency(integer),
-
-
Dosage.timing.repeat.duration (decimal)
-
-
Dosage.timing.repeat.durationUnit (code: s | min | h | d | wk | mo | a - unit of time (UCUM))
-
-
Dosage,timing.repeat.frequencyMax(integer),
-
-
Dosage.timing.repeat.period(decimal),
-
-
Dosage.timing.repeat.periodMax(decimal),
-
-
Dosage.timing.repeat.periodUnit(code: s | min | h | d | wk | mo | a - unit of time (UCUM))
-
-
Dosage.asNeededBoolean(boolean),
-
-
Dosage.timing.repeat.dayofWeek(code: mon | tue | wed | thu | fri | sat | sun)
-
-
Dosage.timing.repeat.when(code: http://hl7.org/fhir/v3/TimingEvent).
Evaluation Results
In our experiment, we tested the results of integrating the MedXN and MedTime NLP pipelines to populate the FHIR resource “MedicationStatement”. The standardization processes were also evaluated as part of this process. Through a comparison of the results of using rule-based classification and our manual annotation (which included the normalized form, if applicable) of each FHIR element within the medication list notes of Mayo Clinic, we evaluated the performance of our NLP pipeline. Evaluation results are shown in Table 2. The evaluation results show that our rules-based method has slightly higher precision, recall, and F-measure comparing with MedXN and MedTime for those 1:1 mapping elements. For example, the element MedicationStatement.medicationCodeableConcept is the case and the reason is probably because the size of our annotated data set is relatively small. However, for those 1:n mapping elements (e.g.,
Table 2.
Evaluation on FHIR resource “MedicationStatement”
| FHIR Elements | Total | Precision | Recall | F-score |
|---|---|---|---|---|
| MedicationStatement.medicationCodeableConcept | 166 | 0.996 | 0.982 | 0.988 |
| Dosage.timing.repeat.frequency | 87 | 0.795 | 0.873 | 0.832 |
| Dosage.timing.repeat.period | 133 | 0.959 | 0.914 | 0.936 |
| Dosage.timing.repeat.periodUnit | 129 | 0.951 | 0.943 | 0.947 |
| Dosage.timing.repeat.duration | 5 | 0.600 | 1 | 0.750 |
| Dosage.route | 143 | 0.957 | 0.816 | 0.878 |
| Medication.ingredient.amount.numerator.quantity.value | 155 | 0.930 | 0.815 | 0.869 |
| Medication.ingredient.amount. numerator.quantity.unit | 135 | 0.926 | 0.899 | 0.911 |
| Medication.form | 127 | 0.871 | 0.704 | 0.779 |
| Dosage.dose.quantity.value | 93 | 0.974 | 0.835 | 0.899 |
| Dosage.timing.repeat.when | 21 | 1 | 0.571 | 0.727 |
| Dosage.asNeededBoolean | 38 | 0.913 | 0.583 | 0.712 |
Medication.ingredient.amount.quantity.value/unit (MedXN:strength)), the overall precision, recall, and F-measure is slightly lower than others, and the reason is because the entity normalization processing may cause the transforming mistakes to some extent..
Discussion
In this study, we proposed an integration framework to support the representation of structured and unstructured EHR data into the FHIR model, leveraging both the NLP-based mapping rules and structured data ETL methods. The FHIR-based type system was used as the integration central-bus for the interchange between multiple information extraction tools. We performed the initial experiment and evaluation on the FHIR resource “MedicationStatement”.
As the FHIR model is very rich and comprehensive with a multi-layered structures and elaborate definitions, in spite of creating mappings rules between NLP output types and FHIR model elements through delicate model analysis, more model elements are still being studied and the current mapping rules are far from complete. However, our results are effective in proving the feasibility of leveraging NLP tools to support the conversion of EHR data into the FHIR model. Currently, in our use case, MedXN and MedTime are used to perform the extraction of medication and temporal entities and attributes from medication lists. In the future, particular entity extraction modules will be developed for the reminder NLP extracted entries from medications list.
Our experiment was implemented using the UIMA architecture, in which we created an FHIR-based type system. We demonstrated that the FHIR type system was effective in facilitating integration of the differing outputs of multiple UIMA-based NLP tools. This mechanism provides powerful extension capability in that other UIMA-based NLP tools not part of this study can be easily integrated. In fact, we are working on the extension implementation on other FHIR resources representation. As part of the next step, we plan to adopt a clinical NLP tool cTAKES to acquire relevant NLP output for the FHIR resource types, such as Condition, Procedure, and Diagnostic Report. The cTAKES type system, based on Intermountain Healthcare’s Clinical Element Models (CEM)12, is currently being studied and mapped into the FHIR elements.
One of ongoing challenges is that the standardization work in FHIR is still in progress: the “code system” list and the “value set” list construction are on progressing22, and more standardization work needs to be done as part of our implementation. These include normalizing Dosage.route using the defined FHIR value set [http://hl7.org/fhir/ValueSet/route-codes], and integrating the standardization service into our pipeline.
Currently, we manually downloaded the FHIR resource in turtle format, and then extracted the resources classes and properties to build an FHIR schema for annotation. In a future design, we plan to automatically generate the FHIR annotation schema using the FHIR RDF specification.
Although the performance of NLP has room for improvement due to the complexity of clinical notes, our mapping rules relying on NLP output were able to populate FHIR elements instances into most elements defined in the FHIR MedicationStatement model. Our FHIR type system-driven integration method provides a generic and scalable framework to support the FHIR modeling of structured and unstructured data.
Conclusion
In this study, we developed a clinical NLP pipeline using an FHIR type system for integrating structured and unstructured EHR data. We demonstrated the feasibility of our approach, focusing on the core elements of the FHIR resource “MedicationStatement”. We are actively working on creating mapping rules and annotation work on other FHIR resource types and on improving the performance of our integration approach. We believe that the standard FHIR modeling method for EHR data, as illustrated in this study, will benefit future semantic data exchange and integration, and rapid generation of computational phenotypes for advancing clinical and translational research.
Acknowledgements
This study is supported in part by NIH grant U01 HG009450.
References
- 1.The HL7 Version 3 Clinical Document Architecture (CDA®) [cited 2017 Aug]. http://www.hl7.org/implement/standards/product_brief.cfm?product_id=7.
- 2.HL7 Reference Information Model. [cited 2017 Aug]. http://www.hl7.org/implement/standards/rim.cfm.
- 3.OHDSI Data Standardization. [cited 2017 Aug]. http://www.ohdsi.org/data-standardization/
- 4.Quality Data Model. [cited 2017 Aug]. http://www.qualityforum.org/QualityDataModel.aspx.
- 5.Informatics for Integrating Biology and the Bedside (i2b2) [cited 2017 Aug]. https://www.i2b2.org/
- 6.FHIR. [cited 2017 Aug]. https://www.hl7.org/fhir/
- 7.Jung S, Kim S, Yoo S, Choi J. Toward the automatic generation of the entry level CDA documents. Journal of Korean Society of Medical Informatics. 2009;15(1):141–51. [Google Scholar]
- 8.Liu S, Wang Y, Hong N, Shen F, Wu S, Hersh W, Liu H. On Mapping Textual Queries to a Common Data Model. Healthcare Informatics (ICHI); IEEE International Conference on; 2017; IEEE. 2017. [Google Scholar]
- 9.Uzuner O, Stubbs A. Practical applications for natural language processing in clinical research: The 2014 i2b2/UTHealth shared tasks. J Biomed Inform. 2015;58 Suppl:S1–5. doi: 10.1016/j.jbi.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong N, Li D, Yu Y, Xiu Q, Liu H, Jiang G. A computational framework for converting textual clinical diagnostic criteria into the quality data model. J Biomed Inform. 2016;63:11–21. doi: 10.1016/j.jbi.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochheiser H, Castine M, Harris D, Savova G, Jacobson RS. An information model for computable cancer phenotypes. BMC Med Inform Decis Mak. 2016;16(1):121. doi: 10.1186/s12911-016-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu ST, Kaggal VC, Dligach D, Masanz JJ, Chen P, Becker L, Chapman WW, Savova GK, Liu H, Chute CG. A common type system for clinical natural language processing. J Biomed Semantics. 2013;4(1):1. doi: 10.1186/2041-1480-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association: JAMIA. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sohn S, Clark C, Halgrim SR, Murphy SP, Chute CG, Liu H. MedXN: an open source medication extraction and normalization tool for clinical text. Journal of the American Medical Informatics Association: JAMIA. 2014;21(5):858–65. doi: 10.1136/amiajnl-2013-002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin YK, Chen H, Brown RA. MedTime: a temporal information extraction system for clinical narratives. J Biomed Inform. 2013;46 Suppl:S20–8. doi: 10.1016/j.jbi.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 16.FHIR: Guide to resources. [cited 2017 Aug]. https://www.hl7.org/fhir/resourceguide.html.
- 17.Solbrig HR, Prud’hommeaux E, Grieve G, et al. Modeling and validating HL7 FHIR profiles using semantic web Shape Expressions (ShEx) J Biomed Inform. 2017;67:90–100. doi: 10.1016/j.jbi.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.HAPI FHIR. [cited 2017 Aug]. http://hapifhir.io/
- 19.Knowtator. [cited 2017 Aug]. http://knowtator.sourceforge.net/index.shtml.
- 20.Consolidated Clinical Document Architecture (CCDA) User Manual. [cited 2017 Aug]. https://www.ihs.gov/RPMS/PackageDocs/BCCD/bccd010u.pdf.
- 21.OHNLP Consortium. [cited 2017 Sep]. http://www.ohnlp.org/index.php/Main_Page.
- 22.FHIR: Using Codes in Resources. [cited 2017 Sep]. https://www.hl7.org/fhir/terminologies.html.


