Abstract
Electronic Health Records (EHRs) have seen a rapid increase in adoption during the last decade. The narrative prose contained in clinical notes is unstructured and unlocking its full potential has proved challenging. Many studies incorporating clinical notes have applied simple information extraction models to build representations that enhance a downstream clinical prediction task, such as mortality or readmission. Improved predictive performance suggests a “good” representation. However, these extrinsic evaluations are blind to most of the insight contained in the notes. In order to better understand the power of expressive clinical prose, we investigate both intrinsic and extrinsic methods for understanding several common note representations. To ensure replicability and to support the clinical modeling community, we run all experiments on publicly-available data and provide our code.
Introduction
Electronic Health Records (EHRs) contain an abundance of data about patient physiology, interventions and treat-ments, and diagnoses. The amount of data can be overwhelming in the intensive care unit (ICU), where patients are severely ill and monitored closely. In this setting, it can be difficult to reconcile data from multiple sources; instead, care staff rely on clinical notes to provide short summaries that capture important events and results. This unstruc-tured, free text thus contains important observations about patient state and interventions, in addition to providing insight from caregivers about patient trajectory.
The secondary use of EHR data in retrospective analyses facilitates a better understanding of factors, such as those contained in clinical notes, that are highly predictive of patient outcomes.1,2,3,4 Additionally, the free-text nature of clinical notes means that data extraction does not rely heavily on each EHR’s implementation, making methods for clinical notes portable across different EHRs. However, there are many ways to represent the information contained in text, and it is unclear how to best represent clinical narratives for the purpose of predicting outcomes.
Many efforts to leverage clinical notes for outcome prediction focus on improving the performance of a final prediction task. 1,3,5,6 Post-hoc feature analysis can assist in discovering those features that are most predictive, but it provides only a partial solution toward improving our understanding. We would like to know what facts and derived features matter most in affecting the predictive abilities of the models we build from them. This will allow us not only to improve performance but to understand what representations of the identified features are most useful.
For example, although a patient’s EHR-coded race and social history may help to identify a Gonorrhea infection accurately,7 if we are trying to use text analysis tools to make such an identification, we would like to know if those tools are able to determine a patient’s race and social history accurately from the notes. While it is seemingly counter-intuitive to predict EHR-coded information using clinical notes, doing so provides insight into what is, and isn’t, reflected in a given note’s representation. Such awareness is important when designing representations for downstream prediction tasks because it exposes assumptions about what sophisticated models may be able to accomplish.
Toward the goal of understanding and improving note representations for downstream prediction performance, we consider several common representations and evaluate them on a variety of tasks. We explore performance on “easy” tasks, such as age, gender, ethnicity, and admission type, each of which are readily accessible as EHR-coded data. Ad-ditionally, we use the same models on common prediction tasks, such as in-hospital mortality and length of stay. We show that 1) no single representation outperforms all others, 2) a simple representation tends to outperform more com-plex representations on “easy” tasks while the opposite is true for “common” prediction tasks, and 3) some seemingly “easy” tasks, such as ethnicity, are difficult for all of the representations considered.
Related Work
Work leveraging clinical notes for prediction can be broadly categorized into those focusing on clinical prediction tasks and those focusing on the representation of text.
Clinical Prediction Tasks: Several existing works have demonstrated the utility of clinical narratives in forecasting outcomes. A standard approach for converting narrative prose to structured vector-based features has used unsuper-vised topic modeling to represent each note as a distribution over various topics. Lehman et al.6 and Ghassemi et al.1,5 use note-derived features in a framework to predict mortality. In recent work, Caballero Barajas and Akella2 use generalized linear dynamic models on top of latent topics to detect an increase in the probability of mortality before it occurs. Luo and Rumshisky8 use a supervised topic modeling approach to improve prediction of 30-day mortal-ity. Grnarova et al.4 use convolutional neural networks (CNNs) to construct document representations for the task of mortality prediction. Although the authors do not perform this prediction task in a time-varying manner, their results show that both doc2vec9 and their CNN approach improve performance relative to a topic representation. Further, Cohen et al.10 explore the use of redundancy-aware topic models in order to combat the prevalent issue of copying notes forward in a patient’s clinical record; however, they do not apply this model in a downstream prediction task. Similarly, Pivovarov et al.11 explore the use of topic models in the discovery of probabilistic phenotypes, but do not use these phenotypes toward predictions.
Text Representations: In the general domain, it has been observed that simple models with access to much data often outperform complex models with access to less data. Banko and Brill12 observe this effect directly in the general natural language processing domain, noting that many methods continue to be optimized on small datasets and prove ineffective when applied to datasets orders of magnitude larger. Similarly, Halevy et al.13 discover that “for many tasks, words and word combinations provide all the representational machinery we need to learn from text.” Previously, limited access to clinical narratives have made this observation less applicable to the clinical domain.
Data
This work uses data from the publicly-available Multiparameter Intelligent Monitoring in Intensive Care (MIMIC-III) database, version 1.4.14 MIMIC-III v1.4 contains de-identified EHR data from over 58,000 hospital admissions for nearly 38,600 adult patients. The data were collected from Beth Israel Deaconess Medical Center from 2001-2012. A typical clinical note might look like the one shown in Figure 1, which shows the radiology report of a 64-year-old patient with poor respiratory status.
Figure 1.
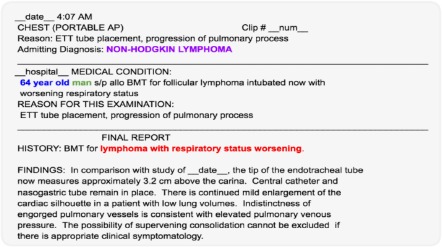
An example clinical note. The age, gender, and admitting diagnosis have been highlighted. Also note, thatdescriptions such as “status worsening” suggest deterioration and possible in-hospital mortality.
We consider only adult patients, older than 15 years old, who were in the ICU for at least 12 hours. Young patients are excluded as they typically exhibit different physiology from an adult population. Further, we include only each patient’s first ICU stay, thus precluding training and testing on data from the same patient. Due to recording and measurement issues in the database, we exclude any ICU stays that do not conform to the common sense ordering of
hosp-admission < icu-intime < icu-outtime < hosp-dischtime
Finally, we consider Nursing and Nursing/Other, Radiology, and Physician notes, because other categories occurred relatively infrequently. For each ICU stay, we extract the first 24 notes (or fewer if the stay has fewer notes). These cri-teria result in 29,979 unique ICU stays, an equivalent number of patients, and 320,855 notes. The dataset is randomly divided into a 7:2 train/test split.
As “easy” prediction tasks, we extract several coded variables for each patient that remain constant throughout the stay, including: age, gender, ethnicity, and admission type. In addition, we also retrieve “common” clinical outcomes and findings during the stay, such as: diagnosis, length of stay, and in-hospital mortality. We then try to predict these characteristics and outcomes from different representations of the text notes.
As observed in replication studies, one of the central obstacles in replicability — even for work done on public datasets — is that descriptions of data cleaning and preprocessing are often inadvertently underspecified.15 Therefore, we make our code publicly available. †
Methods
MIMIC-III v1.4 contains de-identified clinical notes. In preprocessing these notes, tags indicating de-identified pro-tected health information are removed. Phrases written entirely in capital characters are then replaced by a single token, effectively coalescing common structural elements; for example, the section heading “RADIOLOGIC STUD-IES” would be replaced with a single token. Additionally, regular expressions for common age patterns are used to replace all specified ages with symbols binned by decade to ensure relevant age information is not lost. Finally, we remove all non-alphanumeric tokens, and normalize all remaining numbers to a single number token.
For each word, we compute the number of unique patients who have a note containing that word — this is the “doc-ument” frequency. For each note, we compute the term frequency-inverse document frequency, or tf-idf of every word and keep the top-20 words of that document. Thus a patient’s stay is represented as an ordered list of filtered bags-of-words.
The following subsections describe several approaches to aggregate each patient’s multiple note vectors into one fixed-size patient vector that summarizes their stay in the ICU, as illustrated in Figure 2.
Figure 2.
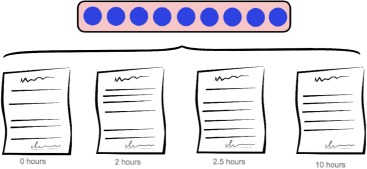
A patient’s time in the ICU generates a sequence of timestamped notes. Each of the methods described transforms the sequence of notes into a fixed-length vector representing the ICU stay.
Bag of Words
Bag of words (BoW) is one the simplest methods for creating vector representations of documents. Using the top-20 tf-idf words from each note produces a vocabulary of size |V| = 17,025 words. In this representation, the patient vector is a |V|-dimensional sparse, multi-hot vector. If a word appears in any of the notes for a given patient, then the corresponding dimension for that word is “on” in the resulting patient vector.
Bag of words presents a strong baseline representation for downstream predictive tasks. In this work, its strength is a result of its high dimensionality relative to other models: by reducing the representations into a smaller, denser space, other models may inadvertently throw out information with predictive value. More specifically, we expect that bag of words will perform well on tasks that involve the prediction of categories which may be directly represented by single words in their notes. For example, we would expect a note which frequently contains the word “male” to correctly identify the patient as male.
Word Embeddings
Due to the immense success of word2vec in recent years, we embed words and documents into a dense space in order to accommodate soft similarities. We train clinical word vectors using the publicly-available word2vec tool‡ on 129 million words from 500,000 notes taken from MIMIC-III. Hyperparameters were specified using Levy et al. 16 as a reference: 300-dimensional SGNS with 10 negative samples, a min-count of 10, a subsampling rate of 1e-5, and a 10-word window. These clinical embeddings are available for public use on the MIMIC-III Derived Data Repository. §
As shown in Figure 3, we create a note representation by aggregating the top tf-idf words in the document. With these top words, we look up each of their word2vec embeddings (blue) and collapse them into a final vector using elementwise -max, -min, and -average. We apply the same aggregation scheme (max, min, and average) to collapse the patient’s list of document vectors into one fixed-length patient vector.
Figure 3.
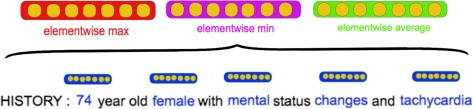
How the embedding for a single document is built by combining constituent word embeddings.
Recurrent Neural Network
One problem with the approaches described above is that they all ignore temporal ordering of the documents. That is to say, they fail to track the progression of a patient’s state over time during the ICU stay. One solution to this limitation is to use a sequence-based model. We use a Bidirectional LSTM network, which has proven to be very effective at modeling temporal sequences.17,18 For a more fair comparison, we build the list of document vectors for each patient in the same way that was done for word embeddings. These document vectors are then fed into the LSTM one document per timestep.
Our LSTM was implemented in Keras19 using an Bidirectional LSTM with 256 hidden units, a dropout rate of 0.5, and a 128-unit fully connected layer immediately before the output label softmax. Models were trained for 100 epochs.
Experimental Setup
The principal aim of this work is to better understand what information is captured by various representations of clinical notes. Because most of the derived representations have non-interpretable dimensions from the embedding process, we cannot look for correlations between individual dimensions and our queries. Instead, we develop a pre-diction framework to determine whether a particular representation has encoded the necessary information to predict the correct query against alternative values.
We consider the following prediction tasks modeling clinical states and outcomes:
Diagnosis. We filter down to patients with one of the 5 most common primary diagnoses and predict: CoronaryArtery Disease, Pneumonia, Sepsis, Intracranial Hemorrhage, and Gastrointestinal Bleed.
In-Hospital Mortality. Binary classification of whether the patient died during their hospital stay.
Admission Type. Binary classification of Urgent or Elective.
Length-of-Stay. Three-way classification of whether patients stayed in the ICU for Less than 1.5 days, Between1.5 and 3.5 days, and longer than 3.5 days.
However, we are also interested in whether the notes are able to capture basic demographic information:
Gender. Binary classification of Male or Female.
Ethnicity. Binary classification of White or Non-White.
Age. Three-way classification of age as Less than 50 years old, Between 50 and 80 years old, or Older than 80years old.
While these tasks reflect those commonly found in research, we use them to evaluate our representations rather than as clinically-actionable targets. For example, it might be noted that a single patient can suffer from multiple conditions, but here we consider only their primary diagnosis. Similarly, the ranges for age and length-of-stay are reasonable, but would need to be tailored in other conceivable applications. In both cases, however, these choices serve to highlight the types of information each representation is capturing.
Binary classification tasks are evaluated using AUC, while multi-way classification tasks are evaluated using the macro-average F1-score of the different labels. Predictions are made for bag-of-words and word embedding repre-sentations using a scikit-learn20 support vector classifier with linear kernel. Predictions are made for the LSTM usinga softmax layer.
Results
Performance for the 7 classification tasks using the 3 representation models are shown in Table 1 (binary classifi-cations) and Table 2 (multi-way classifications). In general, our findings match our expectations: while a complexmodel tends to do well for “downstream” tasks involving reasoning, such as diagnosis and length-of-stay, it strugglesto compete with a simpler model in token-matching tasks like age and gender.
Table 1.
AUCs for the binary classification tasks.
| in-hospital mortality | admission type | gender | ethnicity | |
|---|---|---|---|---|
| BoW | 0.821 | 0.883 | 0.914 | 0.619 |
| Embeddings | 0.814 | 0.873 | 0.836 | 0.580 |
| LSTM | 0.777 | 0.870 | 0.837 | 0.533 |
Table 2.
Macro-average F1 scores for the multi-way classification tasks.
| diagnosis | length of stay | age | |
|---|---|---|---|
| BoW | 0.828 | 0.724 | 0.635 |
| Embeddings | 0.828 | 0.730 | 0.544 |
| LSTM | 0.836 | 0.758 | 0.450 |
Specifically, the bag-of-words (BoW) model performs best at predicting so-called ‘common-sense’ tasks: age, gender, and (less significantly) ethnicity, for which there are words which almost directly predict the labels. In contrast, theLSTM model outperforms BoW on tasks more related to clinical reasoning: diagnosis and length of stay, for which we expect the temporal information to be important in predictions. Embeddings serve as a halfway between BoW and LSTM; while the method does not leverage a temporal sequence, this experiment allows us to untie the pre-trained word vectors from the temporal dynamics of the LSTM. In doing so, we see that the embeddings typically perform very competitively against BoW, but the LSTM is able to leverage them further.
Discussion
As shown in Table 1 and Table 2, the different models exhibit varied performance across tasks with no consistent winner. Bag-of-words tends to do well on tasks where a single word, or a few words, are strongly associated with prediction categories. Notably, bag-of-words is much better at predicting age. This is likely because the normalized, per-decade age tokens created during preprocessing are, of course, strongly associated with predicting age. The LSTM, on the other hand, had a difficult time distinguishing between the age token embeddings since all age tokens fall nearby one another within the embedding space, as shown in Figure 5.
Figure 5.

PCA 2-D projection of the word embeddings. Vectors of the special age tokens are colored red. Note thatthese tokens cluster close together in the embedding.
For these tasks, bag-of-words provide a strong baseline because some standard demographic information, such as age and gender, are typically specified in the notes. However, it is precisely because of their frequency of occurrence that information retrieval methods, such as tf-idf, underestimate their importance. Recall that tf-idf reduces the score of exceedingly common words. While this step is clearly important in the treatment of “stopwords” — words that are so common they provide no additional value — here it inadvertently removes commonly recorded information. This presents a challenge for aggregating the word embeddings of a note into one single document embedding because including too many words in the aggregate statistical values (i.e., averages, maximums, and minimums) drives down the “informativeness” of the representation by adding noise to these aggregate statistics.
Further, all methods achieve high AUCs for mortality, admission type, and gender; similarly, each performs poorly for ethnicity. The highest ethnicity AUC is still 20 points lower than the worst reported AUC for the other tasks. This suggests that predicting ethnicity from notes is an inherently difficult challenge. This is largely because race, while commonly coded elsewhere, is not typically specified in the notes. Additionally, 71% of patients are white in our dataset. This class imbalance may be large enough that a “default” value may be assumed and not recorded. When ethnicity is mentioned, it is usually to denote a language barrier, e.g. “Spanish-speaking” or “required translator.”
In general, interpretability is seen as a desirable feature for machine learning, particularly in the clinical setting: doctors care not only about what decision is made, but what information is used to inform that decision. Here, BoW seems to have a natural advantage over other embedding models, as it is very easy to examine what words have the most predictive power for given tasks.
Indeed, Table 3 clearly demonstrates the interpretability of the features for predicting gender. Words such as ‘man’, ‘male’, ‘wife’, and ‘he’ directly suggest a male patient, and these are shown to have high predictive power for gender. More interestingly, we see words corresponding to gender-correlated conditions and body parts, such as ‘prostate’ for men and ‘breast’ for women. Unsurprisingly, BoW performs better than other methods on this task.
Table 3.
Most predictive words for gender
| (a) Male | |
|---|---|
| man | 1.4012 |
| he | 1.0589 |
| wife | 0.9953 |
| male | 0.7956 |
| his | 0.6772 |
| prostate | 0.2435 |
| prop | 0.1965 |
| ofm | 0.1850 |
| hematuria | 0.1816 |
| esophagectomy | 0.1812 |
| distention | 0.1756 |
| trauma | 0.1748 |
| (b) Female | |
|---|---|
| she | 1.0176 |
| woman | 0.9051 |
| her | 0.7561 |
| husband | 0.7004 |
| breast | 0.3206 |
| daughter | 0.2656 |
| nausea | 0.2309 |
| female | 0.2246 |
| commode | 0.2183 |
| responded | 0.2052 |
| fick | 0.2009 |
| cco | 0.1975 |
Admission type, with features shown in Table 4 is less-easily interpreted, but still provides understandable features. Words such as ‘er’ and ‘ew’ refer to the emergency room or ward, and ‘fracture’ or ‘fall’ refer to traumatic injuries, all of which reasonably suggest an urgent-care admission. Conversely, many of the predictive words for elective admissions suggest chronic conditions or planned surgical procedures (‘artery’, ‘valve’, ‘replacement’). We see thatBoW also performs quite well on this task.
Table 4.
Most predictive words for admission types
| (a) ‘Urgent’ admissions | |
|---|---|
| ew | 0.2639 |
| er | 0.2495 |
| fracture | 0.2258 |
| fx | 0.2248 |
| osh | 0.2235 |
| b | 0.2194 |
| disease | 0.2138 |
| vertebral | 0.2061 |
| cabg | 0.2029 |
| fractures | 0.1971 |
| fall | 0.1893 |
| arteriogram | 0.1877 |
| (b) ‘Elective’ admissions | |
|---|---|
| sda | 0.8048 |
| flap | 0.4646 |
| esophagectomy | 0.4617 |
| artery | 0.4435 |
| epidural | 0.4415 |
| valve | 0.3845 |
| lobectomy | 0.3838 |
| resection | 0.3644 |
| avr | 0.3527 |
| replacement | 0.3324 |
| nephrectomy | 0.2812 |
| whipple | 0.2740 |
However, we see some differences when we examine the predictive features for the length-of-stay task in Table 5. In contrast to gender or admission type, the features for length-of-stay are much more generic, seeming to have little interpretable relation to the prediction task. At the same time, we see that the LSTM achieves a higher F1 score as compared to the BoW model for this task. This suggests that BoW is interpretable for the simple token-matching tasks, but not the harder reasoning tasks. Therefore, more complex and performant models should be used for these harder tasks.
Table 5.
Most predictive words for length-of-stay
| (a) Short stay (0 - 1.5 days) | |
|---|---|
| ml | 0.5295 |
| pt | 0.5086 |
| to | 0.3570 |
| b | 0.3403 |
| sensitivity | 0.2489 |
| meq | 0.2090 |
| atrial | 0.1934 |
| tamponade | 0.1784 |
| valuables | 0.1770 |
| vomited | 0.1738 |
| s | 0.1708 |
| weaning | 0.1676 |
| (b) Medium stay (1.5 -3.5 days) | |
|---|---|
| followed | 0.2014 |
| aps | 0.1888 |
| lifting | 0.1811 |
| of | 0.1796 |
| device | 0.1790 |
| trunk | 0.1747 |
| available | 0.1644 |
| metastatic | 0.1610 |
| the | 0.1603 |
| holes | 0.1576 |
| this | 0.1520 |
| decubitus | 0.1509 |
| (c) Long stay (> 3:5 days) | |
|---|---|
| amio | 0.2470 |
| mn | 0.2393 |
| brain | 0.2172 |
| decreasing | 0.2002 |
| fentanyl | 0.1971 |
| withdrawal | 0.1933 |
| vasospasm | 0.1900 |
| previously | 0.1890 |
| coiling | 0.1811 |
| exercises | 0.1799 |
| dobbhoff | 0.1779 |
| frequently | 0.1776 |
Conclusions
In this work we consider both demographic and clinical prediction tasks in order to “stress test” a variety of common note representations. We show that different representations have different strengths: while complex models can outperform simple ones on reasoning tasks, they struggle to capture seemingly “easy” information. On the other hand, simple word-matching models prove to be very effective and interpretable for tasks that are so simple that complex models tend to overlook their differences. In doing so, we motivate the need for considering multiple representations rather than adopting a one-size-fits-all approach. Finally, to promote open and reproducible research, our code is publicly available, alongside word vectors trained on a very large corpus of clinical notes.
Figure 4.
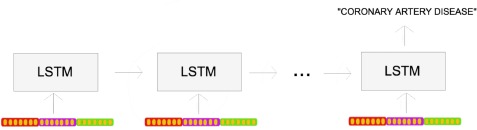
The many-to-one prediction task for the LSTM, in which a document representation is fed in at eachtimestep, and it makes a prediction (e.g. diagnosis) at the end of the sequence.
Acknowledgments
The authors would like to thank Jen Gong for her input and suggestions. This research was funded in part by the Intel Science and Technology Center for Big Data, the National Library of Medicine Biomedical Informatics Research Training grant 2T15 LM007092-22, the National Science Foundation Graduate Research Fellowship Program under Grant No. 1122374, NIH grants U54-HG007963 and R01-EB017205, and collaborative research agreements from Philips Corporation and Wistron Corporation.
Footnotes
Authors contributed equally.
Code available at http://www.github.com/wboag/wian.
Code available at https://github.com/tmikolov/word2vec.
Data available at https://physionet.org/works/MIMICIIIDerivedDataRepository/.
References
- 1.Ghassemi M, Naumann T, Doshi-Velez F, Brimmer N, Joshi R, Rumshisky A, et al. Unfolding physiological state: Mortality modelling in intensive care units. In: Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining; 2014; ACM. pp. 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caballero Barajas KL, Akella R. Dynamically modeling patient’s health state from electronic medical records: A time series approach. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2015; ACM. pp. 69–78. [Google Scholar]
- 3.Rumshisky A, Ghassemi M, Naumann T, Szolovits P, Castro V, McCoy T, et al. Predicting early psychiatric readmission with natural language processing of narrative discharge summaries. Translational Psychiatry. 2016;6(10):e921. doi: 10.1038/tp.2015.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grnarova P, Schmidt F, Hyland S, Eickhoff C. Neural Document Embeddings for Intensive Care Patient Mortality Prediction. In: NIPS 2016 Workshop on Machine Learning for Health Workshop. 2016 [Google Scholar]
- 5.Ghassemi M, Naumann T, Joshi R, Rumshisky A. Topic Models for Mortality Modeling in Intensive Care Units. In: ICML 2012 Machine Learning for Clinical Data Analysis Workshop. 2012 [Google Scholar]
- 6.Lehman Lw, Saeed M, Long W, Lee J, Mark R. Risk stratification of ICU patients using topic models inferred from unstructured progress notes. In: AMIA annual symposium proceedings; American Medical Informatics Association. 2012. p. 505. [PMC free article] [PubMed] [Google Scholar]
- 7.US Centers for Disease Control and Prevention. Health Disparities in HIV/AIDS, Viral Hepatitis, STDs, and TB; [Accessed September 26, 2017]. https://www.cdc.gov/nchhstp/healthdisparities/africanamericans.html.
- 8.Luo YF, Rumshisky A. Interpretable Topic Features for Post-ICU Mortality Prediction. In: AMIA Annual Symposium Proceedings; American Medical Informatics Association. 2016. p. 827. [PMC free article] [PubMed] [Google Scholar]
- 9.Le Q, Mikolov T. Distributed representations of sentences and documents. In: Proceedings of the 31st International Conference on Machine Learning (ICML-14); 2014:1188–1196. [Google Scholar]
- 10.Cohen R, Aviram I, Elhadad M, Elhadad N. Redundancy-aware topic modeling for patient record notes. PloS one. 2014;9(2):e87555. doi: 10.1371/journal.pone.0087555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pivovarov R, Perotte AJ, Grave E, Angiolillo J, Wiggins CH, Elhadad N. Learning probabilistic phenotypes from heterogeneous EHR data. Journal of biomedical informatics. 2015;58:156–165. doi: 10.1016/j.jbi.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banko M, Brill E. Scaling to very very large corpora for natural language disambiguation. In: Proceedings of the 39th annual meeting on association for computational linguistics; Association for Computational Linguistics. 2001. pp. 26–33. [Google Scholar]
- 13.Halevy A, Norvig P, Pereira F. The unreasonable effectiveness of data. IEEE Intelligent Systems. 2009;24(2):8–12. [Google Scholar]
- 14.Johnson AE, Pollard TJ, Shen L, Lehman LwH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Scientific data. 2016:3. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson AE, Pollard TJ, Mark RG. Reproducibility in critical care: a mortality prediction case study. In: Proceedings of Machine Learning for Healthcare 2017. 2017 [Google Scholar]
- 16.Levy O, Goldberg Y, Dagan I. Improving Distributional Similarity with Lessons Learned from Word Embeddings. In: Transactions of the Association for Computational Linguistics. 2015:211–225. [Google Scholar]
- 17.Hochreiter S, Schmidhuber J. Long short-term memory. Neural computation. 1997;8:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 18.Kosko B. Bidirectional Associative Memories. IEEE Trans SystMan Cybern. 1988 Jan;18(1):49–60. [Google Scholar]
- 19.Chollet F, et al. Keras. GitHub. 2015. https://github.com/fchollet/keras.
- 20.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]


