Abstract
Mental health is increasingly recognized an important topic in healthcare. Information concerning psychiatric symptoms is critical for the timely diagnosis of mental disorders, as well as for the personalization of interventions. However, the diversity and sparsity of psychiatric symptoms make it challenging for conventional natural language processing techniques to automatically extract such information from clinical text. To address this problem, this study takes the initiative to use and adapt word embeddings from four source domains – intensive care, biomedical literature, Wikipedia and Psychiatric Forum – to recognize symptoms in the target domain of psychiatry. We investigated four different approaches including 1) only using word embeddings of the source domain, 2) directly combining data of the source and target to generate word embeddings, 3) assigning different weights to word embeddings, and 4) retraining the word embedding model of the source domain using a corpus of the target domain. To the best of our knowledge, this is the first work of adapting multiple word embeddings of external domains to improve psychiatric symptom recognition in clinical text. Experimental results showed that the last two approaches outperformed the baseline methods, indicating the effectiveness of our new strategies to leverage embeddings from other domains.
Introduction
Mental health is increasingly recognized as an important topic in healthcare.1 In recent years, there has been rapid growth in the implementation of electronic health records (EHRs), leading to an unprecedented expansion in the availability of dense longitudinal datasets for clinical and translational research for psychiatric disorders. Psychiatric symptoms, as one type of fundamentally important information, are usually obtained through interpersonal communications and recorded in clinical text in EHRs.2 Information about the nature, severity, and impact of psychiatric symptoms is indispensable for both the diagnosis of mental disorders, and the customization of interventions to treat them.3 Therefore, it is desirable to develop automated approaches to extract psychiatric symptoms from clinical text.
However, psychiatric symptoms often consist of subjective and individualized descriptions, which are present in details of the patient’s experience (Figure 1). Instead of a single word or simple noun phrase, psychiatric symptoms have tremendous syntactic and semantic variability.3 Moreover, symptoms in clinical notes of different institutions, different types of mental disorders (e.g., bipolar disorder vs. substance abuse) and different populations (e.g., adults, teenagers, military veterans) may have their own sub-languages. Therefore, it is quite challenging for traditional natural language processing (NLP) techniques to automatically extract such diverse mentions of psychiatric symptoms from text. On one hand, existing clinical terminologies such as in Unified Medical Language System (UMLS), SNOMED-CT, and ICD-9 code have low coverage of such complex expressions3. On the other hand, conventional supervised learning-based methods (typically the algorithm of conditional random fields-CRF4) are widely employed for clinical concept recognition. Such methods rely heavily on NLP feature engineering, leading to systems not generalizable to other clinical texts with different sub-languages.5
Figure 1.

An example paragraph from psychiatric notes with symptoms. The psychiatric symptoms are highlighted in italic.
Although emerging research activities have used NLP techniques to unlock information in psychiatric text for various applications recently6,7,8,9, only a few efforts have been made to extract psychiatric symptoms. Given that the diversity and sparity of psychiatric symptoms require a vastly larger labeled corpus compared to concepts like diseases and medications, Gorrell et al. applied active learning to alleviate this problem for negative symptom recognition of schizophrenia.2 However, domain experts still need to be heavily involved in this process. In contrast, our previous work employed an unsupervised framework to address this problem by leveraging distributional representation of phrases.10 In their work, symptoms were collected from online knowledge sources and candidate symptoms were identified based on semantic similarity.10 However, our previous study mainly focused on extracting a highquality candidate list of symptoms; further accurate classification of each specific mention in the context of clinical notes is left to future work. In previous research, we have used distributed representations of phrases composed from word embeddings produced using Latent Semantic Analysis (LSA)11 as a basis for supervised learning of the relationship between phrases in psychiatric narrative and diagnostically meaningful categories, such as “mood disorders” and “dangerousness”.12 While this work demonstrated the utility of distributed representations learned from a background corpus for the classification of psychiatric symptoms, it was limited in scope to a small number of symptom categories, and did not evaluate the influence of different source corpora on classifier performance.
Neural networks, or deep learning-based methods, are growing in popularity as approaches to NLP. Deep learning-based methods do not need time-consuming and labor-intensive feature engineering.13,14 Instead, word embeddings pre-trained from large-scale unlabeled corpora are usually used as features.15 As the currently most widely-used distributional semantic representation (i.e., vector representation) of words, neural word embeddings (such as those produced by the word2vec software package16) are assumed to capture the latent syntactic/semantic information of a word, because the resulting vector representations for words will be similar if these words occur in similar local contexts.16 While this is also the case for prior distributional models such as LSA, neural embeddings have shown advantages over other distributional models with optimized parameters in some experiments.17 Deep learning methods are well-suited to leverage these representations, as they both exemplify the parallel distributed representation paradigm.18 Thus, the framework of deep learning-based methods with word embedding features has stronger generalizability to resolve the diversity and sparseness of natural language.16 This is particularly important in psychiatry on account of different ways in which patients describe their experience of illness, which may explain in part the promising performance of deep learning based methods on NLP tasks in this domain. Furthermore, a recent study of Habibi et al. demonstrated that using deep learning-based methods outperformed state-of-the-art entity-specific NER tools and an entity-agnostic CRF implementation by a large margin, by conducting experiments on 33 data sets covering five different entity classes in the biomedical literature and patent domains.13
However, due to the fact that there is no publicly available, large corpus of psychiatric notes, external resources are necessary for building the distributional representations. On the other hand, word embeddings from multiple largescale external resources such as MEDLINE and Wikipedia have been applied and demonstrated their effectiveness on clinical NLP tasks, such as estimation of the semantic similarity and relatedness between clinical concepts19, NER20, and assertion identification20. So far, current works mainly investigated word embeddings derived from external resources by using them directly, or combining the corpora of multiple sources to train a single word embedding model.10,19,20 An issue with the former approach is that word embeddings from different sources may need to be adapted to the clinical domain for optimal performance, and an issue with the latter is that the distributional information from larger general-domain corpora may overwhelm that of a small target domain corpus. Domain adaptation technologies could be a solution to address the first of these problems. Domain adaptation attempts to maximize the use of existing data (source) for the data of interest (target) by learning useful aspects of source data. It has been applied for various NLP tasks in the biomedical domain, such as semantic role labeling in biomedical literature21 and clinical notes,22 automatic discourse connective detection in biomedical text,23 and de-identification of psychiatric text.24 However, few studies have been conducted on domain adaptation of word embeddings, yet.
This study addresses the use and adaptation of word embeddings derived from external domains for symptom recognition from psychiatrist text, using deep learning-based algorithms. Specifically, four domains – intensive care, biomedical literature, Wikipedia and Psychiatric Forum are used as source domains (DS) and adapted to the target domain (DT) of psychiatrist text. Word embeddings of each DS are investigated using four different approaches. First, two basic approaches commonly used in previous works in the clinical domain [cite] are employed: (1) using word embeddings of DS only and (2) directly combining data of DS and DT to generate word embeddings. In addition, two novel approaches are also implemented to adapt word embeddings of DS to DT, by (3) assigning different weights to word embeddings of DS and DT and (4) retraining the word embedding model of each DS using the available dataset of DT. To the best of our knowledge, this is the first work of adapting multiple word embeddings of external domains to psychiatric symptom recognition from clinical text. Experimental results demonstrated that the proposed strategies achieve promising performance, which outperformed the baseline of only using word embeddings of DT and a CRF-based system with fine-tuned features.
Methods
Study design
Figure 2 shows the study design of domain adaptation based psychiatric symptom recognition. For this task, data from four source domains (DS) - intensive care, biomedical literature, Wikipedia and Psychiatric Forum - are employed for domain adaptation. After word embeddings are generated by applying different strategies on data of DS and DT, they are fed into the deep learning-based methods as initial features to train automate systems for psychiatric symptom recognition. The systems are evaluated based on predictions on the test dataset.
Figure 2.
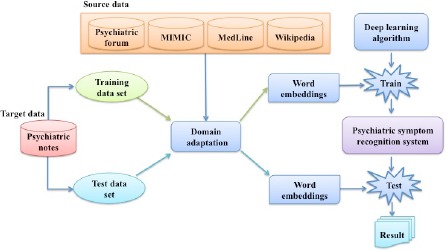
Study design of adapting word embeddings of multiple source domains to psychiatric notes for psychiatric symptom recognition using the deep learning method.
Datasets
We used five unlabeled datasets in our study, four as source domain datasets and one as the target dataset, as described below:
Source domain datasets: (1) MIMIC III25 is a publicly available collection of clinical notes in intensive care unit (ICU), which is commonly used to generate word embeddings for clinical NLP. (2) MEDLINE is a collection of scientific article abstracts in the biomedical domain. While MEDLINE covers clinical concepts such as diseases, medication and treatments, it contains well-formed sentences different from the telegraphic style in clinical notes. We used the MEDLINE data of 2013 in this study. (3) Psychiatric Forum is a collection of forum posts on the WebMD Community. The forum posts are written largely by health consumers, who use similar expressions to those of patients, as recorded in their psychiatric notes. (4) Wikipedia is an online encyclopedia built from crowdsourcing. It has extensive coverage of topics of multiple domains, including medical topics. The text in Wikipedia is usually well formed.
Target domain dataset: The psychiatric notes provided by the CEGS N-GRID 2016 challenge26 organizers are used for as the target domain dataset in this study. This is the first corpus of mental health records released to the NLP research community, which contains about 1,000 initial psychiatric evaluation records. These records were produced by psychiatrists during the course of the elicitation of psychiatric signs and symptoms, disorders, and other medical conditions in order to decide the course of treatment.
Table 1 displays the statistics of the five corpora, including the size of data, number of unique tokens and the percentage of token overlap between each source and the target data.
Table 1.
Corpus statistics for MIMIC, MEDLINE, Psychiatric Forum, Wikipedia and psychiatric notes.
| Corpus | Size of data | #Unique tokens | #Coverage of target tokens | |
|---|---|---|---|---|
| Source | MIMIC III | 1.95G | 257,472 | 3,029 (73.1%) |
| MEDLINE | 1.3G | 251,080 | 3,947(95.2%) | |
| Psychiatric Forum | 78.5M | 36,336 | 3,264 (78.8%) | |
| Wikipedia | 10.4G | 2,448,552 | 4,063 (98.0%) | |
| Target | Psychiatric Notes | 2.4M | 4,144 | # |
Methods of using word embeddings of source domains
Firstly, two basic approaches commonly used in previous works in the clinical domain are employed:
-
(1)
Source_only: directly using word embeddings of each DS to initiate the features of the deep learning-based method;
-
(2)
Source+target: directly combining datasets of each DS with DT to generate the word embeddings;
Moreover, two domain adaptation strategies are investigated, in order to better leverage the word embeddings of DS:
-
(3)
Weighted_concatenate: concatenating the word embedding vectors of DS and DT. Different weights are assigned to the word embeddings of DS, based on the intuition that word embeddings more important to the system performance should be assigned higher weights as features.27
Retrain_source: re-training the word embedding models of DS using DT. In this approach, the final input and output weights of a neural network trained on DS serve as the initial input and output weights for the derivation of embeddings from DT. In this way, the original semantic distributions and representations of words in DS are adjusted in accordance with their distributions in DT, mediating their adaptation to the target domain.
Deep learning algorithm
The deep learning algorithm of long short-term memory network-conditional random field (LSTM-CRF) proposed by Lample et al. is employed to build the psychiatric symptom recognition model.15 This algorithm adds a layer of CRF based prediction model on top of the original bi-directional LSTM structure of recurrent neural network (RNN), and has shown state-of-the-art performance in various NER tasks in open domain and biomedical text.15
Experiments and evaluation
Method comparison: Experiments were conducted to systematically investigate the performance of the Source_only, Source+target, weighted_concatenate, and Retrain_source methods. For the weighted_concatenate method, weight in the range of [1,10] was assigned to word embeddings of DS and the optimal performance was reported. Moreover, to examine the effectiveness of these algorithms, three baseline methods were also developed for comparison: the “Randomize” method without using any pre-trained word embeddings, the “Target_only” method using only word embeddings of DT to train a model, and a CRF-based model using a fine-tuned feature set.28
Parameter setup: The gensim implementation29 of the neural network architectures provided by the word2vec package [cite] was used to train the word embeddings of each domain, because this package permits retraining an existing model. The corpus used for retraining is not necessarily the same as the corpus from which the original model was generated. The parameters of the word2vec model include: (1) the skip-gram architecture was adopted to train the model; (2) the window size was set to 4; (3) in this preliminary study, the dimensionality of embedding vectors was set to 50 for the Target_only, Source_only, Source+target and Retrain_source methods, and 100 for the Weighted_concatenate method; (4) the initial learning rate (i.e., alpha) was 0.025; (5) and all the words with total frequency lower than 5 (i.e., min_count) were ignored. Each word embedding model was trained for 100 epochs. As for the LSTM-CRF algorithm, we followed the optimal parameter setup established in the work of Lample et al.15
Evaluation: 400 psychiatric notes are annotated with symptoms. These were split into training (60%), development (20%) and test datasets (20%) for experiments. Each model was trained for a total of 100 epochs using the training set. The performance on the test dataset was reported using the optimal model evaluated on the development set. The performance of psychiatric symptom recognition was evaluated using precision, recall and the F-measure.
Results
Table 2 illustrates the results for RNN-based psychiatric symptom recognition using word embeddings from multiple domains. The Target_only baseline of using word embeddings generated from psychiatric notes outperformed the model with randomized word embedding features (F-measure: 67.26% vs. 65.87%). Applying word embeddings of the source domains further improved the performance of the Target_only baseline. For the Source_only method, the Wikipedia dataset yielded the highest F-measure of 69.51% among the four source datasets, while MIMIC produced the lowest F-measure of 67.34%.
Table 2.
Results for RNN-based psychiatric symptom recognition using word embeddings from multiple domains (%).
| Corpus | Method | P | R | F-measure |
|---|---|---|---|---|
| None | Randomize | 68.93 | 63.06 | 65.87 |
| Psychiatric Notes | Target_only | 70.82 | 64.05 | 67.26 |
| Psychiatric Forum | Source_only | 67.85 | 67.42 | 67.63 |
| Source+target | 69.34 | 65.52 | 67.38 | |
| Weighted_concatenate | 72.01 | 64.05 | 67.80 | |
| Retrain_source | 71.60 | 64.84 | 68.05 | |
| Mimic | Source_only | 69.66 | 65.16 | 67.34 |
| Source+target | 67.86 | 66.71 | 67.28 | |
| Weighted_concatenate | 69.52 | 66.90 | 68.19 | |
| Retrain_source | 71.25 | 67.87 | 69.52 | |
| Wikipedia | Source_only | 69.99 | 69.04 | 69.51 |
| Source+target | 69.80 | 68.82 | 69.30 | |
| Weighted_concatenate | 71.42 | 66.86 | 69.07 | |
| Retrain_source | 69.50 | 66.19 | 67.80 | |
| MEDLINE | Source_only | 72.18 | 63.85 | 67.76 |
| Source+target | 71.82 | 64.05 | 67.71 | |
| Weighted_concatenate | 68.95 | 67.34 | 68.14 | |
| Retrain_source | 73.62 | 64.81 | 68.49 |
Interestingly, for this task of psychiatric symptom recognition, the performance of Source+target was lower than the Source-only method. The Source-only method performed best when Wikipedia was the source corpus only. With all other corpora, models that considered the target prevailed. This may reflect the importance of general-domain semantic information for the interpretation of patients’ descriptions of their subjective experience of their symptomatology, a point we have argued previously.12,30 Assigning higher weights to word embeddings of the source domains in the concatenation (Weighted_concatenate) generally outperformed the methods of Source+target and Source_only, with the exception of the Wikipedia corpus. Nevertheless, Wikipedia still produced the highest F-measure of 69.07% when using concatenation methods among the four DS. In contrast, the Retrain_source method achieved the highest F-measure for three DS (Psychiatric Forum, MIMIC and MEDLINE), with the optimal performance of 69.52% yielded by retraining on MIMIC. In contract, Wikipedia produced the lowest F-measure of 67.80% with this method.
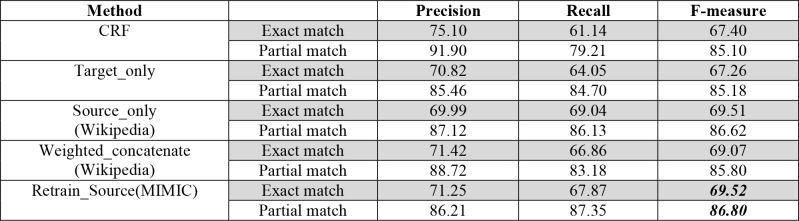
To examine whether our proposed methods could achieve the state-of-the-art performance for psychiatric symptom recognition. We further compared our methods with the CRF algorithm using a fine-tuned feature set. 28 Table 3 listed the performance of using CRF, Target_only, Source_only with Wikipedia as DS, Weighted_concatenate with Wikipedia as DS and Retrain_source with MIMIC as DS. Considering the practical usage of the psychiatric symptom recognition system, performance of both exact match and partial match was reported. As can be seen, the performance of CRF was better than the Target_only method (F-measure: 67.40% vs. 67.26%), demonstrating that CRF was a strong baseline. Notably, by adapting word embeddings from source domains using different methods, the psychiatric symptom recognition systems built in this study achieved better performance than the system based on CRF. Among all the methods, Retrain_Source with MIMIC as DS achieved the best F-measure of 86.80% in terms of partial match.
Table 3.
Performance comparison between the Deep learning based algorithm and the CRF algorithm for psychiatric symptom recognition (%).
Discussion
Recently there has been a rapid growth of deep learning methods, and their application to various NLP tasks. This growth has been promoted by the availability of word embeddings pre-trained from large-scale corpora of multiple domains. However, many clinical sub-domains may not have publicly available, large corpus, and external resources are necessary for building the distributional representations. This study exploits word embeddings of external domains for clinical NLP tasks. Specifically, we take the psychiatric symptom recognition task as a typical use case. Extracting psychiatric symptoms from clinical text suffers from the data sparseness problem and a low coverage of relevant terms in existing biomedical lexicons, making it challenging to apply traditional named entity recognition methods to this task. We evaluated different approaches toward adapting word embeddings from four source domains to recognize entities of psychiatric symptoms using deep learning methods. To the best of our knowledge, this is the first work of adapting multiple word embeddings to psychiatric symptom recognition. Experimental results showed that the proposed domain adaptation strategies could achieve promising performance, outperforming the other strategies evaluated, and a system built with CRF algorithm and fine-tuned features.
To further improve the performance, we looked into the current prediction errors of psychiatric symptom recognition. The major reasons of false positive and false negative errors are illustrated in Table 4. A majority of false positive errors are caused by recognizing terms that may stand for psychiatric symptoms in wrong context, such as the term of “anxiety” in the context of “from a mood and anxiety standpoint”. In addition, some general expressions that do not convey specific psychiatric symptoms are another cause of false positive errors. Moreover, although some authentic clinical symptoms/disorders are recognized, there is no explicit evidence in the context to indicate that they are related to mental disorders, which leads to false positive errors. As for the false negative errors, psychiatric symptoms present in complex syntactic structures such as conjunctive structures are often missed to be identified. Besides, abbreviations of psychiatric symptoms are frequently used in psychiatric text and miss-identified by the system. Some psychiatric symptoms are of rare patterns, whereas the telegraphic writing style in psychiatric text also causes some false negative errors. Furthermore, we also reviewed the symptom predictions partially matched with gold-standard symptoms; some typical examples are illustrated in Table 5. As can be seen, some of the boundary errors were not critical, in terms of recognizing essential symptoms entities. Therefore, we argue that in-exact matching could be reasonable in psychiatrc symptom recognition.
Table 4.
Error analysis of deep learning based psychiatric symptom recognition. False positive/negative errors are highlighted in italic.
| Error type | Example |
|---|---|
| False positive | |
| Term taken out of context | She felt very well from a mood and anxiety stand point prior to pregnancy |
| Non-specific symptomatology | it is in context of needing to work long hours and sometimes can be associated with impulsive incidents in the remote past but no recent episodes |
| Non-psychiatric symptomatology | Patient is legally blind and has had impaired hearing since birth. |
| False negative | |
| Complex syntactic structure | notices light sensitivity, emotional numbness / detachment / lack of interest in usual activities cluster B traits, (rigidity, interpersonal difficulty, degree of perfectionism) |
| Abbreviations | Denies history of symptoms of AH / VH / TH / OH / GH / TI / TB / TW / IOR |
| Rare symptom pattern | At heaviest use was smoking 7 grams cannabis per day (for 5 mths, fall 2082 while at college) |
| Telegraphic writing | Brother bipolar sometimes violent |
Table 5.
Examples of partial matched psychiatric symptoms.
| Symptom annotation | Partially matched symptom |
| Suicidal thoughts | Periods of suicidal thoughts |
| etoh or drug use | drug use |
| Complicated Grief; pain medication abuse | Complicated Grief and pain medication abuse |
| subjective sense of “ brain fogginess | sense of “ brain fogginess |
| had anxiety all my life | anxiety |
Previous works on exploiting word embeddings of multiple DS have obtained different findings in terms of their contribution to specific tasks in DT. For example, Roberts27 assessed the performance of word embedding based features on the i2b2 2010 concept recognition and assertion identification tasks and found that merging multiple corpora to generate word embeddings generally worked best. He also pointed out that the single-best corpus was generally task-dependent.27 Pakhomov et al.25 constructed word embeddings of clinical terms for semantic similarity and relatedness between clinical concept pairs and found that measures computed from biomedical literature were on par with measures computed from clinical reports, while measures from Wikipedia were worse than sources from the biomedical domain. Zhang et al. 31 used embeddings of short text and semantic similarity to identify psychiatric symptom candidates and found that the dataset from Psychiatric Forum contributed the most and the MIMIC corpus decreased the performance when merged with the other corpora. In contrast, our study of using word embedding based features and deep learning-based supervised methods for psychiatric symptom extraction found that the two novel methods of Weighted_concatenate and Retrain_source achieved better performance in general, validating the effectiveness of our proposed approaches. Moreover, directly merging corpora of DS and DT (i.e., Source+target) decreased the performance slightly. Notably, the performance when using Wikipedia was an exception from MIMIC, MEDLINE and Psychiatric Forum in that the Source_only method performed the best, while Retrain_source got the lowest F-measure of 67.78%, a marked contrast to its leading performance with all other corpora.
Limitation and future work: One limitation of our current work is that the performance difference of varying dimensions of word embedding vectors has not been examined. In addition, psychiatric symptom lexicons and syntactic patterns commonly present in the target domain will be incorporated as features of the deep learning methods, in order to tailor the system to the target domain. Besides, as shown in Table 3, the CRF-based system got higher precision than deep learning based systems, while deep learning based systems usually obtained relatively higher recall.
Therefore, we will explore potential approaches to leverage the advantage of these two types of algorithms in a single system. Furthermore, domain adaptation strategies to adapt word embeddings from other domains as features in CRF will also be investigated.
Conclusion
This study takes the initiative to use and adapt word embeddings of four source domains – intensive care, biomedical literature, Wikipedia and Psychiatric Forum – to recognize symptoms in the target psychiatric domain using deep learning-based methods. Experimental results showed that the proposed domain adaptation strategies achieve promising performance, indicating that deep learning-based methods leveraging word embeddings of source domains have the potential to resolve the data diversity and scarcity challenged of clinical NLP tasks.
Acknowledgement:
This study was supported in part by R00LM012104-02.
References
- 1.Proctor EK, Landsverk J, Aarons G, Chambers D, Glisson C, Mittman B. Implementation research in mental health services: an emerging science with conceptual, methodological, and training challenges. Administration and Policy in Mental Health and Mental Health Services Research. 2009;36(1):24–34. doi: 10.1007/s10488-008-0197-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorrell G, Jackson R, Roberts A, Stewart R. Finding negative symptoms of schizophrenia in patient records. Proc NLP Med Biol Work (NLPMedBio), Recent Adv Nat Lang Process (RANLP) 2013:9–17. [Google Scholar]
- 3.Carter M, Matthew Samore M. Sitting on Pins and Needles”: Characterization of Symptom Descriptions in Clinical Notes. 2013 [PMC free article] [PubMed] [Google Scholar]
- 4.Lafferty J, McCallum A, Pereira FC. ICML '01. San Francisco, CA, USA: 2001. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. [Google Scholar]
- 5.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. Journal of biomedical informatics. 2002;35(4):222–235. doi: 10.1016/s1532-0464(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 6.Pestian JP, Grupp - Phelan J, Bretonnel Cohen K, et al. A controlled trial using natural language processing to examine the language of suicidal adolescents in the emergency department. Suicide and life-threatening behavior. 2015 doi: 10.1111/sltb.12180. [DOI] [PubMed] [Google Scholar]
- 7.Patel R, Wilson R, Jackson R, et al. Cannabis use and treatment resistance in first episode psychosis: a natural language processing study. The Lancet. 2015;385:S79. doi: 10.1016/S0140-6736(15)60394-4. [DOI] [PubMed] [Google Scholar]
- 8.Rumshisky A, Ghassemi M, Naumann T, et al. Predicting early psychiatric readmission with natural language processing of narrative discharge summaries. Translational Psychiatry. 2016;6(10):e921. doi: 10.1038/tp.2015.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCoy TH, Castro VM, Roberson AM, Snapper LA, Perlis RH. Improving prediction of suicide and accidental death after discharge from general hospitals with natural language processing. Jama psychiatry. 2016;73(10):1064-1071. doi: 10.1001/jamapsychiatry.2016.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Zhang O, Wu Y, et al. Psychiatric symptom recognition without labeled data using distributional representations of phrases and on-line knowledge. Journal of Biomedical Informatics. 2017 doi: 10.1016/j.jbi.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landauer TK, Dumais ST. A solution to Plato's problem: The latent semantic analysis theory of acquisition, induction, and representation of knowledge. Psychological review. 1997;104(2):211. [Google Scholar]
- 12.Cohen T, Blatter B, Patel V. Simulating expert clinical comprehension: Adapting latent semantic analysis to accurately extract clinical concepts from psychiatric narrative. Journal of biomedical informatics. 2008;41(6):1070–1087. doi: 10.1016/j.jbi.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habibi M, Weber L, Neves M, Wiegandt DL, Leser U. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics. 2017;33(14):i37–i48. doi: 10.1093/bioinformatics/btx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Xu J, Jiang M, Zhang Y, Xu H. A study of neural word embeddings for named entity recognition in clinical text. Paper presented at: AMIA Annual Symposium Proceedings. 2015 [PMC free article] [PubMed] [Google Scholar]
- 15.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural architectures for named entity recognition. arXivpreprint arXiv:160301360. 2016 [Google Scholar]
- 16.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Paper presented at: Advances in neural information processing systems. 2013 [Google Scholar]
- 17.Levy O, Goldberg Y, Dagan I. Improving distributional similarity with lessons learned from word embeddings. Transactions of the Association for Computational Linguistics. 2015;3:211–225. [Google Scholar]
- 18.Rumdhart D, McClctland J. PDP Research Group: Parallel distributed proccssing. MIT Press. 1986 [Google Scholar]
- 19.Pakhomov SV, Finley G, McEwan R, Wang Y, Melton GB. Corpus domain effects on distributional semantic modeling of medical terms. Bioinformatics. 2016;32(23):3635–3644. doi: 10.1093/bioinformatics/btw529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts K. Assessing the Corpus Size vs. Similarity Trade-off for Word Embeddings in Clinical NLP. ClinicalNLP 2016. 2016:54. [Google Scholar]
- 21.Dahlmeier D, Ng HT. Domain adaptation for semantic role labeling in the biomedical domain. Bioinformatics. 2010;26(8):1098–1104. doi: 10.1093/bioinformatics/btq075. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Tang B, Jiang M, Wang J, Xu H. Domain adaptation for semantic role labeling of clinical text. Journal of the American Medical Informatics Association. 2015;22(5):967–979. doi: 10.1093/jamia/ocu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polepalli Ramesh B, Prasad R, Miller T, Harrington B, Yu H. Automatic discourse connective detection in biomedical text. Journal of the American Medical Informatics Association. 2012;19(5):800–808. doi: 10.1136/amiajnl-2011-000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hee-Jin L, Yaoyun Z, Kirk R, Hua X. DC: 2017. Leveraging existing corpora for de-identification of psychiatric notes using domain adaptation AMIA. [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Scientific data. 2016;3 doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filannino M, Stubbs A, Uzuner Ö. Symptom severity prediction from neuropsychiatric clinical records: Overview of 2016 CEGS N-GRID Shared Tasks Track 2. Journal of Biomedical Informatics. 2017 doi: 10.1016/j.jbi.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin W, Schütze H. Learning word meta-embeddings. Paper presented at: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics. 2016;1(Long Papers) [Google Scholar]
- 28.CLAMP | Natural Language Processing (NLP) Software. 2017 http://clamp.uth.edu/ [Google Scholar]
- 29.Le QV, Mikolov T. Paper presented at: ICML2014. Distributed Representations of Sentences and Documents. [Google Scholar]
- 30.Cohen T, Blatter B, Patel V. Exploring dangerous neighborhoods: latent semantic analysis and computing beyond the bounds of the familiar. Paper presented at: AMIA Annual Symposium Proceedings2005. [PMC free article] [PubMed] [Google Scholar]
- 31.Yaoyun Z, Yonghui W, Hee-Jin L, Jun X, Hua X, Kirk R. UTH: An unsupervised framework for psychiatric symptom recognition using distributed representations and seed terms. I2b2 CEGS N-GRID challenge. 2016 [Google Scholar]