Abstract
Medication regimen may be optimized based on individual drug efficacy identified by pharmacogenomic testing. However, majority of current pharmacogenomic decision support tools provide assessment only of single drug-gene interactions without taking into account complex drug-drug and drug-drug-gene interactions which are prevalent in people with polypharmacy and can result in adverse drug events or insufficient drug efficacy. The main objective of this project was to develop comprehensive pharmacogenomic decision support for medication risk assessment in people with polypharmacy that simultaneously accounts for multiple drug and gene effects. To achieve this goal, the project addressed two aims: (1) development of comprehensive knowledge repository of actionable pharmacogenes; (2) introduction of scoring approaches reflecting potential adverse effect risk levels of complex medication regimens accounting for pharmacogenomic polymorphisms and multiple drug metabolizing pathways. After pharmacogenomic knowledge repository was introduced, a scoring algorithm has been built and pilot-tested using a limited data set. The resulting total risk score for frequently hospitalized older adults with polypharmacy (72.04±17.84) was statistically significantly different (p<0.05) from the total risk score for older adults with polypharmacy with low hospitalization rate (8.98±2.37). An initial prototype assessment demonstrated feasibility of our approach and identified steps for improving risk scoring algorithms.
Introduction
Polypharmacy has been characterized as a significant risk factor for adverse drug events in multiple studies. High prevalence of polypharmacy in older adults has been widely reported1. Multiple studies have mentioned that polypharmacy increases the risk of adverse drug reactions, hospitalization, falls, mortality, and other adverse health outcomes in elderly patients2. Polypharmacy represents a significant risk for drug-drug, drug-disease and drug-gene interactions2 and, undoubtedly, has become a significant source of health care utilization and costs.
Previous studies showed high utility of pharmacogenomics for preventing potential side effects of polypharmacy. Pharmacogenomics allow identify how hereditary profile affects an individual response to drugs. As a strategy for optimizing medication usage, pharmacogenomics became an important element of precision medicine. A recent study has demonstrated that precision medicine has significant potential in people with polypharmacy particularly in older adults with history of urgent care utilization3.
However, majority of current pharmacogenomic decision support tools are based only on assessment of single druggene interactions without taking into account complex drug-drug and drug-drug-gene interactions which are prevalent in people with polypharmacy and may result in adverse drug events or suboptimal drug efficacy. Many of the most frequently prescribed medications for older adults are metabolized by multiple cytochrome pathways, each of which, taken alone, insufficiently represents the actual metabolic pharmacokinetics. With increasing number of drugs, growing complexity of multiple contemporaneous drug-drug and drug-gene interactions results in a significant cognitive workload for a practicing provider preventing optimal regimen prescription. Thus, development of comprehensive decision support tools accounting for multiple drug interactions is a crucial step in promoting precision medicine in people with polypharmacy.
The main objective of this project was development of comprehensive pharmacogenomic decision support for medication risk assessment in people with polypharmacy. To achieve this goal, the project addressed two aims: (1) development of comprehensive knowledge repository of actionable pharmacogenes; (2) introduction of scoring approaches reflecting potential adverse effect risk levels of complex medication regimens accounting for pharmacogenomic polymorphisms and multiple drug metabolizing pathways.
Methods
Multiple open database sources and online literature repositories were used in this project to obtain verified drug metabolism information and metabolic phenotypes of cytochrome P450 (CYP) alleles. Available knowledge repositories containing pharmacogenomic information have been reviewed based on literature reports and citation index and their content compared. After a comprehensive review of available pharmacogenomic information resources, the following sources were utilized to build knowledge base for this project: PharmGKB, Indiana University Cytochrome P450 Drug Interaction Table (IU), SuperCyp, UpToDate, and SNPedia (Figure 1). Wikipedia was initially considered however it was eventually excluded from further consideration due to inconsistencies in its article format preventing from automated data extraction. The Indiana University (IU) portal provides information on major P450 drug interactions which were included in our initial database4. SuperCyp contains exhaustive Cytochrome-Drug interaction data5. UpToDate is one of the most trusted clinical decision support resources6. It was used in this project to retrieve information about substrate weight status for particular drug metabolism pathways. CYP450 enzyme activity for various single nucleotide polymorphisms (SNP), deletions and copy number variations (CNV) were imported from PharmGKB and SNPedia7, 8.
Figure 1.
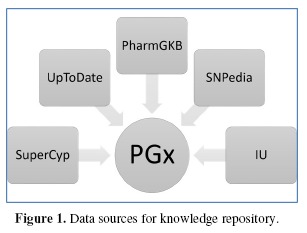
Data sources for knowledge repository.
We created our own relational database to merge complex data from different data sources. The database was designed to be scalable, pluggable, and platform-independent. The key component was represented by two tables consisting of drug names and enzyme names. Extended from that, we had detailed information about the drugs and enzymes, saved in individual tables respectively based on content and source. We had four kinds of detailed tables, with each category shared the same schema. The Interactions table contained drug metabolism effects; the IteractionStatus table included effect weight information; the Allele table stored the allele name of the enzymes and the functionality of those allelic variations, and the DrugNames table had the drug aliases such as brand names or chemical names of the drugs that helped normalize drug names coming from different sources. Figure 2 depicts the relational schema of the database.
Figure 2.
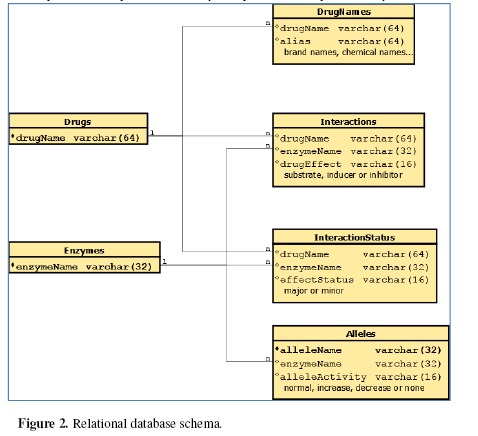
Relational database schema.
Figure 3 represents the structure of computer program that implemented data aggregation and scoring. The program was mostly written in Java to make it platform-independent. For each data source, we created different modules to fetch the data, cleaned them and modified the data format to fit our database. Python scripts were used to perform the batch operations in this step. We wrote a Database class and used JDBC module to connect with the database. Drug, Enzyme and Patient classes were created to bring the data into instances. We built a user-friendly interface using JavaFX in UI class, but one could still use the Batch class to start a batch process, or use the Console program to trace the results.
Figure 3.
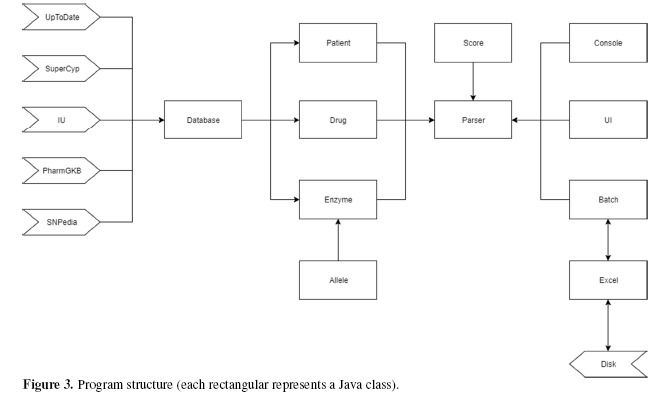
Program structure (each rectangular represents a Java class).
User can easily change parameters through the APIs we provided in Score class, or create their own algorithms by overriding the Parser class. The program is guaranteed to be thread safe and have limited memory usage. We implemented a module for low level Excel access through Java API to Access Microsoft Excel Format Files (POIXSSF).
Model
To account for simultaneous contribution of multiple cytochrome pathways and cytochrome polymorphisms, we introduced a scoring framework for medication risk in people with polypharmacy (Figure 4). The framework reflects contemporaneous impact of cytochrome allele types based on corresponding metabolic phenotypes (i.e. extensive, poor or rapid metabolizer) and individual drug effect on activity of specific cytochrome enzymes (i.e. substrate, inhibitor, or inducer) to account for phenoconversion phenomena.
Figure 4.

A scoring framework for medication risk.
Initially, we experimented with two algorithms, the additive and multiplicative algorithms, to numerically represent the risk on potential drug interactions through CYP450 metabolism in patients. In our previous study, we found that the multiplicative approach had limited ability to distinguish two patients’ drug-gene interactions9. This study represents further development of an additive algorithm.
The overall algorithm for medication risk score calculation is delineated in Figure 5. We divided the medication risk score into three parts: (1) drug-drug interaction representing contemporaneous effect of multiple drugs on cytochrome pathways affecting drug metabolism, (2) drug-gene interaction reflecting impact of individual cytochrome polymorphisms on metabolic phenotypes, and (3) gene function representing deleteriousness of cytochrome polymorphisms in a particular patient. For each drug the patient takes, the drug-drug interaction score Sdd can be represented as
| (1) |
Figure 5.
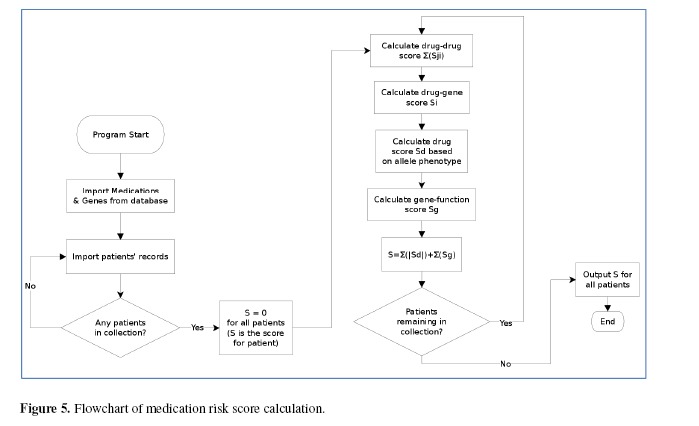
Flowchart of medication risk score calculation.
Where i is the binding enzyme of that drug, n is total number of enzymes; j. is the other drugs that the patient also takes and will have effects on the same enzyme i and m isthe number of drugs the patient is taking.
For each drug, the drug-gene interaction score Sdg is defined as
| (2) |
where si represents the metabolic phenotype of enzyme i.
Drugs are often metabolized through multiple cytochrome pathways, each pathway has different contribution to drug’s pharmacologic action. We collected weight data of the most common drugs from UpToDate.com and saved this information in our database. Then we assigned a weight percentage for each drug. The weighted score for each drug (Sd) is
| (3) |
where Wi is the weight on enzyme i for drug d.
Since allele functionality has decisive effect on drug metabolism, we added a term named gene function in our equation, to represent the impact of pharmacogene mutations. For each patient, the final risk score S is defined as
| (4) |
where Sg is the gene function score. Still, n represents total number of enzymes and m represents the number of drugs the patient is taking.
Results
The resulting knowledge repository contained information on 763 drugs, 11 cytochrome enzymes, and 132 alleles. It included 2076 unique pairs of drug-enzyme interactions reflecting effect of medications on cytochrome activity, 132 allele phenotype characteristics of individual cytochrome polymorphisms, and 721 enzyme-drug associations reflecting multiple metabolizing pathways of individual drugs. Out of 2076 drug-enzyme interactions, 691 were obtained from IU, 1848 – from SuperCyp, with 463 overlapping relationships. All 721 enzyme-drug associations were extracted from UpToDate. Out of 132 allele phenotype characteristics, 54 were obtained from SNPedia, and 132 – from PharmGKB, with 54 characteristics overlapping between these two data sources. A summary of alleles for which metabolic phenotypes were collected is presented in Table 1.
Table 1.
Cytochrome alleles represented in the resulting knowledge repository.
| Cytochrome | Alleles with Metabolic Phenotypes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP1A2 | *1A | *1C | *1F | *1K | *3 | *4 | *6 | *7 | |||
| CYP2B6 | *1A | *4A | *6A | *16 | *26 | ||||||
| CYP2C19 | *1A | *1B | *1C | *2A | *2B | *2C | *2D | *2E | *2F | *2G | *2H |
| *2J | *3A | ||||||||||
| *3B | *4A | *4B | *5A | *5B | *6 | *7 | *8 | *9 | *10 | *11 | |
| *13 | *15 | ||||||||||
| *16 | *17 | *18 | *19 | *22 | *24 | *25 | *26 | *28 | *35 | ||
| CYP2C8 | *1A | *5 | |||||||||
| CYP2C9 | *1A | *2A | *3A | *3B | *4 | *5 | *6 | *8 | *9 | *11A | *12 |
| *13 | *14 | ||||||||||
| *15 | *31 | *59 | *60 | ||||||||
| CYP2D6 | *1A | *1B | *1C | *1XN | *2A | *2XN | *3A | *4A | *4B | *4C | *4D |
| *4K | *4X2 | ||||||||||
| *5 | *6A | *6B | *6C | *7 | *8 | *9 | *10A | *10B | *10X2 | *11 | |
| *12 | *13 | ||||||||||
| *14A | *15 | *17 | *18 | *19 | *20 | *21A | *21B | *29 | *31 | *33 | |
| *35A | *35X2 | ||||||||||
| *36 | *38 | *40 | *41 | *42 | *44 | *45A | *45B | *46 | *68A | *68B | |
| *69 | *92 | ||||||||||
| *100 | *101 | ||||||||||
| CYP2E1 | *1A | *2 | *3 | *4 | |||||||
| CYP3A4 | *1A | *18A | *22 | ||||||||
| CYP3A5 | *1A | *3A | |||||||||
| CYP3A7 | *1A | ||||||||||
We used twelve patients whose pharmacogenetic profile, medication list and hospitalization records were available for building scoring system. Among the twelve cases, pharmacogenomic testing indicated significant genetic polymorphisms in six of them, which affected overall metabolism of their prescribed drugs. These six patients had higher urgent care utilization than those without significant pharmacogenomics polymorphisms10.
To obtain optimal parameters for the scoring algorithm, weights for drug-drug interaction, drug-gene interaction and gene function were modeled in series of iterative experiments.
Figure 6 illustrates the total score for patients over the scaling of gene function score. Patient results from higher hospitalization rate (HHR) group are colored in red. The subject pgx15, which is represented in blue color, is the patient with the highest total score in low hospitalization rate (LHR) group. Patients in LHR group are supposed to have much smaller scores than others. To identify thresholds distinguishing patients in LHR and HHR groups, we used the patient with the lowest score in HHR group and the patient with the highest score in the LHR group (pgx15). Subsequently, the value of gene function factor was determined to be 30.
Figure 6.

Patient general score over gene function score.
Figure 6 shows a graphical view the patients’ average score changes over different factors scaling. We still colored the HHR group with red, and LHR group with blue. The comparison between Figure 6a and Figure 6b indicated that the gene function factor played a key role in distinguishing two groups of patients: the score of HHR group increased rapidly through the increasing of gene function factor. The gene function factor had minor impact on LHR group as expected.
Figure 7 demonstrates the impact of drug-drug interaction, drug-gene interaction more clearly. Since all the test cases had significant polypharmacy, the change of drug-drug interaction factor lead to obvious changes in total score. It also showed that drug-gene interaction factor had more influence in HHR group as well as gene function factor.
Figure 7.
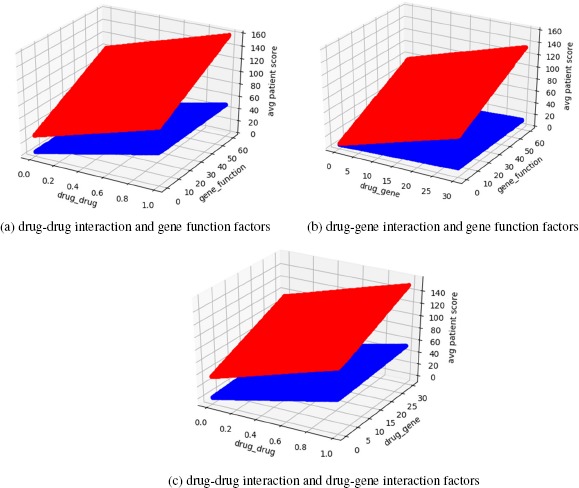
Patient general score over different interaction factors.
The scaling factor calculation revealed that we should set a higher drug-gene interaction and gene function factors, and keep drug-drug interaction factor as low as possible. In our model, the drug-drug interaction factor was eventually set to 0.1 for each competing drug (0: none; -0.1: substrate; -0.1: inhibitor; +0.1: inducer), the drug-gene interaction factor was set to 16 (0: normal/extensive; -16: poor; +16: rapid; -4: intermediate), and the gene function factor was 30 for all significant polymorphisms as mentioned above.
We calculated the total score for two groups of patients. Table 2 shows the percentage of different contributing components in total score depending on patient group (HHR vs. LHR). Depending on the nature of individual polypharmacy and pharmacogenomic profile, there is different contribution of each of three total risk score components.
Table 2.
Percentage of drug-drug interaction, drug-gene interaction and gene function score in patient’s total score.
| Subject ID | drug-drug | drug-gene | gene | total score (value) | |
|---|---|---|---|---|---|
| 1 | 0.106 | 0.690 | 0.204 | 146.88 | |
| 2 | 0.186 | 0.287 | 0.538 | 55.75 | |
| High hospitalization rate (HHR) | 6 7 8 | 0.045 0.038 0.028 | 0.332 0.115 0.000 | 0.623 0.860 0.972 | 96.3 34.88 30.85 |
| 10 | 0.059 | 0.503 | 0.444 | 67.6 | |
| 0 | 1.000 | 0.000 | 0.000 | 9.22 | |
| 11 | 0.314 | 0.686 | 0.000 | 9.72 | |
| Low hospitalization rate (LHR) | 12 13 | 1.000 0.545 | 0.000 0.455 | 0.000 0.000 | 2.15 8.8 |
| 14 | 0.167 | 0.833 | 0.000 | 4.8 | |
| 15 | 0.385 | 0.625 | 0.000 | 19.2 | |
| average | |||||
| HHR | 0.077 | 0.321 | 0.607 | 72.04±17.84 | |
| LHR | 0.569 | 0.433 | 0.000 | 8.98±2.37 | |
| Total | 0.323 | 0.377 | 0.303 | 40.51±12.81 | |
The resulting total risk score for frequently hospitalized older adults with polypharmacy (72.04±17.84) was statistically significantly different (p<0.05) from the total risk score for older adults with polypharmacy with low hospitalization rate (8.98±2.37).
Discussion and Conclusion
Our initial goal was to build a prototype of platform-agnostic medication risk scoring system that can numerically represent the general effects of potential drug-drug and drug-gene interactions based on CYP450 metabolism. Such a system may potentially help identify patients with polypharmacy in need for optimizing their drug regimens and prioritize pharmacogenomics testing in risk populations with polypharmacy. We built a scalable and comprehensive database and a pluggable Java application. The initial prototype testing demonstrated feasibility of our approach and helped identify next steps in improving scoring algorithms.
The main limitation of the study is small data set due to which systematic accuracy evaluation will be performed in the next stage of the project. Our next step is to utilize significant number of verified pharmacogenomics cases for training and testing the scoring algorithms. We plan to obtain sufficient sample from an ongoing study assessing impact of pharmacogenomic polymorphisms on urgent care utilization in older adults with polypharmacy10. We will employ a variety of machine learning techniques11 such as support vector machines, neural networks and classification and regression trees, to maximize the distance between the total scores representing study groups. A Receiver Operating Characteristic (ROC) curve will be used to identify cut-off points which maximize area under the curve (AUC) 12. Additional parameters affecting risk for hospital admissions will be included in the model.
In the next iteration of this work, the granularity of underlying knowledge repository will be significantly expanded. We plan to expand our knowledge base using automated extraction of drug-drug13 and drug-herb14 interactions from published articles and other authoritative sources15 as well as information on metabolic phenotypes of specific pharmacogenomics polymorphisms16. Patient-reported information on drug side effects17 and drug interaction18 may be an additional source of information for our knowledge database. The resulting knowledge base will have to be assessed for completeness in relation to other sources. Resulting scoring algorithms will be compared with other approaches to evaluate medication regimen risk such as recently introduced combinatorial pharmacogenetics approach for treatment of mental health conditions19.
Once sufficient scoring resolution is achieved, the decision support tool will be tested in routine clinical care for identification of individuals with high risk medication regimen. Pharmacogenomic polymorphism has been demonstrated to be a significant predictor of frequent hospitalizations in older adults with polypharmacy20. Clinical decision support systems for medication optimization may significantly improve clinical outcomes21 and reduce medical errors22. Provider23 and patient education24 on use of pharmacogenomic testing via interactive apps coupled with pharmacogenomic support embedded into electronic medical records25 will further facilitate patient safety, improve quality of care, clinical outcomes and patient satisfaction.
References
- 1.Lavan AH, Gallagher P. Predicting risk of adverse drug reactions in older adults. Ther Adv Drug Saf. 2016;7(1):11–22. doi: 10.1177/2042098615615472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brockmöller J, Stingl JC. Multimorbidity, polypharmacy and pharmacogenomics in old age. Pharmacogenomics. 2017;18(6):515–517. doi: 10.2217/pgs-2017-0026. [DOI] [PubMed] [Google Scholar]
- 3.Finkelstein J, Friedman C, Hripcsak G, Cabrera M. Potential utility of precision medicine for older adults with polypharmacy: a case series study. Future Medicine. Pharmgenomics Pers Med. 2016;9:31–45. doi: 10.2147/PGPM.S101474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flockhart DA. Drug Interactions: Cytochrome P450 Drug Interaction Table. Indiana University School of Medicine. 2007 [Google Scholar]
- 5.Preissner S, Kroll K, Dunkel M, Goldsobel G. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res. 2010;38(Database issue):D237–43. doi: 10.1093/nar/gkp970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jobson MD. UpToDate Inc; 2017. Second-generation antipsychotic medications: pharmacology, administration, and side effects. [Google Scholar]
- 7.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. Pharmacogenomics Knowledge for Personalized Medicine. Clin Pharmacol Ther. 2012;92(4):414–7. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cariaso M, Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res. 2012;40(Database issue):D1308–12. doi: 10.1093/nar/gkr798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Finkelstein J. Introducing Pharmacogenomic Decision Support for Medication Risk Assessment in People with Polypharmacy. Proc. of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2017) 2017:1784–89. [Google Scholar]
- 10.Finkelstein J, Friedman C, Hripcsak G, Cabrera M. Pharmacogenetic polymorphism as an independent risk factor for frequent hospitalizations in older adults with polypharmacy: a pilot study. Pharmgenomics Pers Med. 2016;9:107–116. doi: 10.2147/PGPM.S117014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pamer C, Serpi T, Finkelstein J. Analysis of Maryland poisoning deaths using classification and regression tree (CART) analysis. AMIA Annu Symp Proc; 2008 Nov 6; pp. 550–4. [PMC free article] [PubMed] [Google Scholar]
- 12.Finkelstein J, Jeong IC. Machine learning approaches to personalize early prediction of asthma exacerbations. Ann N Y Acad Sci. 2017 Jan;1387(1):153–165. doi: 10.1111/nyas.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams H, Friedman C, Finkelstein J. Automated Determination of Publications Related to Adverse Drug Reactions in PubMed. AMIA Jt Summits Transl Sci Proc. 2015 Mar 25;2015:31–5. [PMC free article] [PubMed] [Google Scholar]
- 14.Lin K, Friedman C, Finkelstein J. An automated system for retrieving herb-drug interaction related articles from MEDLINE. AMIA Jt Summits Transl Sci Proc. 2016;2016:140–9. [PMC free article] [PubMed] [Google Scholar]
- 15.Finkelstein J, Chen Q, Adams H, Friedman C. Automated Summarization of Publications Associated with Adverse Drug Reactions from PubMed. AMIA Jt Summits Transl Sci Proc. 2016 Jul 20;2016:68–77. [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Friedman C, Finkelstein J. Automated Metabolic Phenotyping of Cytochrome Polymorphisms Using PubMed Abstract Mining. AMIA Annu Symp Proc. 2017:535–544. [PMC free article] [PubMed] [Google Scholar]
- 17.Cross RK, Lapshin O, Finkelstein J. Patient subjective assessment of drug side effects in inflammatory bowel disease. J Clin Gastroenterol. 2008 Mar;42(3):244–51. doi: 10.1097/MCG.0b013e31802f19af. [DOI] [PubMed] [Google Scholar]
- 18.Lapshin O, Skinner CJ, Finkelstein J. How do psychiatric patients perceive the side effects of their medications? German Journal of Psychiatry. 2006;9:74–93. [Google Scholar]
- 19.Brown LC, Lorenz RA, Li J, Dechairo BM. Economic Utility: Combinatorial Pharmacogenomics and Medication Cost Savings for Mental Health Care in a Primary Care Setting. Clin Ther. 2017;39(3):592–602. doi: 10.1016/j.clinthera.2017.01.022. e1. [DOI] [PubMed] [Google Scholar]
- 20.Cabrera M, Finkelstein J. A Use Case to Support Precision Medicine for Frequently Hospitalized Older Adults with Polypharmacy. AMIA Jt Summits Transl Sci Proc. 2016 Jul;20:16–21. [PMC free article] [PubMed] [Google Scholar]
- 21.Finkelstein J, Wood J, Crew KD, Kukafka R. Introducing a Comprehensive Informatics Framework to Promote Breast Cancer Risk Assessment and Chemoprevention in the Primary Care Setting. AMIA Jt Summits Transl Sci Proc. 2017;2017:58–67. [PMC free article] [PubMed] [Google Scholar]
- 22.Bedra M, Hill Golder S, Cha E, et al. Computerized Insulin Order Sets Can Lead to Unanticipated Consequences. Stud Health Technol Inform. 2015;213:53–6. [PubMed] [Google Scholar]
- 23.Nyun MT, Aronovitz JR, Khare R, et al. Feasibility of a palmtop-based interactive education to promote patient safety. AMIA Annu Symp Proc. 2003:955. [PMC free article] [PubMed] [Google Scholar]
- 24.Finkelstein J, Cha EM. Using a Mobile App to Promote Smoking Cessation in Hospitalized Patients. JMIR Mhealth Uhealth. 2016;4(2):e59. doi: 10.2196/mhealth.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott SA, Owusu Obeng A, Botton MR, et al. Institutional profile: translational pharmacogenomics at the Icahn School of Medicine at Mount Sinai. Pharmacogenomics. 2017;18(15):1381–1386. doi: 10.2217/pgs-2017-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]


