Abstract
Background
Diarrheagenic Escherichia coli (DEC) are an important cause of acute gastroenteritis in children; however, there is limited information available on the epidemiology, phylogenetics, serotyping, and antibiotic susceptibility of DEC in children in the United States. The aim of this study was to determine the molecular epidemiology of DEC among children with and without acute gastroenteritis in Davidson County, Tennessee.
Methods
This prospective, frequency matched, case-control study recruited subjects 15 days to 17 years of age and detected DEC with polymerase chain reaction from stool samples. Additional testing was done to define phylogenetics and antibiotics resistance.
Results
Among 1267 participants, 857 cases and 410 controls, 5.5 % were positive for at least one subtype of DEC. Enteroaggregative E. coli (EAEC) [n=32 (45%)] was the most common subtype followed by enteropathogenic E. coli (EPEC) [n=30 (43%)], Shiga toxin-producing E. coli [n=4 (6%)], and diffusely adherent E. coli [n=4 (6%)]. No significant difference in prevalence of DEC was found between cases (5%) and controls (7%) [Odds ratio (OR): 0.66 (95 % confidence interval (CI) 0.4–1.07)] and results were similar when data were stratified by subtypes and adjusted for age, sex, race, and ethnicity. Substantial diversity was found among DEC isolates in terms of phylotypes and serotypes and a large proportion was resistant to, at least, one antibiotic.
Conclusions
EAEC and EPEC were frequently found in both cases and controls in this study population. DNA-based methods for detection of these subtypes need further investigation to help differentiate between pathogenic and colonizing strains.
Keywords: E. coli, diarrhea, children, gastroenteritis, EPEC, EAEC, epidemiology
INTRODUCTION
Acute gastroenteritis is a common cause of morbidity during childhood in developing and developed countries (1) and the fourth leading cause of mortality in children <5 years of age globally (1–3). The Centers for Disease Control and Prevention (CDC) estimates that 0.6 episode of diarrhea occurs per person per year in the United States (4), with higher rates in children, averaging 2.2 episodes per child per year (4). Both viruses and bacteria are important etiologic agents (1, 5–9). In addition, there is a significant cost associated with outpatient (10) and inpatient management of acute gastroenteritis in children in the United States (11).
Escherichia coli (E. coli) are common inhabitants of the human gut and usually innocuous to human health (commensal bacteria); however, certain E. coli strains acquire virulence genes by genetic horizontal transfer and cause gastroenteritis. These strains are classified as diarrheagenic E. coli (DEC) (12, 13), and include at least six different subtypes designated: diffusely adherent E. coli (DAEC); enteroaggregative E. coli (EAEC); enteroinvasive E. coli (EIEC); enteropathogenic E. coli (EPEC); enterotoxigenic E. coli (ETEC) and Shiga toxin-producing E. coli (STEC) (12). Among these subtypes, the two toxigenic strains STEC and ETEC tend to be more pathogenic than non-toxigenic pathotypes (13) while non-toxigenic subtypes EPEC, EAEC and DAEC may be present in both symptomatic and asymptomatic participants (6, 13).
In recent years, unique virulence gene amplification by polymerase chain reaction (PCR) has been used to characterize subtypes (14, 15). PCR testing has the advantage of being fairly rapid and highly sensitive (14, 15). There are, however, concerns that the positive predictive values might be low due to high detection of pathogens in asymptomatic patients(13). Additionally, DEC subtypes might be prevalent in certain geographic regions and in certain age groups(1, 5, 16–18), but limited information on epidemiology of DEC from the United States exists (16, 19). This study aimed to define the epidemiology, phylogenetics, serotyping and antibiotic susceptibility of DEC in acute gastroenteritis in a geographically defined pediatric population in the Southern United States.
MATERIAL AND METHODS
Study Design and Enrollment
This study was a prospective, frequency matched, case-control study conducted at the Monroe Carell Junior Children’s Hospital at Vanderbilt, Nashville, Tennessee from July 1, 2012, to June 30, 2013. Children were 15 days to 17 years of age, resided in Davidson County, Tennessee and consent was obtained from parents. A case was defined as a child who had diarrhea with ≥3 loose stools in a 24-hour period, or ≥1 vomiting episode, within the past 10 days. Controls had similar inclusion criteria except that they had neither acute gastroenteritis in the last 10 days nor respiratory symptoms in the last 3 days. Cases were recruited from the emergency department; inpatient and outpatients clinics, while controls were recruited from outpatient clinics only. The frequency matching was done based on the age and the calendar month. Subjects were excluded if they did not live in Davidson County, TN, were immunocompromised, or could not be enrolled due to inability to understand English or Spanish. This study was a subproject from the New Vaccine Surveillance Network (NVSN), a multi-site, active prospective surveillance system of six geographic locations within the US (20).
Data collection
Demographic data, illness characteristics, medication use, and travel history were obtained by parental interviews using standardized questionnaires and/or medical chart review and stored in a secured Research Electronic Data Capture (REDCap) database (21). Microbiologic data were also stored in REDCap. The Vanderbilt University Institutional Review Board approved this study (IRB No. 120099).
Processing of stool specimens
Stool specimens using swabs preserved in Cary-Blair transport medium were collected within 10 days of symptom onset (20). Specimens plated on MacConkey agar (Becton Dickinson & Co., Sparks, MD) and incubated overnight at 37°C were examined for the presence of lactose-fermenting, non-mucoid colonies. Such isolates were sub-cultured onto eosin-methylene-blue (EMB) agar (Remel, Inc., Lenexa, KS) and incubated for 12–24 hours at 37°C. Colonies with characteristic E. coli morphology on EMB agar (metallic green sheen) were inoculated into sulfide-indole-motility (SIM) medium (Neogen Corporation, Lansing, MI) for biochemical testing. Presumptive E. coli isolates (indole positive, motile, gas-producing, and negative for hydrogen sulfide production) were confirmed as E. coli by iauG PCR confirmation as described previously(22). E. coli isolates were subcultured on Luria broth (LB) agar and preserved in LB broth plus 20% glycerol at −80°C until further testing.
DNA preparation and detection of DEC
Each E. coli colonial isolate was expanded in 2 ml LB broth overnight at 37°C with shaking at 225 rpm. A 200 μL aliquot of bacterial culture was suspended in 500 μL of Tris-EDTA (TE) buffer, boiled for 5 minutes, and centrifuged at 13,000xg for 3 min. The supernatant containing crude DNA extract was used as DNA template for PCR.
Two separate multiplex PCR (mPCR) reactions were performed on each DNA sample. Mix-1 detected EAEC, EPEC, and ETEC. Mix-2 detected STEC, DAEC, and EIEC. Primers used in this study were previously reported in Gomez-Duarte, et al. (14) and Panchalingam, et al. (23) (see Table, Supplemental Digital Content 1). A 20 μL reaction mixture containing 18 μL PCR blue master mix (Invitrogen, Carlsbad, CA), 1μL of oligonucleotide mix-1 or oligonucleotide mix-2 and 1μL DNA template was preheated at 94°C for 5 minutes and subjected to 40 cycles of the following thermal profile: 30 seconds at 94°C, 30 seconds at 58°C, and 1 minute at 72°C. Reactions were held at 72°C for 5 minutes for final product extension. PCR products were separated by electrophoresis (110 V, 150 minutes) in a 3% agarose gel containing ethidium bromide. Gel images were captured and analyzed.
All isolates generating positive results in multiplex PCR mix 1 or mix 2 were retested by a confirmatory singleplex PCR following DNA re-isolation(23).
Serotyping
For O and H serotyping, DEC isolates were submitted to The Pennsylvania State University E. coli Reference Center (University Park, PA). O-serotyping was performed using antisera generated against E. coli serogroups designated O1-O187, with the exceptions of O31, O47, O67, O72, O94, and O122 as these are not designated. H-serotyping was performed by PCR amplification of the flagellar fliC gene followed by restriction fragment length polymorphism analysis.
Multilocus sequence typing (MLST) and phylogenetic grouping
MLST of STEC was performed as described online (http://mlst.warwick.ac.uk/mlst/). Internal regions of seven 7 housekeeping genes, adk (adenylate kinase), fumC (fumarate hydratase), gyrB (DNA gyrase), icd (isocitrate/isopropyl maleate dehydrogenase), mdh (malate dehydrogenase), purA (adenylosuccinate dehydrogenase), and recA (ATP/GTP binding motif of recombinase A), were amplified by PCR, and amplification products were sequenced as described previously (14). Bidirectional DNA sequences were aligned for comparison and editing using DNADynamo software (Blue Tractor Software, North Wales, UK). Sequences for the seven target genes were concatenated to produce an alignment sequence of 3423 bp. Alignment of concatamers was performed with ClustalW software (15). Phylogenetic trees of all four STEC isolates and MLST reference sequences were constructed by bootstrapping procedure. E. coli reference MLSTs included three STEC strains previously isolated in the United States and reported in the http://mlst.warwick.ac.uk/mlst/database. E. coli ancestral sequences also were obtained from the same database, including the E. coli Reference (ECOR) collection (16). Phylogenetic grouping used Clermont’s methods based on PCR detection of chuA, yjaA and TspE4.C2 genes (17).
Biofilm formation assay
A quantitative biofilm assay the as described previously (18). In brief, bacterial liquid cultures diluted in Dulbecco’s Modified Eagle Medium (Thermo Fisher Scientific, Waltham, MA) at a ratio of 1:40 in flat-bottom 96-well polystyrene plates (Becton Dickinson, Franklin Lakes, NJ) were incubated at 37 °C for 24 hours. Plates were washed three times with PBS, and 200 μL of 0.1% crystal violet solution was added to each well and incubated at room temperature for 15 minutes. Plates were washed with water and air dried, followed by stain solubilization using 200 μL of 95% ethanol at room temperature for 15 minutes. Solubilized crystal violet was transferred to a 96-well Immulon 2 HB plate, and the optical density (OD) was measured at 590 nm. A strain was considered biofilm-positive if the OD was higher than the mean negative control OD plus two standard deviations.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using BD BBL™ Sensi-Disc™ Susceptibility Test Discs, (Becton, Dickinson and Company Sparks, MD, USA). The following agents were tested: cefazolin, ceftriaxone, ampicillin, amoxicillin/clavulanic acid, ceftazidime, cefuroxime, cefepime, ciprofloxacin, gentamicin, meropenem, trimethoprim/sulfamethoxazole, and piperacillin/tazobactam. Zones of bacterial growth inhibition were interpreted according to 2014 performance standards issued by the Clinical and Laboratory Standards Institute. Controls included E. coli ATCC 29522 (susceptible to all antibiotics) and Klebsiella pneumonia ATCC 700603D-5 (resistant to all beta-lactam antibiotics).
Statistical Analysis
Sample Size Calculations
The sample size was calculated with the assumption that at least 6% of cases would be positive for DEC compared to 2% of controls. These values were based on previous studies showing similar proportions of diarrheagenic E. coli in children with AGE and healthy controls (9, 19). By choosing a 2:1 ratio of cases versus controls, approximately 848 children were required as cases and 424 as controls to achieve 90% power at an alpha level of 5%.
Association analysis
A Pearson Chi-square test or Fisher’s exact test was used to compare the proportion of cases and controls with pathogenic E. coli. Odds ratios and corresponding 95% confidence intervals served to quantify observed differences. Logistic regression was used to adjust the estimated odds ratio for potential confounders. The decision to include variables in the model was a priori and was based on previous studies(5). The variables that were selected a prior included age, sex and race. Matching of number of cases compared to control was based on frequency and therefore no adjustment was done for matching during regression analysis. Missing data were noted and reported accordingly. We decided a priori that we would do multiple imputations for a key variable if data were missing for more than 10 % of the participants for that variable. Data for none of the key variables was missing for more 10 % of the study population, so we did not perform imputations for missing data.
We attempted two regression analyses: First analysis was based on ‘any diarrheagenic E. coli vs. no diarrheagenic E. coli’ irrespective of the type of the E. coli. This analysis was adjusted for different subtypes meaning that subtypes were mutually exclusive. The second analysis was based on types of diarrheagenic E. coli and each subtype was compared against ‘no diarrheagenic E. coli’ irrespective of presence or absence of other subtypes in cases or controls. Statistical significance was designated as a two-tailed p-value <0.05. All statistical analyses were performed using STATA version 14.0.
RESULTS
Study population
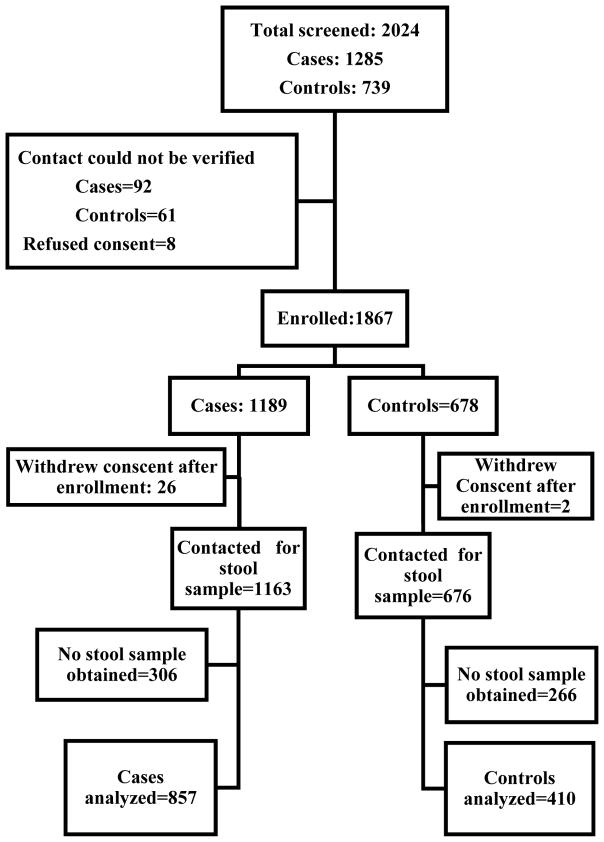
We screened a total of 2024 potentially eligible participants and enrolled 1267 participants, 857 cases, and 410 controls. The study screening and enrollment numbers are shown in Figure 1. Cases and controls were comparable for most of the demographic variables except gender, where the cases had a slightly higher proportion of females (Table 1).
Figure 1. Enrollment of Cases and controls.
a Primary caregiver noted in the medical chart could not be verified.
Table 1.
Demographics of cases and controls.
| Characteristics | Total (n=1267) |
Cases n=857 |
Controls n=410 |
p-value |
|---|---|---|---|---|
| Sex: Female, n (%) | 636 (50) | 448 (52) | 188(45) | 0.03 |
| Age, months, median (IQR) | 24 (11–57) | 24 (11–57) | 24 (10.2–57) | 0.7 |
| Age by category, n (%) | ||||
| 0–12 months | 347(27) | 236 (28) | 111(27) | |
| 13–24 months | 282(22) | 189(22) | 93(23) | 0.9 |
| 25–60 months | 339(27) | 231(27) | 108(26) | |
| > 60 months | 299(23) | 201(23) | 98(23) | |
| Race, n (%) | ||||
| White | 792 (62.5) | 534 (62) | 258 (63) | 0.6 |
| Black | 450 (35.5) | 306 (35) | 144 (35) | |
| Asian | 20 (1.58) | 12 (1.5) | 8 (2) | |
| Hawaiian | 1 (0.08) | 1 (0.08) | 0 (0) | |
| Other | 4 (0.32) | 4 (0.47) | 0 (0) | |
| Ethnicity, n (%) | ||||
| Hispanic | 481 (38) | 321(38) | 160 (40) | 0.59 |
| Non-Hispanic | 786 (62) | 536 (62) | 250 (60) | |
| Annual Household Income a, n (%) | ||||
| Less than $ 25 000, | 711 (56) | 464 (54) | 247(60) | 0.03 |
| Between $ 25 000 to 75 000 | 214 (16) | 127(14) | 87(21) | |
| More than 75 000 | 40 (3) | 35(4) | 5(1) | |
| Maternal Education b, n (%) | ||||
| Less than high school | 374 (29) | 244 (28) | 130 (31) | 0.7 |
| High School | 540 (42) | 364 (42) | 176 (42) | |
| College degree and above | 298 (23) | 200 (23) | 98 (24) | |
| Insurance c n (%) | ||||
| Public | 1071 (84) | 708 (82) | 363 (88) | 0.02 |
| Private | 151 (12) | 113(13) | 38(9) | |
| Both public and private, | 25 (2) | 17 (2) | 8 (2) | |
All percentages in table are in comparison to totals given at the top of each column. P values were obtained by using Fisher exact test for categorical variables and Wilcoxon-rank test for continuous variable.
Data were missing for 23% of participants.
Data were missing for 4% of participants.
Data were missing for 1% of participants and 0.5% of participants had no insurance
DEC among cases and controls
Overall, 48% of stool samples were positive for E. coli, and DEC was isolated from 5.5%. Among DEC, 4 subtypes were found and included EAEC [n=32 (45%)], EPEC [n=30 (43%)], STEC [n=4 (5%)], and DAEC [n=4 (5%)]. No significant difference was found in the prevalence of DEC between cases (5%) and controls (7%) with an odds ratio of 0.66 (95 % confidence interval 0.4–1.07). Results were similar when data were adjusted for age, sex, race, and ethnicity (Table 2). None of the four recovered subtypes of DEC were statistically significantly associated with cases; however, overall numbers were small for STEC (n=4) and DAEC (n=4) (Table 2). The sensitivity analysis when each pathotype was considered individually without consideration for the presence of other pathotypes (not mutually exclusive) revealed similar results (data not shown).
Table 2.
PCR testing results for diarrheagenic E. coli among cases and controls.
| Organism | Total n=1267 |
Cases n=857 |
Controls n=410 |
Odds ratios (95% CI) |
|---|---|---|---|---|
| Any diarrheagenic | ||||
| E. coli, n (%) | 70 (5) | 41 (5) | 29 (7) | Crude OR: 0.66 (0.40–1.07) Adjusted OR: 0.65 (0.39–1.06)* |
| EPEC, n (%) | 30 (2) | 18 (2) | 12 (3) | Crude OR: 0.70 (0.33–1.46) Adjusted OR: 0.69 (0.32–1.4)* |
| EAEC, n (%) | 32 (2) | 17 (2) | 15 (4) | Crude OR: 0.52 (0.26–1.07) Adjusted OR: 0.50 (0.25–1.083* |
| DAEC, n (%) | 4 (0.3) | 3 (0.3) | 1 (0.2) | Crude OR: 1.4 (0.14–13.5) Adjusted OR: 1.5 (0.14–14.0)* |
| STEC, n (%) | 4 (0.3) | 3 (0.3) | 1 (0.2) | Crude OR: 1.4 (0.14–13.5) Adjusted OR: 1.4 (0.14–14.0)* |
All percentages are in comparison to totals at the top of the column.
adjusted for sex, age, gender, race, ethnicity
Abbreviations: DAEC: diffusely adherent E. coli; EAEC: enteroaggregative E. coli; EPEC: enteropathogenic E. coli; and STEC: Shiga toxin-producing E. coli
Genotypic and phenotypic features of DEC
Each DEC isolate was characterized genotypically by evaluating DEC-associated virulence genes and phylogenetic grouping. DEC strains were also evaluated phenotypically for expression of specific serotypes, antibiotic resistance, and among EAEC, for biofilm formation (Online only). No significant differences were observed between cases and controls with respect to the proportion of EAEC biofilm-forming strains. From 32 EAECs, 13 (40%), 11 (34%), and 8 (25%) were positive for gene aggR, aaiC and aaiC plus aggR, respectively. EAEC strains were diverse in terms of phylogenetic ancestral grouping; 25%, 21.8%, 15.6%, 15.6%, and 21.8% belonged to B2, B1, A, D, and untypable phylogenetic groups, respectively. Similarly, serotypic diversity was high among EAEC strains, with the most common serotype, O86: H27, constituting only 12.5% of total. Twenty-five (78%) of EAEC isolates were resistant to at least one antibiotic; 24 (75%) were resistant to ampicillin.
EPEC, detected among 30 subjects, was the second most common DEC pathotype isolated (Online only). Four of the EPECs were typical (i.e., eae plus bfpA positive), and the remaining were atypical EPEC (i.e. eae positive and bfpA negative). All four typical EPEC strains were identified in cases. EPEC phylogenetic groups consisted of B1 (33.3%), B2 (26.6%), D3 (10%), and untypable (33.3%). Seventy-percent of EPEC isolates were resistant to at least one antibiotic, and 16.7% were multi-drug resistant (resistance detected to more than one antibiotic class). (Supplemental Digital Content 2)
Four DAEC isolates were recovered, 3 in cases and one in a control. DAEC strains isolated from cases were resistant to at least two antibiotic classes. One DAEC strain was resistant to beta-lactams, fluoroquinolones, trimethoprim/sulfamethoxazole, and tetracyclines. The DAEC strain isolated from a control was resistant to ampicillin and ampicillin/clavulanic acid. (Supplemental Digital Content 2)
Four STEC clinical isolates were detected in this study, three from cases and one from a control. All STEC serotypes were non-O157 and included O103: H2, O159: H-, O145: H25, and O113: H21. Each STEC from the cases was positive for both eae and stx genes. Two strains were positive for stx1, and two for stx2 (Online only). The case positive for STEC O145: H25 was admitted with bloody diarrhea and developed severe hemolytic uremic syndrome (HUS). Three STEC strains were susceptible to all antibiotic tested and one, isolated from a case, was resistant to ampicillin and ampicillin/clavulanic acid. (Supplemental Digital Content 2)
DISCUSSION
In this single-site, prospective, case-control study, we observed a prevalence of 5.5 % of DEC with almost equal distribution in both acute gastroenteritis cases and healthy controls. Among DEC subtypes, EAEC was the most common subtype followed by EPEC and there were a small number of participants with DAEC and STEC. Diarrheagenic E. coli isolates exhibited substantial diversity in phylotype and serotype for both the EAEC and non-EAEC groups. The most common phylogenetic type was B1 (n=19) followed by B2 (n=17). A large proportion of DEC isolates were resistant to at least one antibiotic.
The epidemiology of DEC varies in different regions of the world and may depend on the host and environmental factors (13). Enteroaggregative E. coli have been recognized as a cause of diarrhea in both developing and developed countries; however, it has also been described to be present in both symptomatic and asymptomatic patients (6, 13, 24). The Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health (MAL-ED) study was conducted in 8 developing countries and it reported that EAEC was detected in similar proportions during symptomatic and asymptotic periods of diarrhea (6). The Global Enteric Multicenter Study (GEMS) reported that EAEC was found only in one of the age group at one of the seven study sites and explained about 9 % of the moderate to severe diarrhea in that population(5). In our study, EAEC was detected commonly in both controls and cases. The pathogenicity of EAEC depends on a set of genes that confer pathogenic characteristics (13, 25) that include adherence to gastrointestinal epithelium, mucus production biofilm formation and toxin production (13, 26). We specifically tested biofilm formation ability of the EAEC isolates and found that the majority demonstrated capacity for biofilm formation; however, these strains were almost equally distributed between cases and controls, therefore, it is likely that pathogenic factors other than biofilm formation may confer virulence for EAEC or the gene targets for PCR used to detect EAEC are not specific to truly pathogenic EAEC strains (27, 28). A study from Mali showed that only a subset of EAEC was associated with diarrhea and that additional virulence genes may be responsible for the pathogenesis of EAEC which are not routinely tested in research studies (27). An alternative hypothesis is that EAEC is an avid colonizer of the human gut and only pathogenic in a small fraction of infected children as seen in studies from Peru and MAL-ED study (6, 24). Further studies to understand the reasons for similar detection frequencies in cases and controls are needed. Research questions to be addressed may include: first, what are the key virulence factors involved in the pathogenesis of EAEC; second, as a consequence of the above, what molecular-based targets are necessary for detection of true pathogenic strains; and third, could EAEC may result in a healthy-carrier state after initial infection.
EAEC clinical isolates in this study were highly diverse based on serotyping, phylogrouping, or MLST typing analysis (13, 26). Among the classic EAEC serotypes (O44, O86, O104, O125, and O126) only O86 was observed in this study with 18.7% among all EAEC isolates. Similar EAEC diversity in clinical isolates was found from children in Colombia (29). EAEC diversity may be explained, at least in part, by horizontal transfer of the EAEC virulence plasmid into E. coli host strains in the environment and within the host intestine (13). Since only a few E. coli hosting the EAEC plasmid belong to known pathogenic EAEC serotypes or phylotypes, we speculate that not all PCR-positive EAEC strains as they lack key virulence genes including, aatA, aap, astA, set1A.
The second most common DEC pathotype in our study was EPEC, which may be associated with fatal infant diarrhea in developing countries (5). In our study, atypical EPEC were more common than typical EPEC and were present in both cases and controls. Typical EPEC were observed less frequently and in only 4 individual cases. We did not perform a separate analysis based on typical and atypical EPEC because the numbers were small; however, our finding of typical EPEC preferentially associated with acute gastroenteritis cases compared to atypical EPEC is consistent with previous studies (18).
STEC is associated with illness in both developing and developed countries, most notably in outbreaks(13). In our study, STEC was present in 4 subjects, 3 cases, and 1 control. None of the STEC isolates were serotype O157: H7. Importantly, one case of O145:H25 STEC infection resulted in a severe case of HUS. This is a significant finding because routine stool cultures in most clinical laboratories do not detect nonO157 STEC strains. Therefore, additional testing for STEC may be warranted in children who present with HUS in the setting of negative culture for O157 STEC strain.
DAEC binds to HEp-2 cell monolayers in a diffuse pattern (24) and has been associated with diarrhea in multiple studies conducted primarily in Latin America (26, 27). In the U.S, Cohen et al. showed that DAEC was isolated more frequently from cases of acute gastroenteritis than controls (17). We found that cases of diarrheal disease had a higher percentage of DAEC detected than controls, but cannot draw conclusions based on this as the overall numbers of isolates were small (n=4). We did not find any EIEC or ETEC subtypes. This may be because the EIEC are mainly associated with outbreaks and ETEC is commonly found in patients with traveler’s diarrhea (13).
Treating DEC with antibiotics is not routinely recommended; however, understanding the antibiotic susceptibility of these pathogens is important as intestinal E. coli strains may serve as antibiotic resistance genes reservoirs (30, 31). Horizontal transfer of these genes among opportunistic Gram-negative and Gram-positive gut organisms may explain, at least in part, the increased number of multidrug-resistant organisms leading to intractable community and nosocomial infections worldwide. In our study, 74.3% of DEC isolates were resistant to at least one antibiotic class, and 24.3% were multi-drug resistant. It is unclear if a similar proportion of antibiotic resistance is also found among commensal E. coli strains, as we did not test non-DEC isolates.
This study has some important limitations. The detection of pathogenic E. coli was based on PCR testing for known targets, but functional testing of associated virulence properties (e.g., confirmation of DAEC and EAEC using HEp-2 cell cultures) was not performed. Also, only classic genetic determinants of pathogenesis were assessed, and additional virulence genes determinants were not performed (19). This might introduce non-differential misclassification and could be the explanation of null results. Samples were collected from just one medical center and the majority of children were <2 years; therefore, these results may not be generalized to other U.S. cities or older children. Stool samples from cases and controls, not obtained at enrollment, were collected up to 10 days from onset of symptoms. Stool samples recovered at the end of the 10 days maximum collection period may have resulted in a decrease number of etiologic agents identified. The sample size calculations were based on overall prevalence of DEC and not on the prevalence of individual pathotypes, therefore the association analyses for individual pathotypes was underpowered. Furthermore, age-related differences in susceptibility to enteropathognes might be important as shown in the GEMS study and the study by Cohen et al (5, 16); our study had limited sample size to detect a difference among age subgroups.
Our study shows that DEC was present in children with and without acute gastroenteritis in Davidson County, TN. The majority of DEC isolates were EAEC and EPEC, neither of which was specifically associated with acute gastroenteritis. Typical EPEC, STEC, and DAEC were found in small numbers – predominantly among AGE cases – but no conclusions can be made due to the low prevalence of these pathogens in this study population. Therefore, larger, multi-center studies are needed to precisely define the role of different subtypes of DEC in AGE causation and identify additional markers of DEC pathogenicity.
Supplementary Material
Acknowledgments
Sources of Funding
This study was funded in part by funds from the Department of Pediatrics, Vanderbilt University School of Medicine, to O.G.G.-D and from Center for Disease Control as part of The New Vaccine Surveillance Network (NVSN) to N.B.H.
This study was funded in part by funds from the Department of Pediatrics, Vanderbilt University School of Medicine, to O.G.G.-D. Surveillance was funded by a cooperative agreement from the US Centers for Disease Control and Prevention to N.B.H.
This study used REDCap online database system to collect data and REDCap project was supported by the grant support (UL1 TR000445 from NCATS/NIH)
Footnotes
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention (CDC).
Conflict of Interest: All authors declare that they do not have any conflict of interest.
References
- 1.Walker CL, Rudan I, Liu L, et al. Global burden of childhood pneumonia and diarrhoea. Lancet. 2013;381:1405–1416. doi: 10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung DT, Chisti MJ, Pavia AT. Prevention and Control of Childhood Pneumonia and Diarrhea. Pediatr Clin North Am. 2016;63:67–79. doi: 10.1016/j.pcl.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of under-5 mortality in 2000–15: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet. 2016 doi: 10.1016/S0140-6736(16)31593-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones TF, McMillian MB, Scallan E, et al. A population-based estimate of the substantial burden of diarrhoeal disease in the United States; FoodNet, 1996–2003. Epidemiol Infect. 2007;135:293–301. doi: 10.1017/S0950268806006765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 6.Platts-Mills JA, Babji S, Bodhidatta L, et al. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED) Lancet Glob Health. 2015;3:e564–575. doi: 10.1016/S2214-109X(15)00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heusinkveld M, Mughini-Gras L, Pijnacker R, et al. Potential causative agents of acute gastroenteritis in households with preschool children: prevalence, risk factors, clinical relevance and household transmission. Eur J Clin Microbiol Infect Dis. 2016;35:1691–1700. doi: 10.1007/s10096-016-2714-9. [DOI] [PubMed] [Google Scholar]
- 8.Chhabra P, Payne DC, Szilagyi PG, et al. Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008–2009. J Infect Dis. 2013;208:790–800. doi: 10.1093/infdis/jit254. [DOI] [PubMed] [Google Scholar]
- 9.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. Foodborne illness acquired in the United States--unspecified agents. Emerg Infect Dis. 2011;17:16–22. doi: 10.3201/eid1701.P21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mughini-Gras L, Pijnacker R, Heusinkveld M, et al. Societal Burden and Correlates of Acute Gastroenteritis in Families with Preschool Children. Sci Rep. 2016;6:22144. doi: 10.1038/srep22144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai R, Curns AT, Steiner CA, Tate JE, Patel MM, Parashar UD. All-cause gastroenteritis and rotavirus-coded hospitalizations among US children, 2000–2009. Clin Infect Dis. 2012;55:e28–34. doi: 10.1093/cid/cis443. [DOI] [PubMed] [Google Scholar]
- 12.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 13.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Duarte OG, Arzuza O, Urbina D, et al. Detection of Escherichia coli enteropathogens by multiplex polymerase chain reaction from children’s diarrheal stools in two Caribbean-Colombian cities. Foodborne Pathog Dis. 2010;7:199–206. doi: 10.1089/fpd.2009.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh KH, Kim SB, Park MS, Cho SH. Development of a one-step PCR assay with nine primer pairs for the detection of five diarrheagenic Escherichia coli types. J Microbiol Biotechnol. 2014;24:862–868. doi: 10.4014/jmb.1312.12031. [DOI] [PubMed] [Google Scholar]
- 16.Cohen MB, Nataro JP, Bernstein DI, Hawkins J, Roberts N, Staat MA. Prevalence of diarrheagenic Escherichia coli in acute childhood enteritis: a prospective controlled study. J Pediatr. 2005;146:54–61. doi: 10.1016/j.jpeds.2004.08.059. [DOI] [PubMed] [Google Scholar]
- 17.Platts-Mills JA, Liu J, Gratz J, et al. Detection of Campylobacter in stool and determination of significance by culture, enzyme immunoassay, and PCR in developing countries. J Clin Microbiol. 2014;52:1074–1080. doi: 10.1128/JCM.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trabulsi LR, Keller R, Tardelli Gomes TA. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002;8:508–513. doi: 10.3201/eid0805.010385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denno DM, Shaikh N, Stapp JR, et al. Diarrhea etiology in a pediatric emergency department: a case control study. Clin Infect Dis. 2012;55:897–904. doi: 10.1093/cid/cis553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowen MD, Mijatovic-Rustempasic S, Esona MD, et al. Rotavirus Strain Trends During the Postlicensure Vaccine Era: United States, 2008–2013. J Infect Dis. 2016;214:732–738. doi: 10.1093/infdis/jiw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez-Duarte OG, Romero-Herazo YC, Paez-Canro CZ, Eslava-Schmalbach JH, Arzuza O. Enterotoxigenic Escherichia coli associated with childhood diarrhoea in Colombia, South America. J Infect Dev Ctries. 2013;7:372–381. doi: 10.3855/jidc.2667. [DOI] [PubMed] [Google Scholar]
- 23.Panchalingam S, Antonio M, Hossain A, et al. Diagnostic microbiologic methods in the GEMS-1 case/control study. Clin Infect Dis. 2012;55(Suppl 4):S294–302. doi: 10.1093/cid/cis754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acosta GJ, Vigo NI, Durand D, et al. Diarrheagenic Escherichia coli: Prevalence and Pathotype Distribution in Children from Peruvian Rural Communities. Am J Trop Med Hyg. 2016;95:574–579. doi: 10.4269/ajtmh.16-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cennimo D, Abbas A, Huang DB, Chiang T. The prevalence and virulence characteristics of enteroaggregative Escherichia coli at an urgent-care clinic in the USA: a case-control study. J Med Microbiol. 2009;58:403–407. doi: 10.1099/jmm.0.005793-0. [DOI] [PubMed] [Google Scholar]
- 26.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boisen N, Scheutz F, Rasko DA, et al. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J Infect Dis. 2012;205:431–444. doi: 10.1093/infdis/jir757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chattaway MA, Harris R, Jenkins C, et al. Investigating the link between the presence of enteroaggregative Escherichia coli and infectious intestinal disease in the United Kingdom, 1993 to 1996 and 2008 to 2009. Euro Surveill. 2013 Sep 12;18 doi: 10.2807/1560-7917.es2013.18.37.20582. pii: 20582. [DOI] [PubMed] [Google Scholar]
- 29.Rugeles LC, Bai J, Martinez AJ, Vanegas MC, Gomez-Duarte OG. Molecular characterization of diarrheagenic Escherichia coli strains from stools samples and food products in Colombia. Int J Food Microbiol. 2010;138:282–286. doi: 10.1016/j.ijfoodmicro.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommer MO, Church GM, Dantas G. The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence. 2010;1:299–303. doi: 10.4161/viru.1.4.12010. [DOI] [PubMed] [Google Scholar]
- 31.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.