Abstract
Availability of high-throughput screening (HTS) data in the public domain offers great potential to foster development of ligand-based computer-aided drug discovery (LB-CADD) methods crucial for drug discovery efforts in academia and industry. LB-CADD method development depends on high-quality HTS assay data, i.e., datasets that contain both active and inactive compounds. These active compounds are hits from primary screens that have been tested in concentration-response experiments and where the target-specificity of the hits has been validated through suitable secondary screening experiments. Publicly available HTS repositories such as PubChem often provide such data in a convoluted way: compounds that are classified as inactive need to be extracted from the primary screening record. However, compounds classified as active in the primary screening record are not suitable as a set of active compounds for LB-CADD experiments due to high false-positive rate. A suitable set of actives can be derived by carefully analysing results in often up to five or more assays that are used to confirm and classify the activity of compounds. These assays, in part, build on each other. However, often not all hit compounds from the previous screen have been tested. Sometimes a compound can be classified as ‘active’, though its meaning is ‘inactive’ on the target of interest as it is ‘active’ on a different target protein. Here, a curation process of hierarchically related confirmatory screens is illustrated based on two specifically chosen protein use-cases. The subsequent re-upload procedure into PubChem is described for the findings of those two scenarios. Further, we provide nine publicly accessible high quality datasets for future LB-CADD method development that provide a common baseline for comparison of future methods to the scientific community. We also provide a protocol researchers can follow to upload additional datasets for benchmarking.
Keywords: HTS, PubChem, Datasets, LB-CADD
Introduction
The development of ligand-based computer-aided drug discovery (LB-CADD) methods for in silico (virtual) high-throughput screening (HTS) shows promising results for identifying potential hit compounds, i.e., compounds that share a biological activity of interest [1]. With the popularity gain of HTS in academia, the need for LB-CADD method development continues to increase [2,3]. The cost of an HTS screen correlates nearly linearly with the number of physically screened compounds. LB-CADD has the potential to reduce these costs in a resource-limited academic environment by helping to prioritize which compounds to include in a screening campaign. However, LB-CADD method development depends on the availability of reliable HTS assay datasets to study the relationship of ligand structure and biological activity. It is a challenge to identify suitable refined datasets for LB-CADD benchmarking that are available to the research community. Frequently in both industry and academia, proprietary datasets are not disclosed to the research community for use in LB-CADD benchmarking and methods development. Therefore, novel methods cannot be directly compared to existing algorithm implementations and scientific progress is difficult to gauge. In other research fields, e.g., machine learning, standardized datasets are available and serve as foundation for evaluation and benchmarking of novel algorithms. Examples are the MNIST database for hand-written digits and UCI Machine Learning Repository [4,5]. These datasets provide a common ground for testing new methods and allowing for easy comparison of novel and previously established approaches.
Compound data repositories host libraries of molecular compounds and associated biological activities
PubChem is a public repository providing HTS experiment results containing biological activities for several hundred thousand of compounds tested against different biological targets [6–8]. It provides a platform to host target-related HTS datasets. PubChem is maintained by the National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine, which is part of the National Institutes of Health (NIH). Over 1,000,000 bioassays for more than 9,000 protein targets can be accessed online contributed by more than 70 small molecule and RNAi screening centers and research laboratories. It is also supported by over 300 small molecule vendors contributing to the growing compound database of PubChem worldwide. Vendors include US government-funded institutions, research laboratories pharmaceutical companies, and collaborators hosting chemical biology databases. Other HTS repositories, such as ChEMBL or BindingDB, are alternatives to PubChem with different philosophies of annotation and evaluation of chemical biology datasets with their respective databases. A review of these HTS repositories can be found here [9–13].
False positive rate in primary HTS experiments is high
Typically, primary HTS experiments categorize small molecules as hit, inactive, or unspecified about the desired biological activity. However, depending on the design of the HTS experiment, there are many other reasons why a compound might be designated as hit ranging from activity of the compound an undeclared target in the cell to optical interference. Therefore, primary screens are only a first iteration that reduces the available compound library to a smaller set that can be interrogated in more detail. As compounds are tested without replication (singleton?) and the cut off for activity is typically loose to minimize the number of false negatives, the false positive rate can be high. Although outliers are common in HTS experiments, statistically robust methods not sensitive to outliers are necessary for hit selection, e.g., z*-score, SSMD*, B-score, and quantile-based methods [14]. Confirmatory screens act as a validation filter by testing hit compounds with multiple replications of the experiment, recording concentration response curves, test hit compounds with an identical assay setup but in the absence of the putative target protein, and sometimes exclude even compounds that act on the target protein but not selectively.
Hierarchical confirmatory screening experiments validate primary hit compounds
The biological assay database of PubChem allows for the deposition of primary as well as confirmatory HTS experiments. Due to the requirement from funding agency on data sharing, primary screening results from NIH funded HTS projects were often deposited to PubChem prior the deposition of confirmatory assays and counter screens. Confirmatory assays seek to establish the relationship between chemical structure and a defined biological outcome (SAR). Confirmatory assays applications range from validating active compounds identified in the primary screen, over the target confirmation through orthogonal assays, and determination of specificity through testing against other subtypes of the target protein or related proteins. For molecular probe development, confirmatory assays are used to investigate a smaller subset of often similar compounds to investigate the SAR around the given scaffold further. A hierarchy of confirmatory assays is established when results of dependent confirmatory screens are analysed. In progressed stages of the hierarchy, concentration response experiments provide values for half maximal effective concentration (EC50) or inhibition (IC50) in addition to the determined binary active/inactive outcome. Despite of multiple update mechanisms provided by the PubChem system, datasets regarding the same HTS assay project but deposited under different time lines are sometimes not sufficiently summarized. Upon completion of the HTS project, a curation process is necessary to incorporate all experimental data from different stages of the assay project and provide a dataset with the ultimate bioactivity outcomes.
Previous studies underline importance of chemical data curation for LB-CADD modelling
In a previous study, we assembled nine datasets from HTS campaigns representing major families of drug target proteins for benchmarking LB-CADD methods see (Table 1). Emphasis was placed on biological target diversity and the high quality HTS activity obtained through confirmatory screen validation. These collated datasets provided the foundation for an extensive LB-CADD benchmarking study using the cheminformatics framework BCL: ChemInfo [15]. For the present manuscript, we collaborate with PubChem to make these datasets easily accessible for all researchers.
Table 1.
Listing of datasets containing curated compounds uploaded to PubChem.
| Protein Target | Target Class | Internal ID | Number of Actives | PubChem AID |
|---|---|---|---|---|
| Orexin1 Receptor | GPCR | SAID_435008 | 234* | 743306 |
| M1 Muscarinic Receptor agonists | GPCR | SAID_1798 | 188 | 652178 |
| M1 Muscarinic Receptor antagonists | GPCR | SAID_435034 | 447* | 1053187 |
| Potassium Ion Channel Kir2.1 | Ion Channel | SAID_1843 | 172 | 743120 |
| KCNQ2 potassium channel | Ion Channel | SAID_2258 | 287* | 1159610 |
| Cav3 T-type Calcium Channels | Ion Channel | SAID_463087 | 703 | 1053190 |
| Choline Transporter | Transporter | SAID_488997 | 256* | 1053196 |
| Serine/Threonine Kinase 33 | Kinase Inhibitor | SAID_2689 | 172 | 743321 |
| Tyrosyl-DNA Phosphodiesterase | Enzyme | SAID_485290 | 292 | 489007 |
| NPY-Y1 Receptor | GPCR | SAID_1040 | 801 | 1159609 |
| NPY-Y2 Receptor | GPCR | SAID_793 | 699 | 1159608 |
These datasets were selected with the goal to cover a wide-range of protein target classes. Each target class is represented by a sampled chemical space, spanned by the screened molecules evaluated within related HTS assay experiments. Primary and confirmatory screens were curated from PubChem and this curation process represent a tool for more systematic benchmarking of novel LB-CADD algorithms. For this manuscript, each curated dataset was re-assembled and aligned by CIDs before being uploaded into PubChem. Datasets marked with an asterisk in Table 1 have been modified with respect to our previous study due to compound alignment by common substructure overlap rather than PubChem identifier (CID).
Significance
LB-CADD is particularly attractive in the resource-limited environment of academia as it reduces the cost and increases quality of drug discovery and/or probe development. Quantitative structure-activity-relationship (QSAR) models developed in LB-CADD are only as good as the data quality used for training such models. Thus, there is a pressing need to develop and systematically employ HTS assay record curation protocols helpful in the pre-processing of any chemical dataset. This manuscript highlights difficulties when working with HTS experimental data in the public domain and illustrates the curation process on two chosen examples targets as well as the re-upload of the new datasets into PubChem.
Establishing a dataset “gold standard” for benchmarking novel LB-CADD methods is important for testing performance of new algorithms in respect to the complexity of the chemical space and for different biological targets. It also counters a trend that newly developed methods are tested on proprietary datasets which creates difficulties when reproducing results and reduces transparency when comparing methodological advances in LB-CADD method development. As chemical space differs in complexity for each protein target, it is imperative for new LB-CADD methods to be benchmarked on representative high quality datasets. The here described curation process has the potential to provide a wide range of higher quality datasets freely accessible to the research field.
Materials and Methods
Curation process based on hierarchy of confirmatory high-throughput screens validates active compounds
The following curation process evaluates the description of PubChem assays, identifies the PubChem assay ID (AID) of the primary screen and discusses the validation and classification of active compounds from confirmatory screens. Confirmatory screens can be subdivided into the categories “confirming” and “descriptive”. “Confirming” assays validated a compound as active at a declared molecular target (e.g., testing the compounds in the presence and absence of the declared target). The application of “confirming” assays results in identification of a set of validated hits.
The second sub-category is “descriptive”. Typically, “descriptive” assays occupy a position in the hierarchy downstream from the confirmatory assays. An example of a descriptive assay is a “counter screen” against another molecular target. Since the compound activity has been validated, it is viewed as a validated hit. Additional data add to our understanding of the compound’s activity. e.g., a compound could be demoted from “active” to “inactive” based on a descriptive assay. However, this would be in the context of a previously declared intent (e.g., antagonists of the NPY Y1 receptor) and a gating criterion (e.g., 50-fold selective against Y1). Such criteria are commonly used but need to be highlighted in the context of curating a data set. Here, validated hits are active at the declared target but can be declared “inactive” within the context of the curated dataset when additional “descriptive” data is taken into consideration. To construct a final dataset, the inactive compounds are taken from the corresponding primary assay. However, the authors would like to emphasize that this manuscript does not endorse or vouch for the applied HTS methods, given assay results or interpretations of the mentioned assays below. The here described curation process merely utilizes the assay outcomes given by the assay providers and the screening facilities.
Results
High-throughput screens validate active compounds associated with NPY – Y1 and Y2 HTS screens
PubChem provides publicly available biological assay results for a diverse set of protein targets. For the scope of this manuscript, we chose neuropeptide Y (NPY) receptor type 1 and 2, (Y1 and Y2). These receptors are members of a larger family of NPY receptors (Y1, Y2, Y4, Y5) which are part of the family of G-protein-coupled receptors (GPCR) [16,17]. As their name suggests, the receptors are effectors of the neuropeptide neurotransmitter NPY, studies have implicated these receptors in diverse biological events, including feeding, alcoholism, anxiety and depression, pain perception, immunity and inflammation, vascular remodeling hypothermia, and bone and energy metabolism [18–25]. Due to the varied role of these receptors in human disease and physiology, the identification of high-affinity selective probes that target each receptor subtype may provide novel tools for the study of NPY-related pathologies.
Case study: curating primary cell-based high-throughput screening assay for antagonists of the Y1 receptor
In this case study, the PubChem assay AID1040 tests compounds for their ability to act as antagonists of the NPY receptor Y1. A cell line transfected with Y1 and a cyclic-nucleotide gated channel (CNGC) was used to measure Y1 antagonism by the test compound. The cells were treated with the β-adrenergic receptor agonist, isoproterenol, to activate adenylate cyclase, thus increasing cytosolic cyclic adenosine monophosphate (cAMP) concentrations, and therefore increasing CNGC activity. Elevated CNGC activity decreases the cell membrane potential, which is measured using a membrane potential-sensitive fluorescent probe. Because the Y1 receptor is Gi-coupled, addition of the NPY counteracts isoproterenol action resulting in a decrease in CNGC activity. A tested compound that is an Y1 antagonist will counteract NPY action, thus the isoproterenol-evoked high level of cAMP will be maintained and high CNGC activity will be preserved. This primary assay AID1040 tested 196,255 compounds and identified 1,990 actives. A subset of 1,195 hit compounds from the set of 1,990 active compounds was investigated further by the following two confirmatory screens. AID1254 repeated the primary screen experiment to validate activity for the hit compounds. AID1255 tested selectivity of hit compounds by removing antagonists of the Y2 receptor. This assay used a cell line transfected with the Y2 receptor and a cyclic-nucleotide gated channel (CNG) was used to measure receptor antagonism through CNGC opening. This assay serves as an elimination of “false positives” in this context that could result from modulation of other biological protein targets. The findings of AID1255 resulted in 332 compounds active against Y2. 252 compounds were ultimately confirmed through AID1254 as active and selective. The following two HTS screens (AID1277 and AID1278) represent a second level of validation and further investigated a smaller fraction of just 63 compounds. AID1277 determines concentration response curves for a subset of compounds identified as active in the previous experiments. Multiple criteria for testing the compounds had to be fulfilled. The compounds were active against the primary screen (AID1040). Compounds confirmed inactive by the confirmatory screen AID1254 were excluded. Additionally, these compounds had to be inactive when assessing Y2 antagonism through AID1255. The final set of active compounds is comprised of 801 active molecules, taken from the actives of AID1040, subtracting inactive compounds from AID1254, subtracting actives from AID1255, subtracting inactive from AID1277, and actives from AID1278 as shown in Figure 1. ‘Active’ compounds within this context are defined as a combination of active and selective compounds for Y1.
Figure 1.
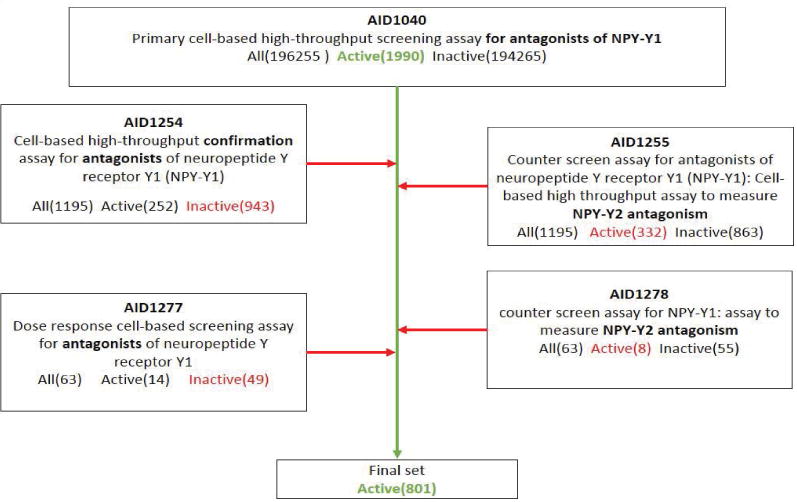
Curation process of AID1040. The center green arrow represents the initial set of active compounds while red arrows symbolize a specific subtraction of compounds.
Case study: curating primary cell-based high-throughput screening assay for agonists of the Y2 receptor
This study investigated small molecules for antagonism of NPY receptor Y2. A cell line transfected with Y2 and CNGC using a primary screening assay similar to the assay described for Y1 receptor, above. This primary screen AID793) tested 140,092 molecules for activity and identified 1,384 hit compounds. The confirmatory screen AID1257 evaluated a subset of 707 from the 1,384 molecules in more detail. It confirmed activity of compounds that were identified as actives in the primary screen AID793 with the same experimental assay setup. 707 compounds were tested in more detail and 479 molecules were confirmed inactive, and thus subtracted from the initial set of active compounds. On the other hand, AID1256 was designed to identify non-selective antagonists among the actives of the primary screen because of inhibition of the Y1 receptor. The same set of 707 compounds was screened and 135 compounds were removed as non-selective. The next stage of confirmatory screens evaluated a more specific subset of 119 compounds. Assay AID1279 determined whether compounds are active against the primary screen (AID793), activity for antagonism towards Y2 had to be confirmed in AID1257, and whether the compound showed activity in the cell-based HTS assay measuring Y1 antagonism (AID 1256). Out of the 119 actives molecules 74 compounds were confirmed and thus excluded from the pool of overall actives. The second assay (AID1272) screened the same 119 compounds as AID1279 but evaluated each molecule by different criteria: The compounds had to be active in the primary screen AID793. This activity had to be confirmed in AID1257. And lastly, these compounds had to be inactive with respect to measuring Y1 antagonism (AID 1256). A total of 119 compounds were screened and 47 inactive compounds were confirmed and removed. Next, a layer of counter screens AID2210, AID2212, AID2224, involved in this series evaluated 89 compounds for cross-findings among actives for agonism of Y1 and antagonism for Y2 and inhibition of cyclic nucleotide gated ion channel (CNGC) activity. Active compounds found through those assays were excluded from the set of final actives. Finally, as assays for late stage results from probe development efforts to identify antagonists of NPY-Y2, AID2211 and AID2220 were set up with the same conditions as AIDs 793, 1256, 1257, 1272, and 1279. Non-selective Y2 agonists and compounds acting as Y1 agonists were excluded. Figure 2 shows a detailed flow chart depicting the individual compound subtractions.
Figure 2.
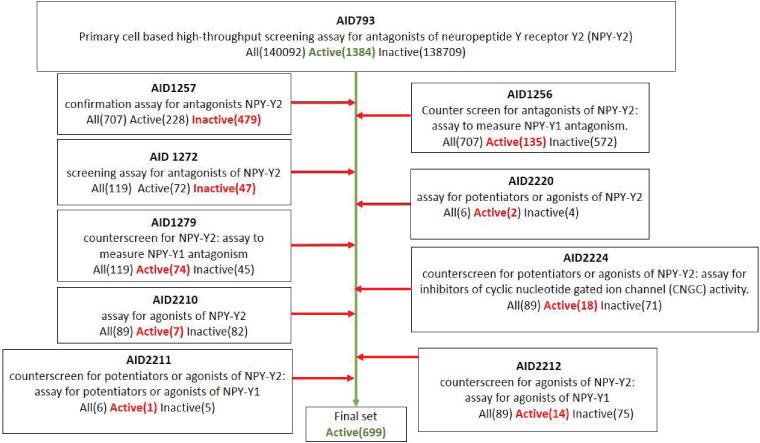
Curation process of AID793. The center green arrow leads to the final set of active compounds while red arrows and numbers and type of compounds in red mark compound subtractions.
In summary, the assembly of the final actives dataset, an ensemble of 699 active compounds was determined by selecting the actives from the primary screen and excluding inactive compounds of AID1257 and AID1272, as well as subtracting actives from AID1256, AID1279, AID2210, AID2211, AID2212, AID2220, and AID222.
Discussion
Uploading of curated datasets into PubChem
PubChem provides access to biological assay data e.g., through its Power User Gateway (PUG) [26]. Data queries can be sent via XML to request AID data for molecule in a specific format (e.g., SDF, SMILES) as well as the associated biological assay data containing metadata, and activity related data. Every compound is uniquely identified by its compound identifier (CID) or substance identifier (SID). Sets of molecules can be downloaded in respect to a given AID. These identifier in conjunction with the activity categorization of a compound allows for the curation of sets of molecules of confirmatory screens as discussed, the two case studies (see above).
Through the hierarchical relationship of primary and confirmatory assay experiments, compounds can be aligned by their respective CID. Dependent on the outcome on each hierarchy level, compounds can be classified as active or inactive depending on the result of the last involved confirmatory screen. The ensemble of molecules that satisfies all levels of the HTS hierarchy represents the final curated dataset.
The PubChem Upload system (pubchem.ncbi.nlm.nih.gov/upload) offers a mechanism to submit the newly curated set of compounds into PubChem. After specifying which compounds are involved by specified by SID identifiers the aligned hierarchy of compound activities through all involved HTS results can be uploaded. Once the submission was successful and approved by a PubChem curator the newly curated dataset is accessible to the public and can be shared with the research community.
Conclusions
High-quality HTS datasets are important for LB-CADD method development. However, results of various validation experiments for an assay project are often reported separately in PubChem and final set of inactive, inconclusive, and conformed active compounds is mostly lacked in the database. The goal of this work is to provide an overview of a curation process, starting from primary screens and their associated confirmatory screens, building a hierarchical structure through multiple related assay experiments. It needs to be emphasized that the applied HTS methodologies, the given assay results, and interpretations are taken ‘as is’. Thus, curation process relies on a high-quality standard for experimental data given by assay providers and the screening facilities. The assembly and upload of the curated dataset to the PubChem database is discussed based on two specifically chosen protein target use-cases. The upload of curated datasets into PubChem is described and thus supports the development of a publicly available database for benchmarking LB-CADD methods. Ultimately, availability of such datasets will eliminate the need to test LB-CADD methods on proprietary datasets allowing ready reproduction and comparison of results. Furthermore, such curation projects help to enhance the utility of HTS data in the PubChem database by summarizing and excluding false positives and experimental artifacts at various assay stages, and thus to highlight confirmed biological compounds.
Acknowledgments
Work in the Meiler laboratory is supported through NIH (R01 GM080403, R01 GM099842, R01DK097376) and NSF (CHE 1305874).
Footnotes
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Sliwoski G, Kothiwale S, Meiler J, Lowe EW. Computational methods in drug discovery. Pharmacol Rev. 2014;66:334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vlaar CP, Hernandez L. Symposium review: drug discovery, development and clinical research in academia. P Health Sci J. 2009;283:268–273. [PubMed] [Google Scholar]
- 3.Verkman AS. Drug discovery in academia. Am J Physiol Cell Physiol. 2004;28:465–474. doi: 10.1152/ajpcell.00397.2003. [DOI] [PubMed] [Google Scholar]
- 4.LeCun Y, Cortes C. MNIST handwritten digit database 2010 [Google Scholar]
- 5.Frank A, Asuncion A. UCI Machine Learning Repository. Irvine, CA: University of California, School of Information and Computer Science; 2010. [Google Scholar]
- 6.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids. 2009;37:623–633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, et al. PubChem’s BioAssay database. Nucleic Acids Res. 2012;40:400–412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Suzek T, Zhang J, Wang J, He S, et al. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014;42:1075–1082. doi: 10.1093/nar/gkt978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overington JP. ChEMBL: large-scale mapping of medicinal chemistry and pharmacology data to genomes. American Chemical Society; 2009. p. 238. [Google Scholar]
- 10.Papadatos G, Overington JP. The ChEMBL database: a taster for medicinal chemists. Future Med Chem. 2014;6:361–364. doi: 10.4155/fmc.14.8. [DOI] [PubMed] [Google Scholar]
- 11.Willighagen EL, Waagmeester A, Spjuth O, Ansell P, Williams AJ, et al. The ChEMBL database as linked open data. J Cheminformatics. 2013;5:1–12. doi: 10.1186/1758-2946-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:198–201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiikkainen P, Franke L. Analysis of Commercial and Public Bioactivity Databases. J Chem Inf Model. 2012;52:319–326. doi: 10.1021/ci2003126. [DOI] [PubMed] [Google Scholar]
- 14.Zhang XD. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. Biomol Screen. 2011;16:775–785. doi: 10.1177/1087057111405851. [DOI] [PubMed] [Google Scholar]
- 15.Butkiewicz M, Lowe EW, Mueller R, Mendenhall JL, Teixeira PL, et al. Benchmarking ligand-based virtual high-throughput screening with the PubChem database. Molecules. 2013;18:735–756. doi: 10.3390/molecules18010735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumont Y, Martel JC, Fournie A, St-Pierre S, Quirion R. Neuropeptide Y and neuropeptide Y receptor subtypes in brain and peripheral tissues. Prog Neurobiol. 1992;38:125–167. doi: 10.1016/0301-0082(92)90038-g. [DOI] [PubMed] [Google Scholar]
- 17.Bettio A, Beck-Sickinger AG. Biophysical methods to study ligand-receptor interactions of neuropeptide Y. Pept Sci. 2001;60:420–437. doi: 10.1002/1097-0282(2001)60:6<420::AID-BIP10183>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 18.Heilig M, Thorsell A. Brain Neuropeptide Y (NPY) in Stress and Alcohol Dependence. Rev Neurosci. 2002;13:85–94. doi: 10.1515/revneuro.2002.13.1.85. [DOI] [PubMed] [Google Scholar]
- 19.Heilig M. The NPY system in stress, anxiety and depression. Neuropeptides. 2004;38:213–224. doi: 10.1016/j.npep.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Hokfelt T, Brumovsky P, Shi T, Pedrazzini T, Villar M. NPY and pain as seen from the histochemical side. Peptides. 2007;28:365–372. doi: 10.1016/j.peptides.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Wheway J, Herzog H, Mackay F. NPY and receptors in immune and inflammatory diseases. Curr Top Med Chem. 2007;7:1743–1752. doi: 10.2174/156802607782341046. [DOI] [PubMed] [Google Scholar]
- 22.Kuo LE, Zukowska Z. Stress, NPY and vascular remodeling: Implications for stress-related diseases. Peptides. 2007;28:435–440. doi: 10.1016/j.peptides.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe K, Tilan JU, Zukowska Z. NPY and NPY receptors in vascular remodeling. Curr Top Med Chem. 2007;7:1704–1709. doi: 10.2174/156802607782340948. [DOI] [PubMed] [Google Scholar]
- 24.Jaszberenyi M, Bujdoso E, Kiss E, Pataki I, Telegdy G. The role of NPY in the mediation of orexin-induced hypothermia. Regul Pept. 2002;104:55–59. doi: 10.1016/s0167-0115(01)00339-1. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen AD, Herzog H, Sainsbury A. Neuropeptide Y and peptide YY: important regulators of energy metabolism. Curr Opin Endocrinol Diabetes Obes. 2011;18:56–60. doi: 10.1097/MED.0b013e3283422f0a. [DOI] [PubMed] [Google Scholar]
- 26.Kim S, Thiessen PA, Bolton EE, Bryant SH. PUG-SOAP and PUG-REST: web services for programmatic access to chemical information in PubChem. Nucleic Acids Res. 2015:396. doi: 10.1093/nar/gkv396. [DOI] [PMC free article] [PubMed] [Google Scholar]


