Summary
Background
Von Willebrand Factor (VWF) is tightly regulated by the metalloproteinase ADAMTS13, which cleaves VWF to reduce VWF multimer size and binding affinity for collagen and platelets.
Objective
This study examines two VWF mutations, R1597W (enhanced cleavage), and Y1605A-M1606A (decreased cleavage) to determine their impact on VWF in addition to ADAMTS13-mediated cleavage.
Methods
In vitro mouse ADAMTS13 digestions were performed on recombinant proteins. VWF knockout mice received hydrodynamic injections of mouse Vwf cDNA, following which VWF antigen, multimer profile, and VWF propeptide levels were determined. A ferric chloride injury model of thrombosis was also evaluated.
Results
In vitro ADAMTS13 digestion of full-length mouse VWF required >97-fold higher ADAMTS13 levels for Y1605A/M1606A, and 68% lower ADAMTS13 levels for R1597W compared to wild type. In vivo, R1597W had reduced VWF:Ag and both mutations exhibited increased VWF propeptide/VWF:Ag ratios. R1597W multimers show a lower molecular weight profile compared to wild type and Y1605A/M1606A mice. When co-injected with Adamts13 cDNA, Y1605A/M1606A multimers were larger compared to wild type, and R1597W showed only a single multimer band and decreased clearance via VWFpp/VWF:Ag ratio. R1597W was associated with reduced thrombus formation but normal platelet accumulation in a ferric chloride injury model while Y1605A/M1606A had a loss of occlusive thrombi but increased platelet accumulation compared to wild type.
Conclusions
This study demonstrates that mutations that alter ADAMTS13 cleavage also can affect VWF clearance, VWF antigen level, multimer structure, and thrombotic potential in the VWF knockout hydrodynamic injection model.
Introduction
The large multimeric glycoprotein von Willebrand Factor (VWF) is critical to normal hemostasis through mediating platelet-subendothelial interactions as well as binding to platelets to mediate their aggregation at the site of endothelial damage [1]. In addition, it serves as a molecular chaperone for FVIII [1]. When activated, endothelial cells release ultra-high molecular weight VWF. This highly adhesive VWF is tightly regulated by the metalloproteinase, ADAMTS13 (the thirteenth member of A disintegrin-like and metalloprotease with thrombospondin type 1 motif family), which cleaves the A2 domain of VWF at Y1605/M1606, thereby reducing VWF multimer size and binding affinity [2].
In type 2A group II von Willebrand disease (2A VWD) mutations, the largest VWF multimers are absent due to an increased susceptibility for cleavage by ADAMTS13 [3]. The remaining lower molecular weight VWF has a decreased binding affinity for collagen and platelets, and results in a bleeding phenotype [4]. In contrast, an uncontrolled increase in ultra-large VWF multimers results in spontaneous platelet binding and microvascular thrombotic occlusions in thrombotic thrombocytopenic purpura [5].
The type 2A group II VWD mutation R1597W is one of the most commonly reported forms of this subtype [6]. This mutation is located in the VWF A2 domain, near the ADAMTS13 cleavage site [6,7]. Patients with the R1597W mutation exhibit a loss of high molecular weight VWF multimers due to increased ADAMTS13-mediated cleavage. This, in turn, results in an increased bleeding time and lower VWF ristocetin-cofactor values [4]. R1597W has been previously demonstrated in vitro to have an enhanced susceptibility to ADAMTS13-mediated cleavage [3,8,9], but near normal synthesis compared to wild type recombinant protein [3,10].
The cleavage site knockout Y1605A/M1606A has previously been demonstrated to greatly decrease the rate of ADAMTS13-mediated cleavage on both full-length multimerized recombinant VWF, and an A2 domain VWF substrate [8,11]. In vivo, this should increase multimer size, and increase the thrombogenic potential of VWF, similar to the situation in ADAMTS13 deficiency. ADAMTS13 knockout mice possess a prothrombotic phenotype with increased VWF multimer size, but require another pathological challenge to produce TTP-like symptoms [12,13].
The VWF knockout mouse can be used to examine different human VWD mutations through the establishment of mouse VWF expression by hydrodynamic delivery of the mouse Vwf cDNA. The homogeneous, inbred genotype of the mice, relatively low cost to introduce new mutations, and the ability to evaluate larger study populations than would be available for human subjects makes this a valuable and convenient methodology to examine different VWD mutations. This strategy also enables the evaluation of discrete VWF variants without the potential confounding influence of additional coding region polymorphisms. This experimental approach has already recapitulated the human disease phenotypes for defective binding to collagen and GPIIbIIIa [14], type 1 VWD [15], and type 2B VWD [16,17]. Until now, this methodology has not been used to evaluate VWF sequence changes that are thought to only affect ADAMTS13-mediated cleavage.
This study examines the role of ADAMTS13-mediated cleavage on VWF clearance rate, multimer size, and function in the VWF knockout mouse via both recombinant protein infusions and VWF gene transfer through hydrodynamic tail vein injection.
Materials and Methods
Plasmid construction and mutagenesis
The mouse Vwf cDNA (courtesy of Peter Lenting) was cloned into the pCIneo plasmid for recombinant full-length mouse VWF protein (mVWF) and into the pSC11 plasmid with the liver-specific enhanced murine transthyretin (ET) promoter (courtesy of Luigi Naldini) for hydrodynamic delivery [16]. mVWF115, a GST and histidine tagged 115 amino acid fragment of the VWF A2 domain, G1554-T1668, was constructed from the pGEX-6P-1 backbone (GE Healthcare Life Sciences, Piscataway, NJ, USA) and mouse Vwf cDNA [18]. Site-directed mutagenesis was performed using the Quikchange XL II kit (Stratagene, La Jolla CA). Both R1597 and Y1605/M1606 are conserved between human and mouse VWF sequences. Mouse Adamts13 cDNA (courtesy of Dr. F. Scheiflinger, Baxter, Austria) was cloned into the pSC11 plasmid with the liver-specific ET promoter for hydrodynamic delivery.
Recombinant protein production and cell culture
HEK293T cells were transiently transfected using the calcium phosphate method as previously described [8]. VWF was secreted into serum-free OptiMEM containing 100 U/ml penicillin, 100 µg/ml streptomycin, 1× Insulin/Selenium/Transferrin G (Invitrogen, Carlsbad, CA, USA). Medium was harvested after 72 hours and recombinant VWF was concentrated using Amicon Ultra-15 or Ultra-70 100K MWCO units (Millipore, Billerica, MA, USA).
Mouse ADAMTS13 (mADAMTS13) derived from the pcDNA3.1-mADAMTS13 expression vector was produced via stable transfection in HEK293T cells similar to the recombinant VWF, using G418 selection [8]. ADAMTS13 activity was determined using the ADAMTS13 Activity ELISA Kit (Japan Clinical Laboratories, Kyoto, Japan) [19].
The mVWF115 proteins were produced in BL21-Gold E. coli (Stratagene) and purified via Ni-NTA agarose (QIAGEN, Valencia, CA) [8].
VWF antigen, propeptide, Western blots and multimer quantitation
Mouse VWF protein concentration (VWF:Ag) was determined by enzyme-linked immunosorbent assay (ELISA) using polyclonal VWF antibodies A0082 and P0226 (DAKO, Carpenteria, CA, USA). Mouse VWF propeptide (VWFpp) concentration was determined using the 349.3 capture antibody, and detected via the horseradish peroxidase-linked 349.2 antibody, produced by Robert Montgomery and provided by Sandra Haberichter [14]. Mouse VWF, VWFpp, and ADAMTS13 activity concentrations were determined using a normal C57Bl/6 plasma pool, derived from 25 normal, mixed sex, eight week old C57Bl/6 mice, with the means arbitrarily determined to be 1 U/mL mVWF, mVWFpp, mADAMTS13 respectively.
Western blots were performed in parallel on betamercaptoethanol-reduced day 2 plasma samples from hydrodynamic-injected mice (1 u/ml, 5 ul each) using NEXT Gel SDS-PAGE 7.5% acrlyamide (Amresco, USA) and transferred to PVDF membrane (BioRad). Gels were visualized with either 1:5000 diluted VWF antibody P0226 (DAKO) or 1:5000 diluted Ab47139 (Abcam) with 1:10,000 diluted P0448 secondary antibody (DAKO).
Mouse VWF multimers were analyzed by electrophoresis using a 1.4% separating sodium dodecyl sulfate (SDS) agarose gel and visualized using the VWF antibody P0226 (DAKO)[8]. Each hydrodynamic injection VWF multimer was from plasma from a single mouse loaded at 1 U/ml VWF. Lanes were analyzed for multimer distance and band number using AlphaEaseFC version 3.1.2 (Alpha Innotech, San Leandro, CA, USA) [8].
In vitro ADAMTS13 digests
For full length mVWF digestion, recombinant mouse ADAMTS13 (mADAMTS13) was diluted two-fold in 5 mM Tris (tris(hydroxymethyl)aminomethane), pH 8.0 and activated with 10 mM BaCl2 for 5 minutes at 37°C. 25 µL of diluted mADAMTS13 was added to 25 µL of mVWF (1U/mL in 1.5M urea, 5mM Tris) and incubated for 24 hours at 37°C. EDTA at a final concentration of 50 mM was added to stop the reaction [8,20]. The relative multimer migration was analyzed as previously described [8,20].
The mVWF115 ADAMTS13 digestion ELISA was modified from the ADAMTS13 Activity ELISA [19]. Reacti-bind Anti-GST coated plates (Pierce, Rockford, IL, USA) were incubated with 1.25 mg/ml mVWF115 in PBS, pH 7.2 for one hour, and washed with PBS/0.05% Tween-20 (PBS/Tween). 2-fold dilutions of ADAMTS13 were made in 5 mM acetate, 5mM MgCl2, pH 5.5 and added to each well for 4 hours at 37°C, the wells were washed with PBS/Tween, and 1 µg/ml HISProbe (Pierce) in PBS/Tween was added for 1 hour, and plates were washed. 1× OPD reagent (Sigma-Aldrich) was used for visualization, the reaction stopped at 10 minutes with 2.5M H2SO4, and absorbance was read at 492 nm.
von Willebrand Factor Studies in VWF Knockout Mice
C57Bl/6 wild type mice or VWF knockout mice [21] on a C57Bl/6 background (The Jackson Laboratory, Bar Harbor, ME, USA) aged eight to ten weeks, were used in all experiments. All mouse experiments were reviewed and approved by the Queen’s University Animal Care Committee.
Hydrodynamic injections
Plasmid DNA (100 µg pSC11-ET-mVWF) was diluted in a 10% body weight volume of lactated Ringer’s solution and injected via tail vein in less than 7 seconds using a 27 gauge needle and 3 ml syringe. VWF and ADAMTS13 co-injections received 50 µg pSC11-ET-mVWF and 20 µg pSC11-ET-mADAMTS13 plasmid. VWF knockout mice were 8–9 weeks of age at the time of injection.
Blood collection
Blood was collected using a 70 µl untreated glass capillary tube via the retrorbital plexus under isofluorane/oxygen anesthetic using 10% buffered citrate as anticoagulant. Blood was centrifuged at 11,000g for 5 minutes to generate platelet poor plasma, and samples stored at −80°C until tested.
Recombinant Protein Infusions
Recombinant mouse VWF was infused into mice at 0.2 U/g weight via tail vein in saline. Each mouse was sampled once for the experiment, with a minimum of 4 animals per time point.
Intravital Microscopy for the Ferric Chloride Injury Model of Thrombosis
Intravital microscopy was performed using a trinocular Wild-Leitz ELR-intravital microscope (Leica Microsystems Canada, Willowdale, ON, Canada), fitted with transmitted (50W halogen) and fluorescence (50W mercury incidence) light accessories. Images of thrombosis formation were captured by a Hamamatsu ORCA ER video camera (Bridgewater, NJ, USA) with fluorescent light. Analysis of the formed thrombi and the accumulated fluorescence intensity was performed using Image Pro Plus 6.0 (Media Cybernetics, Bethesda, MD, USA).
Ferric chloride injury was induced as described previously [16] in VWF knockout mice expressing only Vwf cDNA. At the time of these experiments, the plasma VWF levels were between 0.5 and 1.7 U/mL, between 14–39 days post-injection. Male mice were anaesthetized via intraperitoneal injection of ketamine/xylazine/atropine. The jugular vein was cannulated for injection of rhodamine 6G (40 ng; Sigma-Aldrich, Oakville, ON, Canada) to fluorescently label platelets in vivo and the cremaster was exteriorized. Throughout the experiment, the cremaster muscle was superfused with 37°C saline. Arterioles ranging in size from 7.5 to 18 µm were selected, and after rhodamine 6G infusion, injury was induced through the application of 10% ferric chloride-soaked filter paper (1×1 mm) for 3 minutes. Following injury, the injured area in a single arteriole was observed for 40 minutes. The time to vessel occlusion with thrombus and accumulated fluorescence intensity from images captured at 5 minute intervals were examined. Occlusion times exceeding 40 minutes were recorded as 40 minutes.
Data Presentation and Statistical Analysis
All data and statistical analysis was performed using GraphPad Prism 4.03 for Windows, GraphPad Software, San Diego, CA, USA, www.graphpad.com. Data are presented as mean values ± standard error of the mean. Statistical analyses were performed using the Student’s unpaired t-test, one-way analysis of variance (ANOVA) or two-way ANOVA.
Results
ADAMTS13 digests of full-length mouse VWF
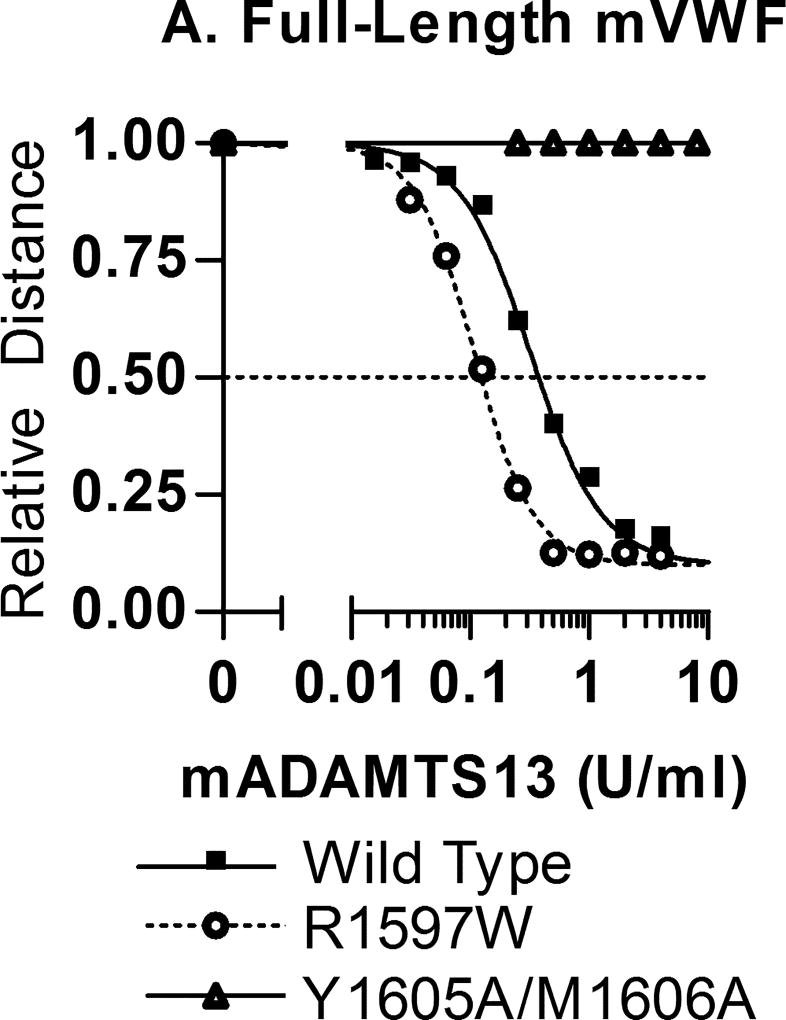
Full-length mVWF digests were performed in the presence of 1.5 M urea using a two-fold dilution series of mouse ADAMTS13 in duplicate (Fig. 1A). Multimers were measured for relative migration distance compared to undigested mVWF and plotted using a four parameter curve fit to determine EC50, the mADAMTS13 enzyme activity required to result in a 50% loss of relative multimer migration distance. Wild type mVWF had a mean EC50 value of 0.341 U/ml mADAMTS13. R1597W had an EC50 of 0.109 U/ml, a significant 68% decrease in mADAMTS13 requirement, while the EC50 of Y1605A/M1606A was greater than 32 U/ml mADAMTS13, the maximum concentration tested, almost 100-fold that of wild type VWF.
Figure 1.
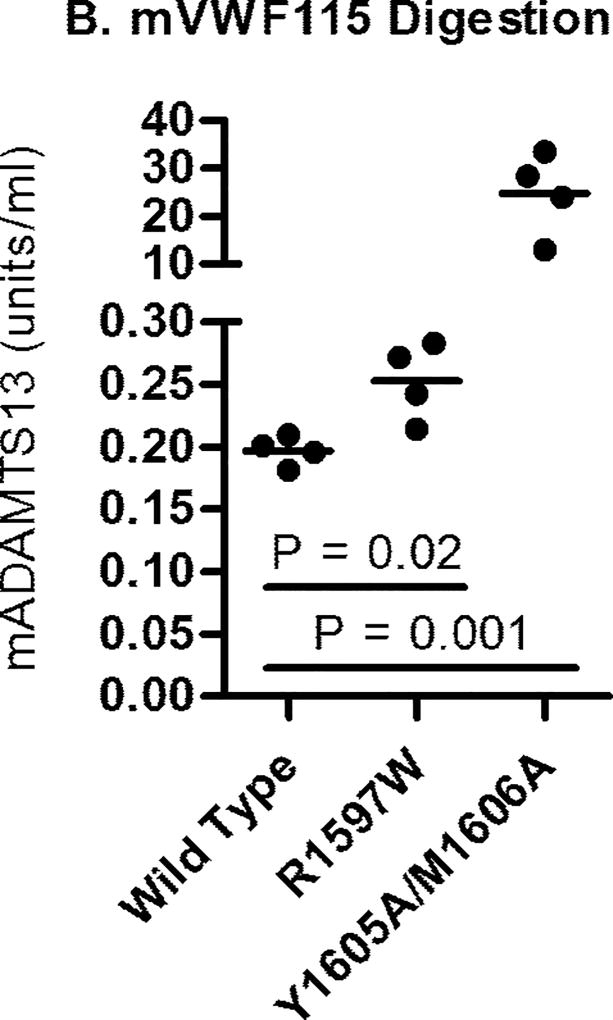
mADAMTS13 digestion of full-length mVWF and mVWF115. Recombinant mADAMTS13 digests were performed as outlined under methods. (A) Varying concentrations of mADAMTS13 were incubated with 1 U/mL full-length mVWF for 24 h with 1.5 M Urea. Multimer graphs for wild-type, R1597W and Y1605A/M1606A mouse VWF were plotted using a four-parameter curve fit. The concentration of ADAMTS13 required to cause a 50% loss of multimer height was determined. Symbols are representative of two independent experiments. (B) Comparison of the mADAMTS13 concentrations necessary for 50% loss of intact mVWF115. Varying mADAMTS13 concentrations were incubated with mVWF115 for 4 h under non-denaturing conditions. The assay was run four times for each construct. Bars represent the mean values.
Mouse ADAMTS13 digestion of mVWF115 substrates
Mouse VWF115 digests were performed under non-denaturing conditions with four replicates per mVWF115 substrate. Cleavage was determined indirectly by measuring the loss of the C-terminal histidine tag, since the Vwf mutations examined alter the epitopes at and around the mADAMTS13 cleavage site. Fig. 1B shows the EC50 values, the concentrations of mADAMTS13 required to cleave the mVWF115 by 50%. Wild type mVWF115 required a mean of 0.200 U/ml mADAMTS13. Y1605A/M1606A had a mean EC50 with a mADAMTS13 concentration of 23.8 U/ml (P = 0.0013), a factor of 119-fold higher than wild type, and R1597W had an EC50 of 0.253 U/ml mADAMTS13 (P = 0.015), an increase of 26.5% compared to wild type.
Recombinant protein infusions
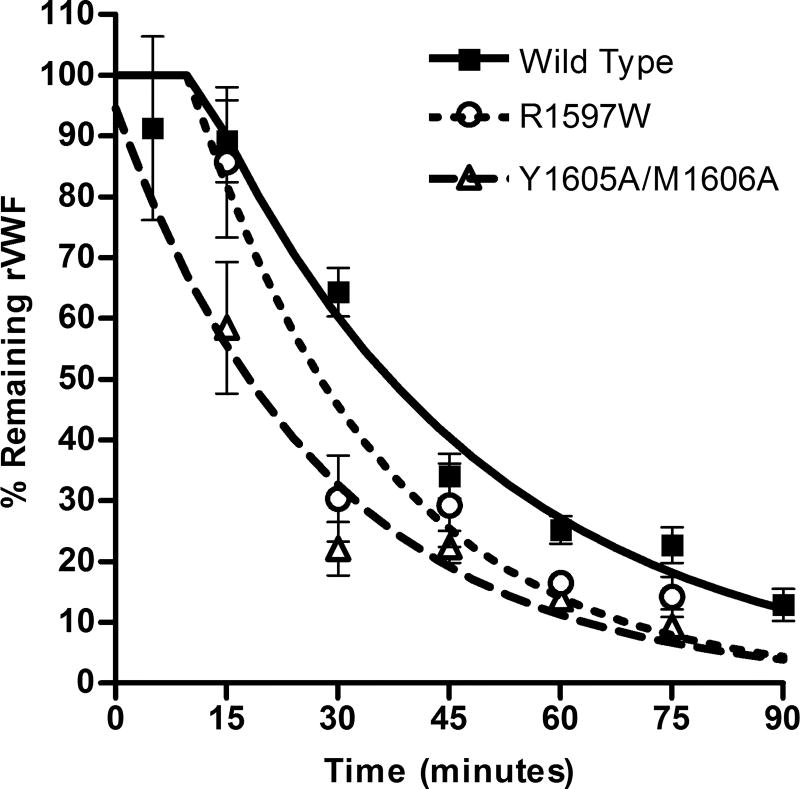
Mice were infused with recombinant mouse VWF, and a minimum of 4 mice were sampled per time point (Fig. 2). Data were fit to a one-phase exponential decay with plateau phase, the model of best fit. The half-life for wild type protein was 35.1 minutes, R1597W was 27.5 minutes (P = 0.025) and Y1605A/M1606A was 18.4 minutes (P < 0.001).
Figure 2.
Recombinant mouse VWF protein clearance. Recombinant mouse VWF protein (200 U kg)1) was injected via the tail vein into VWF knockout mice. Mice were sampled once, n ‡ 4 per time-point. Half-life was determined for each recombinant VWF protein using a one-phase decay model with plateau phase.
VWF expression via hydrodynamic transgene delivery
VWF knockout mice received plasmid DNA containing the liver specific ET promoter and mouse Vwf cDNA. Complete blood counts were performed with the injection of all Vwf cDNAs examined, and only transient thrombocytopenia was observed on day 2, with full recovery following all injections by day 5 (data not shown). The thrombocytopenia is probably a response to the diffuse vascular trauma caused by the injection. No other adverse events or alterations in complete blood count values were observed (data not shown).
VWF antigen levels
VWF knockout mice were sampled post-hydrodynamic injection (N=16/group), and antigen levels determined by VWF:Ag ELISA. Some samples were excluded due to clotting or limited sampling due to concern for animal health due to blood sampling volume and frequency. Each time point has a minimum of 10 animals in each group.
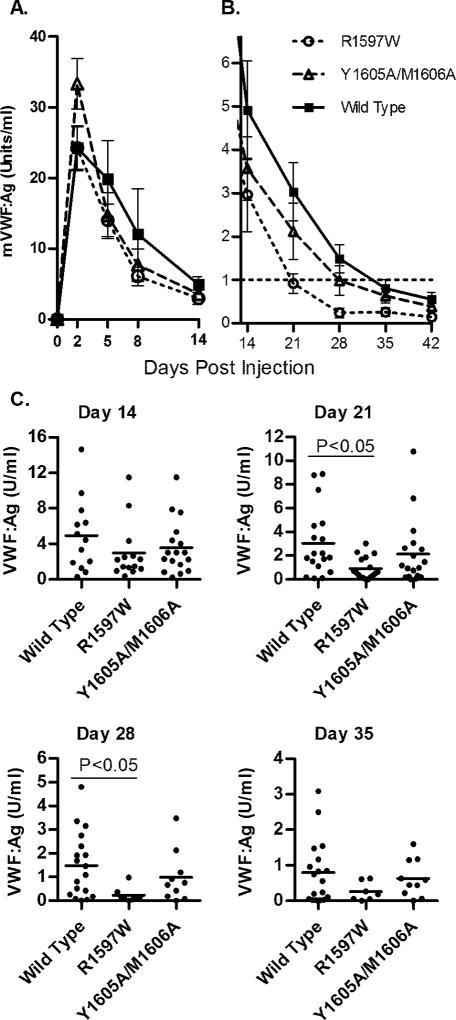
The peak levels of VWF:Ag were observed at day 2 (wild type 25.3U/ml, R1597W 24.4 U/ml, Y1605A/M1606A 33.3 U/ml, p > 0.05 for each), with a rapid decrease between days 5 and 8, and a more slowly decreasing expression phase from days 14 to 42 (Fig. 3A and B). The R1597W cDNA produced less VWF:Ag compared to wild type (p < 0.01), and two way ANOVA for all cDNAs was significantly different (p = 0.02) (Fig. 3B and C). R1597W was statistically significantly lower than wild type at day 21 (0.98 U/ml versus 3.04 U/ml, p < 0.05) and day 28 (0.23 U/ml versus 1.49 U/ml, p < 0.05). Y1605A/M1606A mVWF levels dropped below 1 U/ml by day 28 (0.98 U/ml) and wild type mVWF dropped below 1 U/ml by day 35 (0.79 U/ml).
Figure 3.
Mouse VWF antigen levels after hydrodynamic injection. VWF knockout mice expressing wild-type, R1597W or Y1605A/M1606A mVWF were sampled after hydrodynamic injection (n ‡ 10). VWF:Ag levels were determined viaELISA. (A) VWF:Aglevels from day 2 to 14 after hydrodynamic injection. Symbols represent means with SEM error bars. (B) VWF:Ag levels from day 14 to 42 after hydrodynamic injection. R1597Wwas statistically lower from wild type via paired t-test. (C) Data by day for days 14, 21, 28 and 35. Each circle represents data from a single mouse. No statistical differences were found with the exceptions of wild type vs. R1597 Won days 21 and 28, using one-way ANOVA, Tukeys post test.
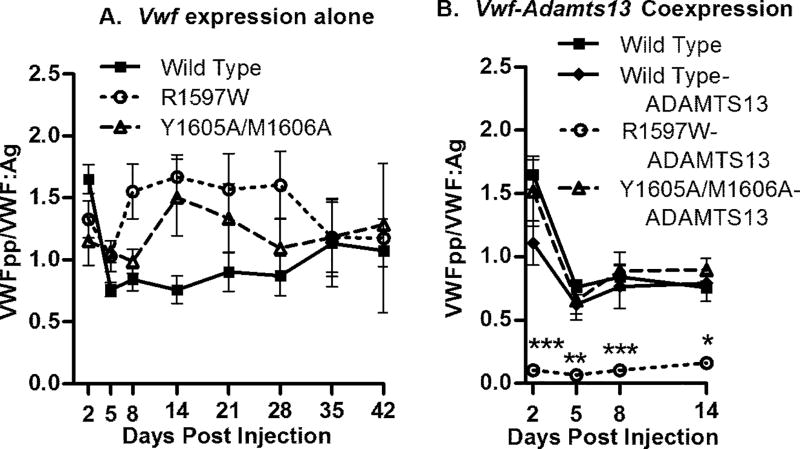
VWFpp/VWF:Ag ratio determination
VWF knockout mice were sampled post-hydrodynamic injection (N ≥ 10/group) and VWF:Ag and VWFpp levels were determined via ELISA. Values were normalized to a mean VWFpp/VWF:Ag ratio so that the overall average ratio for all WT cDNA injected mice from days 2 to 42 inclusive was equal to 1.0 (Fig. 4A). This value is 6.5 fold lower than that measured in the plasma of normal C57Bl/6 mice (data not shown). The R1597W mutant showed an overall average ratio of 1.8 greater than wild type (P < 0.001). The VWFpp/VWF:Ag ratio for the Y1605A/M1606A mutant was an overall average of 1.6, also higher than wild type (P < 0.01). Thus, both mutations demonstrate a higher VWFpp/VWF:Ag ratio, which is a surrogate marker for accelerated VWF clearance from plasma.
Figure 4.
VWFpp/VWF:Ag ratio determination. VWF:Ag and VWFpp levels were determined via ELISA. Values are normalized to a mean wild type ratio equal to 1.0, which is 6.5-fold lower than that in normal C57BL/ 6 mouse plasma. (A) VWF knockout mice expressing wild-type, R1597W or Y1605A/M1606A cDNA were sampled after hydrodynamic injection (n ‡ 10). (B) VWF knockout mice co-expressing mADAMTS13 and mVWF were examined (n ‡ 5). *P < 0.05, **P < 0.01, ***P < 0.001 vs. wild type.
Animals receiving co-injections of mADAMTS13 were also evaluated, to determine if the elevated ADAMTS13 levels altered protein clearance. Mouse ADAMTS13 activity in these mice was determined in a static assay using m73 substrate with an antibody that detects intact A2 domain substrate (data not shown). Following injection of the Adamts13 cDNA, mouse ADAMTS13 activity levels were increased to an average of 5.2 U/ml on day 2, 4.7 U/ml on day 5, 2.8 U/ml on day 8 and 2.4 U/mL on day 14. There was no significant difference in VWFpp/VWF:Ag from the wild type animals for either wild type or Y1605A/M1606A VWF co-expressing mADAMTS13 (Fig. 4B). However, mice expressing R1597W-mADAMTS13 had VWFpp/VWF:Ag ratios that decreased to 10.9% of the ratios documented in wild type VWF animals (P < 0.0001). These results were consistent for all 5 animals tested at all 4 time points, suggesting that R1597W VWF had an extended circulating half life in the presence of full length mADAMTS13.
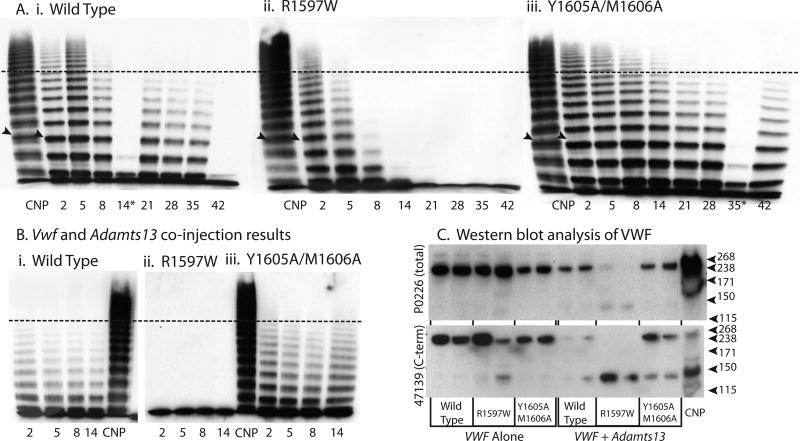
Multimer Analysis
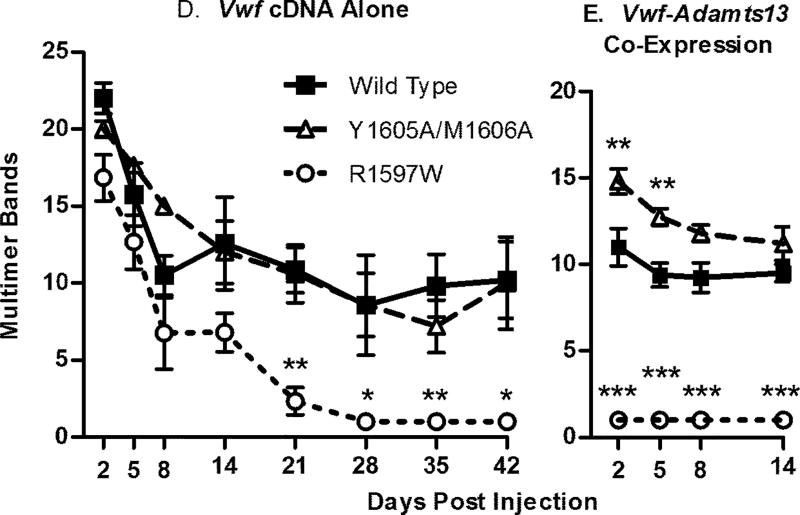
Multimeric profiles of the hydrodynamically-expressed mVWF were examined to determine if the mutations caused altered mVWF multimer structure. Plasma from individual mice was loaded on the multimer gels at 1 U/ml VWF:Ag. Samples that had clotted lost most of their multimer bands, as seen for the wild type sample on day 14. The mouse VWF produced from the liver post-hydrodynamic injection had an altered multimer appearance compared to that of normal mouse plasma pool (Fig. 5A). Single multimer bands rather than a normal triplet structure were observed, with a slightly lower molecular weight for each multimer band. In addition, the multimer profile was skewed toward lower molecular weight multimers compared to normal mouse plasma. Multimer band number counts were used to quantify differences in multimeric structure (Fig. 5D). Wild type cDNA had a peak level of 22.0 bands at day 2 (N = 7), and a low of 8.6 bands at day 28, with an average multimer band number of 12.5. Y1605A/M1606A had an average of 12.6 bands, an increase of 0.1 bands compared to wild type (P > 0.05, N = 5). In contrast, R1597W had an average of 6.1, a decrease of 6.5 bands (P < 0.001, N = 5).
Figure 5.
VWF knockout mice expressing wild-type, R1597W and Y1605A/M1606A mouse VWF were sampled after hydrodynamic injection and evaluated for multimeric structure. Plasma was run on a 1.4% agarose gel at a VWF concentration of 1.0 U mL) 1. (A) Exemplar multimers from individual mice are shown for (i) wild type, (ii) R1597W and (iii) Y1605A/M1606A. The dotted line represents high-molecular-weight multimers, greater than 10 multimer bands. *A multimer from a clotted sample. (B) Exemplar multimers from individual VWF knockout mice expressing mVWF and mADAMTS13 after hydrodynamic injection for for (i) wild type, (ii) R1597W and (iii) Y1605A/M1606A. (C) Western blot analysis of plasma from mice 2 days after hydrodynamic injection. Plasmas from two mice for each condition are shown using two different anti-VWF antibodies for detection of the cleavage product. Hydrodynamic VWF multimer structure. (D) Multimer analysis was performed on VWF knockout mice expressing mVWF after hydrodynamic injection by counting the total number of resolved multimer bands in each lane. n ‡ 4 mice per condition. (E) Multimer analysis on VWF knockout mice expressing mVWF and mADAMTS13 after hydrodynamic injection. n ‡ 4 mice per condition. Symbols represent means with standard deviation error bars for D and E. *P < 0.05, **P < 0.01, ***P < 0.001 vs. wild type.
To address the partial loss of ADAMTS13 function due to C-terminal retrotransposition event in C57Bl/6 mice, mice received co-injection of Vwf and full length Adamts13 cDNAs. The co-injected mice had significantly altered multimer patterns (Fig. 5B and E). With the co-injected Adamts13 cDNA, wild type mVWF had an average band number of 9.8 from days 2–14, which was significantly different compared to the two mutations. Y1605A/M1606A had a higher molecular weight multimer structure with 12.6 bands (P < 0.01). In marked contrast, after Adamts13 co-injection, R1597W resolved only a single multimer band for all time points, a decrease of 8.8 bands (P < 0.001), demonstrating that the addition of full-length, fully active mADAMTS13 to the VWF knockout mouse exaggerates the cleavage profile differences for these two mutant mVWF proteins.
Western Blot Analysis for ADAMTS13 cleavage products
Western blots from plasma samples from individal mice (N=5/condition) at 2 days post-hydrodynamic injection were evaluated (Fig. 5C). Wild type plasma showed faint presence of cleavage product in 2 of 5 mice, but all animals had some cleavage product with co-injection of Adamts13 cDNA. R1597W animals had cleavage product in 3 mice, and all animals had high amounts of cleavage product with Adamts13 cDNA. Y1605A/M1606A had no observable cleavage product for Vwf alone, but all had some cleavage product with co-injection of Adamts13 cDNA, which was less than or equivalent to that of the wild type Vwf with Adamts13. These results demonstrate that ADAMTS13-mediated cleavage does occur in the hydrodynamic mouse model.
Evaluation of in vivo thrombogenesis
For the intravital microscopy studies, relevant values are presented in table 1. The only statistically significant difference was the multimer band number for R1597W compared to wild type (p < 0.01) and Y1605A/M1606A (p < 0.001).
Table 1.
Relevant characteristics of mice on the day of in vivo thrombogenesis.
| Days Post-Injection | VWF:Ag (U/ml) | Multimer Bands | |
|---|---|---|---|
| Wild Type | 24.0 ± 7.8 (14–35, N=8) | 1.13 ± 0.34 (0.81–1.72, N=8) | 10.8 ± 1.9 (8–12, N=4) |
| Y1605A/M1606A | 22.3±8.1 (15–39, N=9) | 0.95 ± 0.37 (0.48–1.42, N=9) | 11.3 ± 1.5 (10–13, N=4) |
| R1597W | 23.0 ±6.3 (14–29, N=7) | 0.91 ± 0.36 (0.54–1.37, N=7) | 6.0 ± 1.3 (5–8, N=6)* |
Values reported are the average ± SD (minimum-maximum range, N).
: the number of multimer bands for R1597W was significantly different compared to wild type (p < 0.01) and Y1605A/M1606A (p < 0.001).
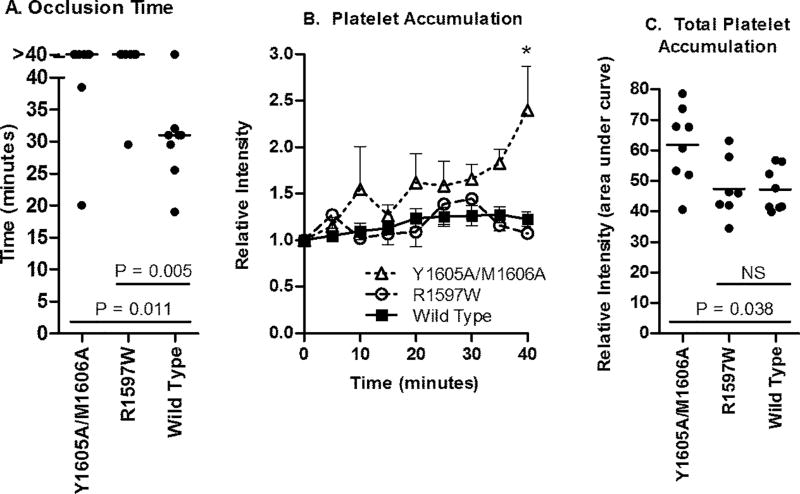
Performance of the ferric chloride injury model of thrombosis on cremaster arterioles demonstrated a difference in mean occlusion times between the two mutations studied and that of wild type mVWF, despite the fact that all mice were expressing VWF:Ag levels between 0.5 and 1.7 U/ml mVWF at the time of these studies (Fig. 6). Occlusion occurred significantly later with the two mutants: Wild type animals had a median time of 31 minutes (N=8). R1597W (n=7) only had one animal occlude at 29.5 minutes and Y1605A/M1606A (n=8) had occlusion events at 20 and 38 minutes. Survival curve comparisions were significantly different between these groups and wild type (P = 0.011 for Y1605A/M1606A and 0.005 for R1597W using the log-rank (Mantel-Cox) test).
Figure 6.
Thrombogenesis in response to ferric chloride injury. VWF knockout mice expressing only Vwf cDNA after hydrodynamic injection with VWF:Ag levels between 0.5 and 1.7 U mL) 1 were evaluated using 10% ferric chloride injury to the cremaster. (A) Time to a stable occlusive thrombus. Each circle represents an individual mouse, bars represent median occlusion time. The experiment was stopped at 40 min; mice that failed to occlude were recorded as 40 min. (B) Platelet accumulation via mean accumulated fluorescence. Mean with SEM was graphed for each mutation. *P < 0.05 vs. wild type and vs. R1597W. (C) Total platelet accumulation. Area under the curve was determined. Each circle represents a single mouse, bars indicate mean.
Platelet accumulation via fluorescence intensity, measured at 5 minute intervals throughout the study, demonstrated that Y1605A/M1606A was associated with increased platelet accumulation compared to wild type and R1597W (Fig. 6 B and C). At 40 minutes, platelet accumulation for Y1605A/M1606A was significantly higher (P < 0.001 for wild type and R1597W comparisons). Total relative platelet accumulation, represented as area under the curve for wild type VWF was 47.11, R1597W 47.4 (P = 0.94) and Y1605A/M1606A was significantly higher at 61.8 (P = 0.038).
Discussion
We selected two contrasting A2 domain mutations that affect ADAMTS13-mediated proteolysis for this study to carry out a broad and comprehensive characterization of the effects that altered ADAMTS13-mediated cleavage has upon VWF protein function and clearance mechanisms. R1597W, a commonly reported type 2A group II VWD mutation, is well recognized to result in increased ADAMTS13 proteolysis in both patients and in vitro studies, with minimal effects on biosynthesis and release [3,8,10]. Y1605A/M1606A, a double alanine substitution of the ADAMTS13 cleavage site, has been shown to greatly decrease ADAMTS13-mediated proteolysis in vitro [8,11].
Mouse ADAMTS13 has poor activity on the human VWF substrate in both full-length and A2 domain VWF73 assays [23]. There is 81% sequence identity and 91% homology between the human and mouse VWF amino acid sequences [24]. This could lead to changes in function, despite sequence conservation for R1597 and Y1605/M1606 between humans and mice. Therefore, it was necessary to confirm that our mutations of interest behaved similarly in an experimental system employing exclusively murine sequences. Full-length mouse ADAMTS13 digests appeared similar to that of previously reported in vitro human ADAMTS13 data [3,8] (Fig. 1). There was a 78% decrease in the mADAMTS13 concentration required to achieve 50% cleavage in R1597W compared with wild type mVWF in the full-length material. In contrast, Y1606A/M1606A did not show any cleavage with the maximum mADAMTS13 concentration tested, 97-fold higher than that used for wild type mVWF cleavage. The mVWF115 assay also showed a 126-fold higher value in mADAMTS13 concentration with the Y1605A/M1606A cleavage site knockout. However, in contrast to the results with the full-length substrate, R1597W showed a 28% increase in mADAMTS13 concentration compared to the wild type mVWF115. This inconsistent result is likely due to the removal of part of the A2 domain in the mVWF115 substrate, which consists only of G1554-T1668. The crystal structure of the human VWF A2 domain indicates that R1597 is in a short helix in a flexible extended loop and interacts with L1497, D1498, S1534, and S1593 (PDB ID 3GXB) [7]. Because three of these four amino acids are absent in the mVWF115 substrate, this mutation will not be accurately modeled in this structure. This is an important caveat to keep in mind when evaluating mutations in the A2 domain with the minimal substrates for ADAMTS13.
Since in vitro mouse full-length VWF digestion by mouse ADAMTS13 appears similar to that of the human system, we investigated the multimer structure of the hydrodynamic injection mice expressing the VWF proteins in vivo over several weeks (Fig. 5). The multimer structure in Y1605A/M1606A showed a statistically insignificant population of higher molecular weight bands than wild type VWF (average increase of 0.11 bands). R1597W multimer structure was significantly reduced, with a loss of half the multimer bands.
In the C57Bl/6 mouse strain, there is a retrotransposon insertion in the mouse Adamts13 gene that leads to the premature truncation of ADAMTS13, resulting in a protein lacking the C-terminal 2 Tsp1 domains and CUB domains [25]. In vitro, there is little change in cleavage under static conditions, and C57Bl/6 mice exhibit normal VWF multimer patterns. However, the truncated enzyme results in increased thrombotic occlusions under shear stress conditions, since it is not fully active in vivo [26]. This could be a factor in the lack of major differences in the multimer patterns between Y1605A/M1606A and the wild type proteins. To mitigate any artifacts caused by the partially functional ADAMTS13 in our mice, we performed co-injections with a full-length, fully functional mouse Adamts13 cDNA to observe any influences on the VWF multimer profile (Fig. 5B and E). Based on previous studies with hydrodynamic injection transgene delivery, it is extremely likely that mice produced both transgenes in the same hepatocytes at the DNA concentrations delivered to the animals [27].
With the co-injected Adamts13 cDNA, the Y1605A/M1606A multimer profile was significantly higher than that of wild type VWF (12.6 versus 9.8 bands). In marked contrast, after co-injection of the Adamts13 cDNA, R1597W resolved only a single multimer band for all time points, demonstrating that the addition of full-length ADAMTS13 to the VWF knockout mouse exaggerates the cleavage profile differences for these two mutant VWF proteins.
Protein clearance studies were performed to determine if these mutations altered the half-life of the VWF protein. The recombinant proteins were cleared rapidly, with wild type half-life being 35.1 minutes (Fig. 2). The rapid clearance of all of the recombinant proteins likely relates to altered glycosylation modifications derived in the HEK293T cells. In addition, in these studies both ADAMTS13 cleavage mutants had significantly shorter half-lives, with R1597W showing a 22% decrease and Y1605A/M1606A a 48% decrease. Previous work in the VWF knockout mouse showed no influence of multimer structure or ADAMTS13 cleavage on protein clearance [28]. This rapid clearance could be due in part to alterations in the folding of the A2 domain [7]. This in turn could then lead to changes in VWF interactions with other plasma proteins, or clearance receptors.
Long term VWF expression was achieved using hydrodynamic tail vein injection of the mouse Vwf cDNA with a strong liver-specific promoter. A strategy utilizing genetic knock-in technology has not been used here due to the need to evaluate several constructs and the associated practical (time and budgetary) limitations. Hydrodynamic gene transfer results in hepatocyte-specific expression of the mouse VWF protein, which presumably has an altered glycan content compared to normal endothelial and platelet-derived mVWF. In addition, the secretion of VWF from hepatocytes may well not result in transiently tethered protein, and thus, the early cleavage of nascent VWF by ADAMTS13 may not occur as efficiently. Nevertheless, our Western blot studies in the hydrodynamic injected mice do show evidence of cleavage products consistent with ADAMTS13 proteolysis. Furthermore, the sensitivity to cleavage is consistent with our expectations from the examination of human pathological states. This transgene delivery method replaces only the plasma component of mVWF in the knockout animals. There were significant although minimal differences in the VWF:Ag plasma levels of the two mutations compared to wild type, although R1597W levels were slightly lower in the slowly decreasing phase of expression from day 14 onward (Fig. 3). Based on a comparison to normal C57Bl/6 mouse plasma, the VWFpp/VWF:Ag ratio in hydrodynamic injected mice is 6.5-fold lower, suggesting a longer circulating time for VWF or shorter time for the VWFpp in these animals. VWFpp/VWF:Ag ratios were mildly increased for both mutations, suggesting increased clearance of the mutant proteins, and supporting the results of the recombinant protein infusions (Fig. 4). This is similar to one report of group 2 type 2A VWD plasmas having an average VWFpp/VWF:Ag ratio of 2.7 in humans.[29]. However, the addition of the Adamts13 cDNA to that of the R1597W Vwf cDNA resulted in a highly decreased VWFpp/VWF:Ag ratio (Fig 4B). This could be explained by the fact that in a monomeric form, VWF does not interact correctly with clearance receptors, increasing the circulating time of the aberrant VWF protein. However, this result could also be an artefact of the experiment, since VWF and ADAMTS13 are not normally co-expressed in hepatocytes.
We have finally evaluated the pro-coagulant function of the two mutant proteins in an established in vivo thrombosis model in the hydrodynamic-injection mice (Fig. 6). The timeframe elements of VWF:Ag levels and multimer composition must be examined and considered when evaluating thrombosis and hemostasis evaluations in this animal model. In this study, we focused on the physiologic range of VWF:Ag concentrations, and observed the multimer structures at the time of the study. Wild type cDNA produced stable occlusive thrombi, demonstrating that hydrodynamically produced protein functions relatively normally. R1597W exhibited prolonged times to vessel occlusion when expressed at physiologic concentrations between 0.5 and 1.7 U/ml at the time of the thrombogenic injury. Surprisingly, platelet accumulation with R1597W was not reduced, suggesting that multimer length may not influence initial platelet adhesion but that the presence of normal length multimers may be more important for stable platelet plug development and eventual occlusive clot formation. With Y1605A/M1606A, increased platelet accumulation was observed with a lack of vessel occlusion. This suggests that other factors are influential in this system, such as thrombospondin 1 [30] and leukocyte proteases [31], or possibly altered interactions with ADAMTS13, which prevent the development of occlusive thrombi despite the increase in platelet accumulation. Finally, these studies have been performed in the context of the injured cremaster arteriolar circulation and caution should be exercised in extrapolating the findings to other vascular beds.
Interestingly, even very high plasma levels of the Y1605A/M1606A mutant (>30 U/mL) were not associated with any laboratory evidence for TTP in these mice. There were no changes in the CBC, including platelet counts, and no fragmented red cells were seen in blood smears (data not shown). This finding supports previous evidence suggesting that a combination of pathologic challenges is required in these mice to trigger the TTP phenotype [12,13].
This study demonstrates that changes in the human VWF gene that result in alterations in ADAMTS13-mediated cleavage will also affect the mouse VWF molecule. R1597W was more easily cleaved by mADAMTS13 in a full-length digest assay and exhibited a lower multimeric profile compared to wild type VWF protein when expressed in the VWF knockout mouse. In addition, R1597W was less thrombogenic and had a shorter half-life and a slightly increased VWFpp/VWF:Ag ratio, suggesting that this variant protein is cleared from plasma more rapidly. This mechanism likely contributes to the lower VWF:Ag levels observed in days 14–42 post-hydrodynamic injection. Overall, these findings in the mouse model recapitulate the human type 2A disease phenotype and extend current knowledge of the effects on VWF pathobiology of this mutant.
The Y1605A/M1606A mutant demonstrated a marked decrease in mADAMTS13-mediated cleavage in vitro. In the hydrodynamic injection mice, Y1605A/M1606A shows minor multimer profile differences compared to wild type VWF. However, when full-length mADAMTS13 was co-expressed in the mice, the cleavage site mutant mice showed a significant population of higher molecular weight multimers, although ADAMTS13 cleavage product was observed. In addition, an accelerated clearance phenotype is suggested by the results of the recombinant protein infusion and VWFpp/VWF:Ag ratio studies. This could be due to a conformation change in the A2 domain due to the loss of stabilizing bonds with the alanine substitutions, which could promote the A2 domain to unfold more easily.[7,32] This alteration in A2 domain stability could explain the enhanced platelet accumulation, but loss of occlusive thrombi in the ferric chloride injury model [33] and increased protein clearance [29].
This study establishes that VWF mutations alter ADAMTS13-mediated cleavage in the mouse model. In addition, this leads to significant alterations to the multimer structure of the VWF present in animals receiving co-hydrodynamic injection of full-length normal Adamts13 cDNA. The altered VWF structure is only problematic under a secondary stress, such as the ferric-chloride induced injury model used here. This study illustrates the importance of examining potentially pathogenic sequence changes in a whole animal system, rather than relying on individual in vitro methodologies to explain the complete pathological phenotype.
Acknowledgments
The authors thank Peter Lenting for provision of the mouse Vwf cDNA, Friedrich Scheiflinger (Baxter Innovations GmbH, Vienna, Austria) for the mouse ADAMTS13 expression vector, Luigi Naldini (Vita-Salute San Raffaele University, Milan, Italy) for the synthetic ET promoter, Robert Montgomery (Blood Research Institute, Blood Centre of Wisconsin, Milwaukee, USA), for the development of the VWF propeptide antibodies, and Jeff Mewburn and Jalna Meens (Queen’s Cancer Research Institute) for help with the intravital microscopy studies. We thank Erin Burnett, Kimberly Laverty, and Kate Sponagle for mouse injections and sampling, and Colleen Notley for performing the mVWF115 digestions. This work was supported by funds from the Canadian Institutes of Health Research operating grant MOP-97849(D.L.) and from NIH program project grant HL081588 (D.L. and S.H.). C.M.P. is the recipient of an Ontario Graduate Studentship and Heart and Stroke Foundation of Ontario Master’s Studentship. D.L. holds a Canada Research Chair in Molecular Hemostasis.
Footnotes
C.M.P performed experimental design, data collection and analysis, and manuscript preparation. M.G. performed research and data analysis. A.B. and C.H. performed research. S.H. provided the mouse VWF propeptide antibodies. D. L. performed experimental design, oversight, and edited the manuscript preparation.
References
- 1.Sadler JE. Biochemistry and genetics of von willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 2.Plaimauer B, Zimmermann K, Volkel D, Antoine G, Kerschbaumer R, Jenab P, Furlan M, Gerritsen H, Lammle B, Schwarz HP, Scheiflinger F. Cloning, expression, and functional characterization of the von willebrand factor-cleaving protease (ADAMTS13) Blood. 2002 Nov 15;100(10):3626–32. doi: 10.1182/blood-2002-05-1397. [DOI] [PubMed] [Google Scholar]
- 3.Hassenpflug WA, Budde U, Obser T, Angerhaus D, Drewke E, Schneppenheim S, Schneppenheim R. Impact of mutations in the von willebrand factor A2 domain on ADAMTS13-dependent proteolysis. Blood. 2006 Mar 15;107(6):2339–45. doi: 10.1182/blood-2005-04-1758. [DOI] [PubMed] [Google Scholar]
- 4.Baronciani L, Federici AB, Cozzi G, Canciani MT, Mannucci PM. Von willebrand factor collagen binding assay in von willebrand disease type 2A, 2B, and 2M. J Thromb Haemost. 2006 Sep;4(9):2088–90. doi: 10.1111/j.1538-7836.2006.02069.x. [DOI] [PubMed] [Google Scholar]
- 5.Moake JL. Von willebrand factor, ADAMTS-13, and thrombotic thrombocytopenic purpura. Semin Hematol. 2004 Jan;41(1):4–14. doi: 10.1053/j.seminhematol.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Von willebrand disease international database [homepage on the Internet] Available from: http://www.shef.ac.uk.vwf/
- 7.Zhang Q, Zhou YF, Zhang CZ, Zhang X, Lu C, Springer TA. Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von willebrand factor. Proc Natl Acad Sci U S A. 2009 Jun 9;106(23):9226–31. doi: 10.1073/pnas.0903679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruss CM, Notley CR, Hegadorn CA, O'Brien LA, Lillicrap D. ADAMTS13 cleavage efficiency is altered by mutagenic and to a lesser extent, polymorphic sequence changes in the A1 and A2 domains of von willebrand factor. Br J Haematol. 2008 Nov;143(4):552–8. doi: 10.1111/j.1365-2141.2008.07266.x. [DOI] [PubMed] [Google Scholar]
- 9.Zanardelli S, Crawley JT, Chion CK, Lam JK, Preston RJ, Lane DA. ADAMTS13 substrate recognition of von willebrand factor A2 domain. J Biol Chem. 2006 Jan 20;281(3):1555–63. doi: 10.1074/jbc.M508316200. [DOI] [PubMed] [Google Scholar]
- 10.Lyons SE, Bruck ME, Bowie EJ, Ginsburg D. Impaired intracellular transport produced by a subset of type IIA von willebrand disease mutations. J Biol Chem. 1992 Mar 5;267(7):4424–30. [PubMed] [Google Scholar]
- 11.de Groot R, Lane DA, Crawley JT. The ADAMTS13 metalloprotease domain: Roles of subsites in enzyme activity and specificity. Blood. 2010 Oct 21;116(16):3064–72. doi: 10.1182/blood-2009-12-258780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motto DG, Chauhan AK, Zhu G, Homeister J, Lamb CB, Desch KC, Zhang W, Tsai HM, Wagner DD, Ginsburg D. Shigatoxin triggers thrombotic thrombocytopenic purpura in genetically susceptible ADAMTS13-deficient mice. J Clin Invest. 2005 Oct 1;115(10):2752–61. doi: 10.1172/JCI26007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banno F, Kokame K, Okuda T, Honda S, Miyata S, Kato H, Tomiyama Y, Miyata T. Complete deficiency in ADAMTS13 is prothrombotic, but it alone is not sufficient to cause thrombotic thrombocytopenic purpura. Blood. 2006 Apr 15;107(8):3161–6. doi: 10.1182/blood-2005-07-2765. [DOI] [PubMed] [Google Scholar]
- 14.Marx I, Christophe OD, Lenting PJ, Rupin A, Vallez MO, Verbeuren TJ, Denis CV. Altered thrombus formation in von willebrand factor-deficient mice expressing von willebrand factor variants with defective binding to collagen or GPIIbIIIa. Blood. 2008 Aug 1;112(3):603–9. doi: 10.1182/blood-2008-02-142943. [DOI] [PubMed] [Google Scholar]
- 15.Pruss CM, Golder M, Bryant A, Hegadorn CA, Burnett E, Laverty K, Sponagle K, Dhala A, Notley C, Haberichter S, Lillicrap D. Pathologic mechanisms of type 1 von willebrand disease mutations R1205H and Y1584C through in vitro and in vivo mouse models. Blood. 2011 Apr 21;117(16):4358–66. doi: 10.1182/blood-2010-08-303727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golder M, Pruss CM, Hegadorn C, Mewburn J, Laverty K, Sponagle K, Lillicrap D. Mutation-specific hemostatic variability in mice expressing common type 2B von willebrand disease substitutions. Blood. 2010 Jun 10;115(23):4862–9. doi: 10.1182/blood-2009-11-253120. [DOI] [PubMed] [Google Scholar]
- 17.Rayes J, Hollestelle MJ, Legendre P, Marx I, de Groot PG, Christophe OD, Lenting PJ, Denis CV. Mutation & ADAMTS13-dependent modulation of disease severity in a mouse model for von willebrand disease type 2B. Blood. 2010 Jun 3;115(23):4870–7. doi: 10.1182/blood-2009-11-254193. [DOI] [PubMed] [Google Scholar]
- 18.Kokame K, Matsumoto M, Fujimura Y, Miyata T. VWF73, a region from D1596 to R1668 of von willebrand factor, provides a minimal substrate for ADAMTS-13. Blood. 2004 Jan 15;103(2):607–12. doi: 10.1182/blood-2003-08-2861. [DOI] [PubMed] [Google Scholar]
- 19.Kato S, Matsumoto M, Matsuyama T, Isonishi A, Hiura H, Fujimura Y. Novel monoclonal antibody-based enzyme immunoassay for determining plasma levels of ADAMTS13 activity. Transfusion. 2006 Aug;46(8):1444–52. doi: 10.1111/j.1537-2995.2006.00914.x. [DOI] [PubMed] [Google Scholar]
- 20.Bruno K, Volkel D, Plaimauer B, Antoine G, Pable S, Motto DG, Lemmerhirt HL, Dorner F, Zimmermann K, Scheiflinger F. Cloning, expression and functional characterization of the full-length murine ADAMTS13. J Thromb Haemost. 2005 May;3(5):1064–73. doi: 10.1111/j.1538-7836.2005.01246.x. [DOI] [PubMed] [Google Scholar]
- 21.Denis C, Methia N, Frenette PS, Rayburn H, Ullman-Cullere M, Hynes RO, Wagner DD. A mouse model of severe von willebrand disease: Defects in hemostasis and thrombosis. Proc Natl Acad Sci U S A. 1998 Aug 4;95(16):9524–9. doi: 10.1073/pnas.95.16.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P, Pan W, Rux AH, Sachais BS, Zheng XL. The cooperative activity between the carboxyl-terminal TSP1 repeats and the CUB domains of ADAMTS13 is crucial for recognition of von willebrand factor under flow. Blood. 2007 Sep 15;110(6):1887–94. doi: 10.1182/blood-2007-04-083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varadi K, Rottensteiner H, Vejda S, Weber A, Muchitsch EM, Turecek PL, Ehrlich HJ, Scheiflinger F, Schwarz HP. Species-dependent variability of ADAMTS13-mediated proteolysis of human recombinant von willebrand factor. J Thromb Haemost. 2009 Jul;7(7):1134–42. doi: 10.1111/j.1538-7836.2009.03453.x. [DOI] [PubMed] [Google Scholar]
- 24.Chitta MS, Duhe RJ, Kermode JC. Cloning of the cDNA for murine von willebrand factor and identification of orthologous genes reveals the extent of conservation among diverse species. Platelets. 2007 May;18(3):182–98. doi: 10.1080/09537100600938816. [DOI] [PubMed] [Google Scholar]
- 25.Banno F, Kaminaka K, Soejima K, Kokame K, Miyata T. Identification of strain-specific variants of mouse Adamts13 gene encoding von willebrand factor-cleaving protease. J Biol Chem. 2004 Jul 16;279(29):30896–903. doi: 10.1074/jbc.M314184200. [DOI] [PubMed] [Google Scholar]
- 26.Banno F, Chauhan AK, Kokame K, Yang J, Miyata S, Wagner DD, Miyata T. The distal carboxyl-terminal domains of ADAMTS13 are required for regulation of in vivo thrombus formation. Blood. 2009 May 21;113(21):5323–9. doi: 10.1182/blood-2008-07-169359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sebestyen MG, Budker VG, Budker T, Subbotin VM, Zhang G, Monahan SD, Lewis DL, Wong SC, Hagstrom JE, Wolff JA. Mechanism of plasmid delivery by hydrodynamic tail vein injection. I. hepatocyte uptake of various molecules. J Gene Med. 2006 Jul;8(7):852–73. doi: 10.1002/jgm.921. [DOI] [PubMed] [Google Scholar]
- 28.Lenting PJ, Westein E, Terraube V, Ribba AS, Huizinga EG, Meyer D, de Groot PG, Denis CV. An experimental model to study the in vivo survival of von willebrand factor. basic aspects and application to the R1205H mutation. J Biol Chem. 2004 Mar 26;279(13):12102–9. doi: 10.1074/jbc.M310436200. [DOI] [PubMed] [Google Scholar]
- 29.Haberichter SL, Castaman G, James PD, Christopherson PA, Roheghiero F, Lillicrap D, Montgomery RR. Clearance of von Willebrand Factor (VWF) in type 2 von Willebrand Disease (VWD): the implications of VWF propeptide (VWFpp) and VWF:Ag levels. J Thromb Haemost. 2009 Dec;7(Suppl. 2):10. [Google Scholar]
- 30.Bonnefoy A, Hoylaerts MF. Thrombospondin-1 in von willebrand factor function. Curr Drug Targets. 2008 Oct;9(10):822–32. doi: 10.2174/138945008785909329. [DOI] [PubMed] [Google Scholar]
- 31.Raife TJ, Cao W, Atkinson BS, Bedell B, Montgomery RR, Lentz SR, Johnson GF, Zheng XL. Leukocyte proteases cleave von willebrand factor at or near the ADAMTS13 cleavage site. Blood. 2009 Aug 20;114(8):1666–74. doi: 10.1182/blood-2009-01-195461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von willebrand factor. Science. 2009 Jun 5;324(5932):1330–4. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auton M, Cruz MA, Moake J. Conformational stability and domain unfolding of the von willebrand factor A domains. J Mol Biol. 2007 Feb 23;366(3):986–1000. doi: 10.1016/j.jmb.2006.10.067. [DOI] [PubMed] [Google Scholar]