Abstract
Current therapeutics for hepatitis B virus (HBV) patients such as nucleoside analogs (NAs) are effective; however, new antiviral drugs against HBV are still desired. Since the interaction between the epsilon (ε) sequence of HBV pregenomic RNA and viral polymerase (Pol) is a key step in the HBV replication cycle, we aimed to identify small compounds for its inhibition, and established a pull-down assay system for the detection of ε-RNA-binding-Pol. Screening showed that 5 out of 3,965 compounds inhibited ε-Pol binding, and we identified rosmarinic acid, which exhibited specificity, as a potential antiviral agent. In order to examine the anti-HBV effects of rosmarinic acid, HBV-infected primary human hepatocytes from a humanized mouse liver were treated with rosmarinic acid. The rosmarinic acid treatment decreased HBV components including the amounts of extracellular HBV DNA with negligible cytotoxicity. We also investigated the combined effects of rosmarinic acid and the NA, lamivudine. rosmarinic acid slightly enhanced the anti-HBV activity of lamivudine, suggesting that the HBV replication step targeted by rosmarinic acid is distinct from that of NA. We analyzed an additional 25 rosmarinic acid derivatives, and found that 5 also inhibited ε-Pol. Structural comparisons between these derivatives implied that the “two phenolic hydroxyl groups at both ends” and the “caffeic acid-like structure” of rosmarinic acid are critical for the inhibition of ε-Pol binding. Collectively, our results demonstrate that rosmarinic acid inhibits HBV replication in HBV-infected cells by specifically targeting ε-Pol binding.
Introduction
Hepatitis B virus (HBV) infection is a major health issue worldwide, with approximately 248 million chronically infected individuals (CHB) [1]. Approximately 686,000 HBV-related deaths occur annually [2]. Interferon-α (IFN-α), pegylated IFN-α (PEG-IFN-α), and six nucleos(t)ide analogues (NAs), including lamivudine, entecavir, adefovir dipivoxil, tenofovir disoproxil fumarate, tenofovir alafenamide, and telbivudine, are currently approved for use in the clinical treatment of CHB patients [3]. Treatments with IFN have the potential to achieve HBsAg seroclearance by immunomodulation; however, not all patients respond to IFN. Although NAs more strongly suppress HBV replication than IFN by inhibiting reverse transcription (RT) with less side effects, the discontinuation of NAs may result in the relapse of HBV. Thus, life-long treatments with NAs are required, but may result in the emergence of resistant virus variants [4]. Since current therapeutics for CHB are insufficient, novel anti-HBV drugs are urgently required.
cccDNA serves as a template for all transcripts of HBV; therefore, it represents an attractive target for chronic HBV infection. Studies on zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and RNA-guided clustered regulatory interspaced short palindromic repeats (CRISPR)-Cas endonucleases were performed in order to specifically eliminate hepadnaviral cccDNA [5–10]. The small compounds, CCC-0975 and CCC-0346, were identified as inhibitors of the conversion from rcDNA to cccDNA [11]. However, their use as a clinical trial approach is difficult because the strategy of targeting cccDNA is associated with the serious risk of side effects due to off-targeting. Although many types of inhibitors targeting different replication steps, including AT-61 and AT-130 (pregenomic RNA (pgRNA) encapsidation), Bay 41–4109 (capsid formation), nucleic acid polymers (multistep including secretion), and Myrcludex-B (entry), have been identified [12–17], a novel drug has not yet been applied to clinical anti-HBV therapy.
HBV Pol functions in many essential steps of HBV replication including RT, DNA synthesis, and RNA degradation. In addition to RT, the RNase H activity of Pol is a possible therapeutic target. Tavis and colleagues identified specific inhibitors for the RNase H activity of HBV Pol [18–21]. Besides its enzymatic roles, HBV Pol is crucially involved in pgRNA encapsidation. HBV Pol interacts with the ε sequence of pgRNA, and the ε-Pol interaction is an indispensable step for encapsidation [22]. Previous studies revealed that porphyrin compounds including hemin suppressed the ε-Pol interaction and subsequent protein-priming reaction [23]. Carbonyl J acid derivatives, known as HIV-1 RT inhibitors, have also been identified as ε-Pol binding and protein-priming inhibitors [24].
Since the interaction between ε and Pol is a distinct step from RT, ε-Pol binding inhibitors may be promising agents for combination therapy with NAs. However, large-scale screening to identify ε-Pol binding inhibitors has not yet been conducted. We herein established a screening system to search for ε-Pol binding inhibitors, and performed it using three chemical libraries including United States Food and Drug Administration (FDA)-, European Medicines Agency (EMA)-, and other agency-approved compounds. As a result of screening, rosmarinic acid and quercetin were identified as novel and specific inhibitors of ε-Pol binding. Quercetin has been reported to inhibit HBV replication. Therefore, we analyzed the anti-HBV effects of rosmarinic acid. Furthermore, an analysis of several types of rosmarinic acid derivatives revealed the critical structural features of rosmarinic acid for the inhibition of ε-Pol binding.
Materials and methods
Cell culture and reagents
HEK-293T [25] and HepG2 [26] cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Nacalai Tesque, 08459) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. Hep38.7-Tet cells [27] were maintained in DMEM containing 5% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 400 μg/ml G418, and 400 ng/ml tetracycline. PXB-cells were purchased from PhoenixBio (PPC-P01) and cultured in dHCGM (DMEM containing 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 20 mM HEPES, 44 mM NaHCO3, 15 μg/ml L-proline, 250 ng/ml human recombinant insulin, 50 nM dexamethazone, 5 ng/ml human recombinant epidermal growth factor (EGF), 100 μM ascorbic acid, and 2% DMSO).
Plasmids
p3×Flag CMV/pol (YE), an expression plasmid of 3×FLAG-Pol, was generated by Dr. Sakaguchi and Dr. Chayama (Hiroshima University, Japan). p3×FLAG-ISG20 was constructed as described below. p3×Flag CMV/pol (YE) was digested using HindIII and EcoRI to remove the cDNA of Pol, and subcloned ISG20 cDNA was then inserted into the cleaved vector. p3×FLAG-ISG20 D94G was generated using the KOD -plus- Mutagenesis Kit (TOYOBO, SMK101).
Nucleic acids
ε-biotin RNA, ε RNA, control-biotin RNA, and DNA-biotin were purchased from Japan Bio Services. The sequence of ε RNA was 5'-ucucaUGUUCAUGUCCUACUGUUCAAGCCUCCAAGCUGUGCCUUGGGUGGCUUUGGGGCAUGGACAuugac-3', and that of control RNA was 5'-ucucaUGACGAGGCUGGGUGCUCCUUCUACGUGUUUUCGUUGUAGGUAGGACCCAAAUUGCCGCUCuugac-3' (Length and composition are the same as ε, whereas the sequence is different [randomized]). The 5' terminal “ucuca” and 3' terminal “uugac” were 2' O-Methyl RNA (for stabilization). ε-biotin and control-biotin were biotinylated at the 3' terminus. The sequence of DNA-biotin was 5'-TCTCATGTTCATGTCCTACTGTTCAAGCCTCCAAGCTGTGCCTTGGGTGGCTTTGGGGCATGGACATTGAC-3', and was biotinylated at the 3' terminus.
Chemical libraries and compounds
The Tocriscreen compound library (Tocris Bioscience), Pharmakon1600 drug library (Microsource Discovery Systems), and Prestwick chemical library (Prestwick chemical) were used for screening. The Pharmakon1600 drug library was generously provided by Dr. Koh Takeuchi (Advanced Industrial Science and Technology, Ibaraki, Japan). The Prestwick chemical library was kindly gifted by Dr. Masayuki Shimojima and Dr. Masayuki Saijo (National Institute of Infectious Diseases, Tokyo, Japan). Rosmarinic acid, quercetin, merbromin, hemin, calcomine orange 2RS, and salvianolic acid A were purchased from Sigma-Aldrich (536954, Q4951, M7011, 51280-1G, C9519, and SML0045, respectively). Erythrosine B, and lamivudine were purchased from Tokyo Chemical Industry (T0557 and L0217, respectively). Verteporfin was purchased from Cayman Chemical (17334). Rosmarinic acid derivatives were synthesized as previously described [28].
Compound treatment to cell lines stably expressing HBV
Hep38.7-Tet cells were seeded on collagen-coated 96-well plates at 1.0×104 cells/well. After cell attachment (2 to 3 h), culture medium was replaced with fresh FBS-free medium containing compounds 0, 1, 2, 3 days after seeding. FBS was added to culture medium 2 to 3 h after it had been changed. On day 5, extracellular HBV DNA was extracted from the culture supernatant and quantified by qPCR, as described below, and cells were then subjected to the WST-1 cell proliferation assay (Takara, MK400) following the manufacturer’s instructions. CC50 in HepG2 cells was calculated from results at least 7 kinds of concentration of compound (Table 1).
Table 1. Citotoxicity of the compounds.
| Compound | CC50 in HepG2 (μM) |
|---|---|
| Rosmarinic acid | 116±34 |
| Quercetin | >500 |
| Erythrosine B | 115±42 |
| Merbromin | 156±43 |
| Verteporfin | 1.11±0.35 |
| Hemin | 111±23 |
| Salvianolic acid A | 86±4 |
| Compound 2 | 258±14 |
| Compound 12c | 239±89 |
| Compound 13c | 229±10 |
| Compound 21a | 356±105 |
Compound treatment to primary human hepatocytes infected with HBV
PXB-cells were infected at 5 Geq/cell using dHCGM containing serum from HBV-infected chimeric mice with humanized livers (PhoenixBio, PPC-BC), and 4% polyethylene glycol 8,000. Culture medium was replaced with fresh dHCGM containing compounds on days 1, 2, and 7 post-infection. On days 7 or 12, extracellular HBV DNA was extracted from the culture supernatant and quantified by qPCR, HBV RNA was extracted from cells and quantified by RT-qPCR, and SHBs were measured by ELISA as described below.
Quantitative real-time PCR (qPCR)
Extracellular HBV DNA was extracted using the SMITEST EX-R+D KIT (Medical and Biological Laboratories, GSJ0201) or Geno Plus Genomic DNA Extraction Miniprep System (Viogene, GG2002). HBV DNA levels were monitored with the StepOnePlus Real Time PCR System using the Fast SYBR PCR Master Mix (Applied Biosystems—Thermo Fisher Scientific), and the following primer set: qPCR primers for HBV DNA (Forward 5'-TTCACCTCACCATACAGCACTC-3', Reverse 5'- ATAGGGGCATTTGGTGGTCTG-3'). The copy number/μl of HBV DNA was measured using the HBV DNA fragment, which was amplified by the following primers: PCR primers for HBV DNA fragment amplification (Forward 5'-TTCACCTCACCATACAGCACTC-3', Reverse 5'- ATAGGGGCATTTGGTGGTCTG-3').
Quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA was isolated using TRIzol reagent (Invitrogen—Thermo Fisher Scientific, 15596018) with DNase I (Roche, 4716728), and subjected to RT using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems—Thermo Fisher Scientific, 4368813). mRNA levels were quantified by the StepOnePlus Real Time PCR System using Fast SYBR PCR Master Mix, and the following primer set: qRT-PCR primers for HBV RNA (Forward 5'-TTCACCTCACCATACAGCACTC-3', Reverse 5'- ATAGGGGCATTTGGTGGTCTG-3') and GAPDH (Forward 5'- ACTGCCAACGTGTCAGTGGT-3', Reverse 5'- TTACTCCTTGGAGGCCATGT-3').
Enzyme-linked immunosorbent assay (ELISA)
Culture supernatants were analyzed using the HBs S Antigen Quantitative ELISA Kit, Rapid-II (Beacle, Inc., BC013).
Electrophoresis mobility shift assay (EMSA)
EMSA was performed as described previously [29]. Briefly, a total of 30 pmol of the recombinant RIG-I protein was mixed with 5 pmol of synthetic dsRNA (25/25c) [29] and in a reaction mixture (20 mM Tris-HCl (pH 8.0), 1.5 mM MgCl2, and 1.5 mM DTT) in the presence of compounds. After an incubation at room temperature for 15 min, the reaction mixture was applied to a 15% acrylamide gel (TBE buffer) and dsRNA was detected by EtBr staining and RIG-I by Coomassie Brilliant Blue staining.
Helicase assay
The RIG-I helicase assay was performed as described previously [29].
Pull-down assay
HEK-293T cells were transfected with p3×Flag CMV/pol (YE) or p3×FLAG-ISG20 D94G. At 48 h post-transfection, cells were harvested and stored at -80°C. The cell pellet was lysed in lysis buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 1 mM DTT, 2 μg/ml leupeptin, 1 mM PMSF, 1 mM vanadate, and 20 units of an RNase inhibitor) and centrifuged. The cleared lysate was incubated in the presence of compounds on ice for 20 min, mixed with RNAs (ε-biotin, ε, or control-biotin) and equilibrated with streptavidin sepharose (GE Healthcare, 17511301, GE Healthcare, Buckinghamshire, UK), incubated at 4°C for 2 h with rotation, collected by centrifugation, washed three times using lysis buffer, resuspended in SDS sample buffer (125 mM Tris-HCl (pH 6.8), 4% SDS, 20% glycerol, 0.01% BPB, and 10% 2-mercaptoethanol), incubated at 95°C for 3 min, and then analyzed by SDS-PAGE using a 7.5% acrylamide gel and immunoblotting using an anti-FLAG-Peroxidase (HRP) antibody (Sigma-Aldrich, A8592). The biotinylated DNA pull-down assay was performed as follows. One microgram of DNA-biotin was incubated in the presence of compounds in buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1% NP-40), equilibrated with streptavidin sepharose at 4°C for 2 h with rotation, collected by centrifugation, washed three times, and DNA was recovered by extraction with TRIzol reagent. Recovered DNA was analyzed by 15% acrylamide gel electrophoresis, followed by staining with EtBr.
Results
Screening of ε-Pol inhibitors by a pull-down assay
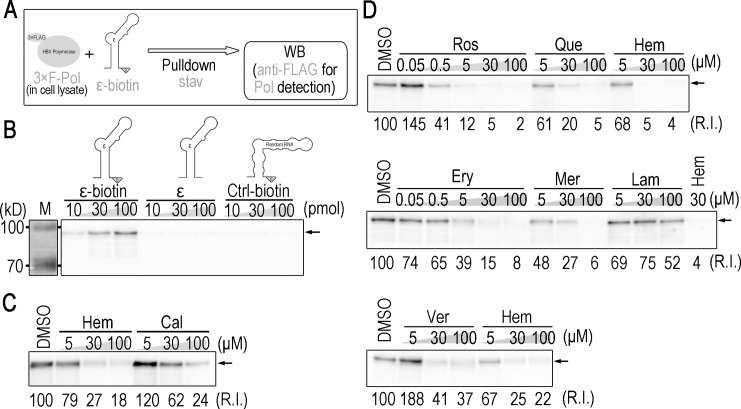
In order to identify novel inhibitors of ε-Pol binding, we established an in vitro screening system based on a pull-down assay, as shown in Fig 1A. Briefly, chemically synthesized ε-biotin RNA was mixed with chemicals in the lysates of human cell lines expressing 3×FLAG-tagged-Pol, pulled down by streptavidin beads, and the ε-binding of 3×FLAG-Pol was then detected by Western blotting. As expected, 3×FLAG-Pol was pulled down with ε-biotin, but not with non-biotinylated ε or biotinylated RNA with an unrelated sequence (Fig 1B). Conditions were optimized further by using the previously reported inhibitors, hemin and calcomine orange 2RS [23, 24] (Fig 1C), and we screened for novel inhibitors using chemical libraries.
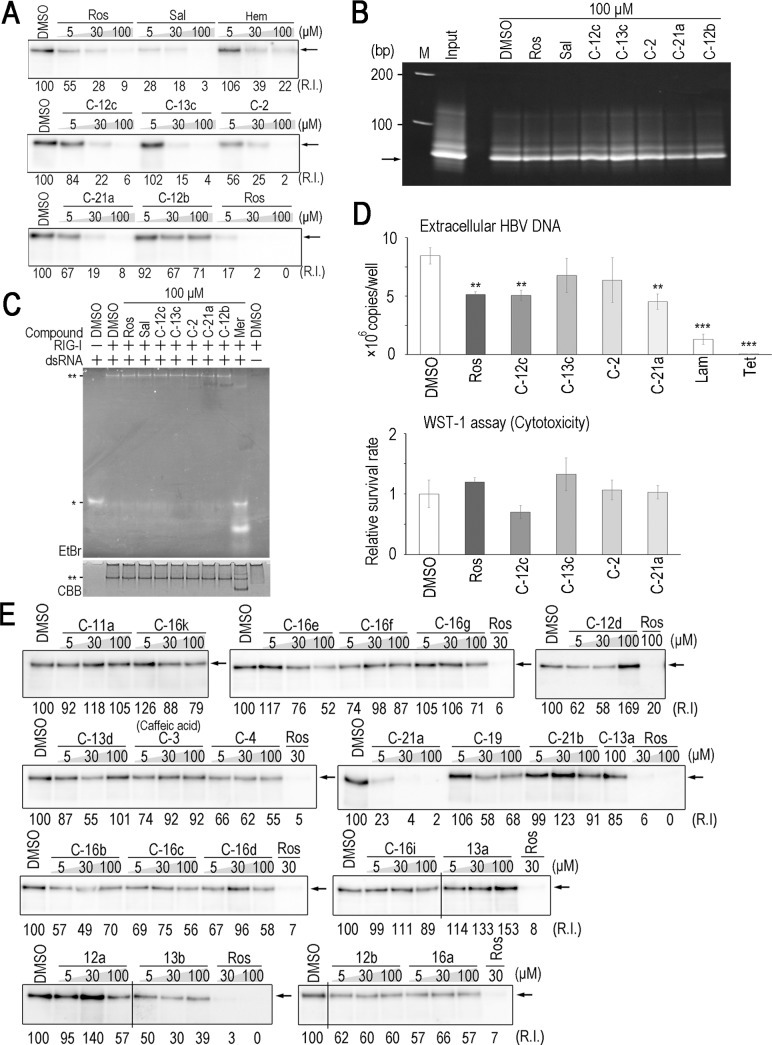
Fig 1. Five ε-Pol binding inhibitors selected by pull-down assay-based screening.
(A) Scheme of the detection of ε-Pol binding by pull-down assays. The lysate of HEK-293T cells expressing 3×FLAG-Pol was incubated with ε-biotin, and pulled down using streptavidin sepharose. Precipitated 3×FLAG-Pol was detected by a Western blot analysis using an anti-FLAG antibody. (B) A Western blot analysis for Pol pulled-down by the indicated RNAs (ε RNA with biotin, ε RNA without biotin, and control RNA with biotin). (C and D) A Western blot analysis for Pol pulled-down by 10 pmol ε-biotin in the presence of the indicated compounds. Lamivudine was used as a negative control. Arrows: bands detected at a position of the estimated mass of full-length 3×FLAG-Pol.
Primary screening was performed using compounds in a 10-drug mix, and candidate inhibitors were eventually identified by secondary screening using a single compound in the candidate 10-drug mix (S1A and S1B Fig). As a result, 5 out of 3,965 compounds were identified as inhibitors of ε-Pol binding. All 5 compounds: rosmarinic acid, quercetin, erythrosine B, merbromin, and verteporfin, exhibited inhibitory activities in dose-dependent manners (Fig 1D). Of note, verteporfin is a porphyrin, similar to hemin.
Rosmarinic acid and quercetin are specific inhibitors of ε-Pol
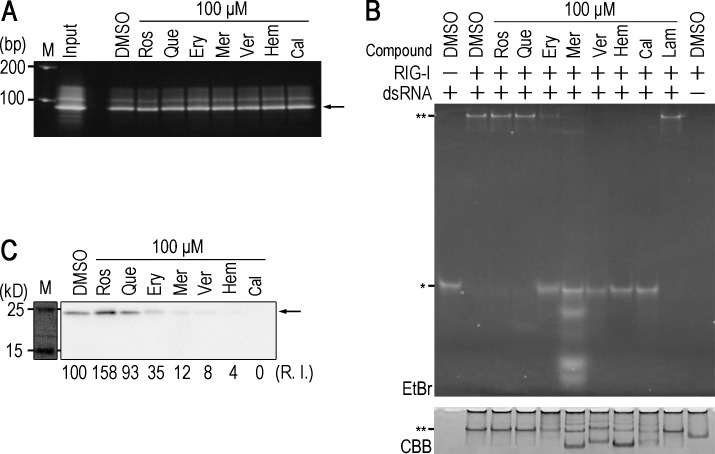
We examined the specificities of the 5 compounds for the inhibition of ε-Pol. In order to exclude the possibility that these compounds inhibit biotin-avidin binding, biotinylated DNA was pulled down by streptavidin sepharose in the presence or absence of the compounds. None of the 5 compounds affected binding between DNA-biotin and streptavidin sepharose (Fig 2A), suggesting that these compounds target the protein-RNA interaction. A previous study reported that RIG-I recognizes dsRNAs, such as the ε sequence in HBV pgRNA, and subsequently exhibits anti-HBV activity [30]. In order to examine whether the compounds identified affect RIG-I-dsRNA binding activity, we conducted an electrophoresis mobility shift assay (EMSA) to investigate the interaction between dsRNA and RIG-I in the presence or absence of the compounds. Rosmarinic acid and quercetin did not affect dsRNA-RIG-I binding, whereas the other compounds, erythrosin B, merbromin, and verteporfin, as well as the previously reported chemicals, hemin and calcomine orange 2RS, inhibited dsRNA-RIG-I binding (Fig 2B). We also tested whether the 5 compounds affect RIG-I helicase enzymatic activity, and found that only rosmarinic acid and quercetin did not affect the helicase activity of RIG-I, suggesting that RIG-I signaling is not influenced by treatments with rosmarinic acid and quercetin (S2A Fig). In order to further validate specificity, another RNP complex was tested. Liu and colleagues recently reported that ISG20 interacted with ε in an enzymatic activity (exonuclease)-independent manner [31]. 3×FLAG-tagged ISG20 D94G, an exonuclease inactive mutant, was pulled down with ε-biotin in the presence or absence of the compounds. Rosmarinic acid and quercetin did not affect ε-ISG20 binding, whereas erythrosin B, merbromin, verteporfin, hemin, and calcomine Orange 2RS exerted inhibitory effects (Fig 2C). These results suggest that rosmarinic acid and quercetin specifically inhibit the ε-Pol interaction.
Fig 2. Rosmarinic acid and quercetin are specific inhibitors of ε-Pol binding.
(A) Seventy-one-base ssDNA-biotin was pulled-down in the presence of the indicated compounds, and detected by EtBr staining. Arrow: 71-base ssDNA. (B) dsRNA-RIG-I EMSA in the presence of the indicated compounds. *: monomeric dsRNA, **: dsRNA-RIG-I complex. (C) A Western blot analysis for ISG20 D94G pulled-down by ε-biotin in the presence of the indicated compounds. Arrow: 3×ISG20 D94G.
Rosmarinic acid suppresses HBV replication
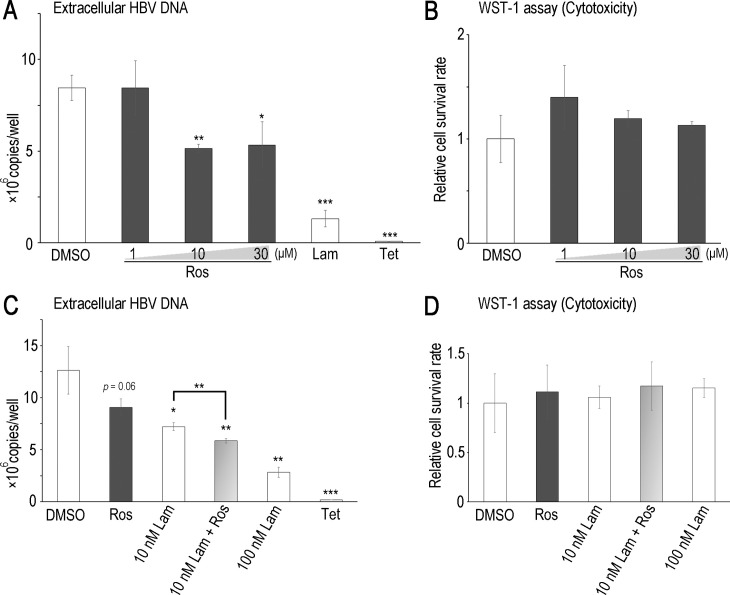
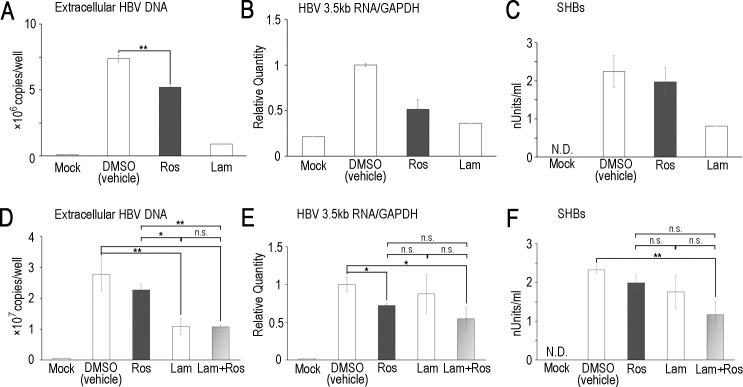
Quercetin was previously reported to inhibit HBV replication in HepG2.2.15 cells [32], a HepG2-derived cell line stably expressing HBV. Thus, we focused on the anti-HBV activity of rosmarinic acid using cell lines stably expressing HBV. No obvious cytotoxicity of rosmarinic acid was observed up to 30 μM in HepG2 cells and Hep38.7-Tet cells, a tetracycline inducible HBV-expressing cell line [27] (Fig 3B and 3D). The cytotoxicity of rosmarinic acid in HepG2 cells was also evaluated (Table 1). Hep38.7-Tet cells were treated with rosmarinic acid, and the amount of HBV DNA secreted into the culture medium was quantified. The rosmarinic acid treatment significantly decreased extracellular HBV DNA with rough estimation of EC50 30 μM (Fig 3A). We then performed a combined treatment with rosmarinic acid and lamivudine. Although a low concentration of lamivudine (10 nM) exerted weak anti-HBV effects, the combined treatment enhanced inhibition (Fig 3C). PXB-cells, primary human hepatocytes isolated from an immunodeficient mouse with a humanized liver, were subjected to HBV infection and the effects of rosmarinic acid on its replication were examined. Rosmarinic acid showed no toxicity up to 100 μM (S3 Fig). Extracellular HBV DNA levels was significantly reduced and intracellular HBV RNA and extracellular HBsAg levels were slightly reduced by rosmarinic acid, suggesting that rosmarinic acid suppresses HBV replication in infected cells (Fig 4A–4C). In PXB-cells, the combined treatment with rosmarinic acid and lamivudine did not further enhance inhibition (Fig 4D–4F). Collectively, these results suggest that the rosmarinic acid treatment suppresses HBV replication in infected cells. Of note, HBV-infected PXB-cells were also treated with quercetin, and its inhibitory activity on HBV replication was confirmed (S4A–S4C Fig). We analyzed the rosmarinic acid derivatives listed in Table 2. Salvianolic acid A, Compound 2, 12c, 13c, and 21a [28] exhibited inhibitory activities against ε-Pol binding in dose-dependent manners (Fig 5A). Similar to rosmarinic acid, these 5 derivatives did not affect biotin-avidin binding, dsRNA-RIG-I binding, or RIG-I helicase activity (Fig 5B, 5C and S2B Fig). Furthermore, these derivatives inhibited HBV DNA production at similar levels to that of rosmarinic acid with low cytotoxicity (Fig 5D and Table 1). On the other hand, other derivatives did not exhibit inhibitory activities (Fig 5E).
Fig 3. Rosmarinic acid inhibits HBV DNA production in cell lines stably expressing HBV.
Hep38.7-Tet cells were treated with rosmarinic acid at the indicated concentrations, 100 nM lamivudine, or 400 ng/ml tetracycline (A and B), or with 30 μM rosmarinic acid and/or 10 nM lamivudine, 100 nM Lam, or 400 ng/ml tetracycline (C and D). Five days after the induction of HBV expression, extracellular HBV DNA was quantified by qPCR (A and C), and cell viability was measured using WST-1 reagent (B and D). Data are from one representative of at least two independent experiments; the means and S.D. of triplicate experiments are shown (*: p < 0.05, **: p < 0.01, ***: p < 0.001).
Fig 4. Rosmarinic acid suppresses HBV replication in HBV-infected primary human hepatocytes.
(A-C) PXB-cells were infected with HBV, and treated with 30 μM rosmarinic acid or 500 nM lamivudine. Seven to 12 days post-infection, extracellular HBV DNA was quantified by qPCR, intracellular HBV 3.5 kb RNA was quantified by RT-qPCR, and SHBs were measured by ELISA. (D-F) PXB-cells were infected with HBV, and were treated with 30 μM rosmarinic acid and/or 20 nM lamivudine. Extracellular HBV DNA, intracellular HBV 3.5-kb RNA, and SHBs were measured as in (A-C). Data are from one representative of at least two independent experiments; the means and S.D. of duplicate or triplicate experiments are shown (* p < 0.05, ** p < 0.01). N.D.: not detected, n.s.: not significant.
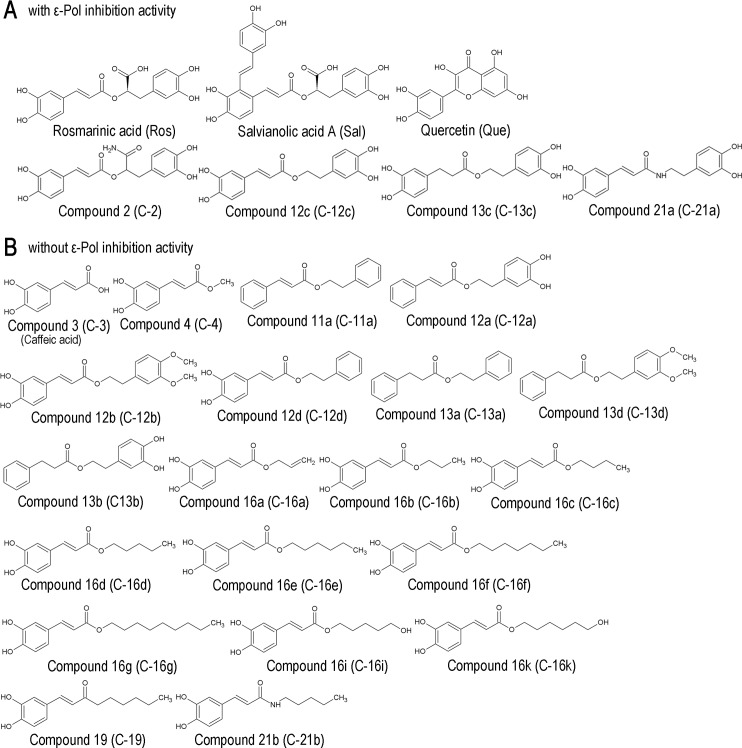
Table 2. Rosmarinic acid derivatives.
| Compound 2 | (R)-2-(3,4-Dihydroxyphenyl)-1-carbamoylethyl 3-(3,4-dihydroxyphenyl)acrylate |
| Compound 3 | Caffeic acid |
| Compound 4 | Methyl caffeate |
| Compound 11a | 2-Phenylethyl cinnamate |
| Compound 12a | 2-(3,4-Dihydroxyphenyl)ethyl cinnamate |
| Compound 12b | 2-(3,4-Dimethoxyphenyl)ethyl caffeate |
| Compound 12c | 2-(3,4-Dihydroxyphenyl)ethyl caffeate |
| Compound 12d | Phenylethyl caffeate |
| Compound 13a | Phenylethyl 3-phenylpropanoate |
| Compound 13b | 2-(3,4-Dihydroxyphenyl)ethyl 3-phenylpropanoate |
| Compound 13c | 2-(3,4-Dihydroxyphenyl)ethyl 3-(3,4-dihydroxyphenyl)propanoate |
| Compound 13d | 2-(3,4-Dimethoxyphenyl)ethyl 3-phenylpropanoate |
| Compound 16a | 2-propenyl caffeate |
| Compound 16b | Propyl caffeate |
| Compound 16c | Butyl caffeate |
| Compound 16d | Pentyl caffeate |
| Compound 16e | Hexyl caffeate |
| Compound 16f | Heptyl caffeate |
| Compound 16g | Nonyl caffeate |
| Compound 16i | 5-Hydroxypentyl caffeate |
| Compound 16k | Hydroxyhexy caffeate |
| Compound 19 | 1-(3’,4’-Dihydroxyphenyl)non-1-en-3-one |
| Compound 21a | N-caffeoyldopamine |
| Compound 21b | Pentyl 1-(3’,4’-dihydroxyphenyl)propen amide |
Fig 5. Rosmarinic acid derivatives also inhibit ε-Pol binding.
(A) Western blot analysis for Pol pulled-down by ε-biotin in the presence of the indicated compounds. Arrows: 3×FLAG-Pol. (B) ssDNA-biotin was pulled-down in the presence of the indicated compounds, and detected by EtBr staining. Arrow: 71-base ssDNA. (C) dsRNA-RIG-I EMSA in the presence of the indicated compounds (100 μM). *: monomeric dsRNA, **: the dsRNA-RIG-I complex. (D) Hep38.7-Tet cells were treated with 10 μM rosmarinic acid, compounds 12c, 13c, 2, and 21a, 100 nM lamivudine, or 400 ng/ml tetracycline. Extracellular HBV DNA and cell viability were measured as in (Fig 3A and 3B). Data are from one representative of at least two independent experiments; the means and S.D. of triplicate experiments are shown (* p < 0.05, ** p < 0.01, *** p < 0.001). (E) A Western blot analysis for Pol pulled-down by ε-biotin in the presence of the indicated compounds. Arrows: 3×FLAG-Pol. Spliced images are indicated with a dividing line.
Discussion
In the present study, we established a pull-down assay to screen for inhibitors of HBV ε-Pol binding. 3×FLAG-tagged full-length Pol was expressed in human cell lines, and cell lysates were used in the assay. It was not necessary to include an exogenous chaperone protein to detect the ε-Pol interaction. The ε-Pol interaction was detected in liver-derived (HepG2) and non-liver (HEK-293T) cells. Due to its high transfection efficiency, we used HEK-293T cells for screening. In order to test a larger number of chemicals, we mixed 10 compounds in primary screening. Although mixed chemicals may exhibit unexpected interference from each other, we identified 5 out of 3,965 chemicals with inhibitory activities. Rosmarinic acid and quercetin appeared to specifically inhibit the ε-Pol interaction.
In Fig 4E, PXB cells treated with rosmarinic acid had significantly lower intracellular 3.5kb HBV RNA levels than DMSO-treated cells. Also, combined treatment with rosmarinic acid and lamivudine had lower 3.5kb HBV RNA level. This observation was unexpected if rosmarinic acid solely targets ε-Pol interaction. However, HBV progenies produced from PXB cells are known to have the potential to re-infect the cells. When rosmarinic acid specifically blocks the epsilon-polymerase binding, the treatment is estimated to reduce the production level of virus progenies and the rate of the secondary infection to PXB cells as well as to inhibit cccDNA synthesis from the pre-genome 3.5 kb HBV RNA. In such a context, we assume that HBV RNA levels in PXB cells were decreased by the treatment of rosmarinic acid.
Rosmarinic acid and quercetin are structurally related and both have two dihydoxybenzene moieties (Fig 6A). We tested the derivatives of rosmarinic acid and found that some did not exhibit inhibitory activities against the ε-Pol interaction. The examination with rosmarinic acid and its derivatives revealed a relationship between its chemical structure and ε-Pol inhibition (Figs 5 and 6). Compounds with inhibitory activities commonly had two dihydoxybenzene moieties (Fig 6A); however, caffeic acid and its derivatives (C-4, -16a-k, -19, and -21b) did not exhibit inhibitory activity. Comparisons between C-12c and C-11a revealed that the two phenolic hydroxyl groups in the catechol structure were critical and the presence of 4 complete hydroxyl groups was required for inhibitory activity (compare C-12c with C-12a and C12d). Similarly, modifications to the hydroxyl groups into–OCH3 groups (hydroxyl group masked with CH3) resulted in the loss of inhibitory activity (compare C-12c with C-12b). On the other hand, the linker structure connecting the two catechol structures appeared to be flexible (compare C-12, C-2, C-13c, and C-21a). These results suggest that the 4 hydroxyl groups in the two catechol structures at both side chain ends are critical “active domains” for recognizing Pol. Whereas the linker structure was less critical for the specific interaction but the linker may determine critical distance between the two active domains.
Fig 6. Chemical structures.
Chemical structures of compounds with ε-Pol inhibitory activity (A), and without ε-Pol inhibitory activity (B).
RIG-I is a viral RNA sensor for eliciting several antiviral responses [33, 34]. Previous studies reported that innate immune signaling is important for anti-HBV responses [26, 30]. ISG20 was shown to inhibit HBV replication in nuclease-dependent and -independent manners [31, 35]. Since rosmarinic acid and quercetin inhibit ε-Pol binding without affecting dsRNA-RIG-I binding, RIG-I helicase activity, or ε-ISG20 binding (Figs 1D, 2B and 2C and S2A Fig), they presumably do not interfere with the host antiviral immune response.
Despite ε-Pol binding being strongly abolished by the rosmarinic acid treatment in vitro, the suppression of HBV replication in cells by rosmarinic acid was less efficient than the nucleotide analogue, lamivudine. One of the reasons for insufficient inhibition is permeability. Another potential reason is chemical stability in cells. These limitations may be overcome in future studies. Rosmarinic acid is a natural compound (abundant in Lamiaceae herbs including spearmint, sage, peppermint, and perilla) and is utilized as a dietary supplement as well as Chinese herbal medicine. To date, no detrimental effects on humans have been reported, suggesting lower toxicity in vivo. Therefore, if we optimize drug administration, we may be able to evaluate the anti-HBV effects of rosmarinic acid in vivo.
Supporting information
(A) A Western blot analysis for Pol pulled-down by ε-biotin in the presence of a chemical mix containing 10 kinds of compounds. The final concentration of each compound was 30 μM. A total of 3,965 compounds were screened. (B) Candidate mixes were divided into single compounds, and analyzed as in (A). The final concentration of the compounds was 30 μM. Arrows: 3×FLAG-Pol.
(TIF)
The RIG-I helicase assay was conducted in the presence of the indicated compounds. #: monomeric ssRNA, ##: annealed dsRNA, (n.s.): non-specific band.
(TIF)
PXB cells were infected with HBV, and treated with Rosmarinic acid at indicated concentrations. On day 12, cells were subjected to WST-1 cell proliferation assay. Data are from one representative of at least two independent experiments; means and S.D. of duplicate experiments are shown.
(TIF)
PXB cells were infected with HBV, and treated with 30 μM Quercetin. Extracellular HBV DNA, intracellular HBV 3.5 kb RNA, and SHBs were measured as in Fig 4A–4C. Data are from one representative of at least three independent experiments; means and S.D. of duplicate experiments are shown (* p < 0.05).
(TIF)
Uncropped and unadjusted gels and Western blots.
(TIF)
Acknowledgments
We thank Dr. Koh Takeuchi (Advanced Industrial Science and Technology, Ibaraki, Japan), and Drs. Masayuki Shimojima and Masayuki Saijo (National Institute of Infectious Diseases, Tokyo, Japan).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by The Ministry of Health, Labour and Welfare/Japan Agency for Medical Research and Development (AMED) for research on the innovative development and the practical application of new drugs for hepatitis B, Grant number: 17fk0310107h0001 to TF and 17fk0310109h0001 to HK (URL: https://www.amed.go.jp/). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386(10003):1546–55. Epub 2015/08/02. doi: 10.1016/S0140-6736(15)61412-X [pii]. . [DOI] [PubMed] [Google Scholar]
- 2.Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385(9963):117–71. Epub 2014/12/23. doi: 10.1016/S0140-6736(14)61682-2 [pii]. ; PubMed Central PMCID: PMC4340604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarin SK, Kumar M, Lau GK, Abbas Z, Chan HL, Chen CJ, et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int. 2016;10(1):1–98. Epub 2015/11/14. doi: 10.1007/s12072-015-9675-4 [pii]. ; PubMed Central PMCID: PMC4722087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137(5):1593–608 e1-2. Epub 2009/09/10. doi: 10.1053/j.gastro.2009.08.063 S0016-5085(09)01556-X [pii]. . [DOI] [PubMed] [Google Scholar]
- 5.Cradick TJ, Keck K, Bradshaw S, Jamieson AC, McCaffrey AP. Zinc-finger nucleases as a novel therapeutic strategy for targeting hepatitis B virus DNAs. Mol Ther. 2010;18(5):947–54. Epub 2010/02/18. doi: 10.1038/mt.2010.20 [pii]. ; PubMed Central PMCID: PMC2890117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom K, Ely A, Mussolino C, Cathomen T, Arbuthnot P. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Mol Ther. 2013;21(10):1889–97. Epub 2013/07/26. doi: 10.1038/mt.2013.170 S1525-0016(16)32255-9 [pii]. ; PubMed Central PMCID: PMC3808145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Zhang W, Lin J, Wang F, Wu M, Chen C, et al. An efficient antiviral strategy for targeting hepatitis B virus genome using transcription activator-like effector nucleases. Mol Ther. 2014;22(2):303–11. Epub 2013/09/13. doi: 10.1038/mt.2013.212 [pii]. ; PubMed Central PMCID: PMC3916035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3:e216 Epub 2014/12/17. doi: 10.1038/mtna.2014.68 [pii]. ; PubMed Central PMCID: PMC4272409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833 Epub 2015/06/04. doi: 10.1038/srep10833 [pii]. ; PubMed Central PMCID: PMC4649911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids. 2014;3:e186 Epub 2014/08/20. doi: 10.1038/mtna.2014.38 [pii]. ; PubMed Central PMCID: PMC4221598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai D, Mills C, Yu W, Yan R, Aldrich CE, Saputelli JR, et al. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrob Agents Chemother. 2012;56(8):4277–88. Epub 2012/05/31. doi: 10.1128/AAC.00473-12 [pii]. ; PubMed Central PMCID: PMC3421587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaney WEt, Edwards R, Colledge D, Shaw T, Furman P, Painter G, et al. Phenylpropenamide derivatives AT-61 and AT-130 inhibit replication of wild-type and lamivudine-resistant strains of hepatitis B virus in vitro. Antimicrob Agents Chemother. 2002;46(9):3057–60. Epub 2002/08/17. doi: 10.1128/AAC.46.9.3057-3060.2002 ; PubMed Central PMCID: PMC127422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katen SP, Chirapu SR, Finn MG, Zlotnick A. Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem Biol. 2010;5(12):1125–36. Epub 2010/09/18. doi: 10.1021/cb100275b ; PubMed Central PMCID: PMC3003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, et al. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299(5608):893–6. Epub 2003/02/08. doi: 10.1126/science.1077215 299/5608/893 [pii]. . [DOI] [PubMed] [Google Scholar]
- 15.Stray SJ, Zlotnick A. BAY 41–4109 has multiple effects on Hepatitis B virus capsid assembly. J Mol Recognit. 2006;19(6):542–8. Epub 2006/09/29. doi: 10.1002/jmr.801 . [DOI] [PubMed] [Google Scholar]
- 16.Volz T, Allweiss L, Ben MM, Warlich M, Lohse AW, Pollok JM, et al. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J Hepatol. 2013;58(5):861–7. Epub 2012/12/19. doi: 10.1016/j.jhep.2012.12.008 S0168-8278(12)00956-7 [pii]. . [DOI] [PubMed] [Google Scholar]
- 17.Schoneweis K, Motter N, Roppert PL, Lu M, Wang B, Roehl I, et al. Activity of nucleic acid polymers in rodent models of HBV infection. Antiviral Res. 2018;149:26–33. Epub 2017/11/12. doi: S0166-3542(17)30596-X [pii] doi: 10.1016/j.antiviral.2017.10.022 ; PubMed Central PMCID: PMC5743593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tavis JE, Cheng X, Hu Y, Totten M, Cao F, Michailidis E, et al. The hepatitis B virus ribonuclease H is sensitive to inhibitors of the human immunodeficiency virus ribonuclease H and integrase enzymes. PLoS Pathog. 2013;9(1):e1003125 Epub 2013/01/26. doi: 10.1371/journal.ppat.1003125 PPATHOGENS-D-12-02267 [pii]. ; PubMed Central PMCID: PMC3551811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Cheng X, Cao F, Huang A, Tavis JE. beta-Thujaplicinol inhibits hepatitis B virus replication by blocking the viral ribonuclease H activity. Antiviral Res. 2013;99(3):221–9. Epub 2013/06/26. doi: 10.1016/j.antiviral.2013.06.007 S0166-3542(13)00164-2 [pii]. . [DOI] [PubMed] [Google Scholar]
- 20.Cai CW, Lomonosova E, Moran EA, Cheng X, Patel KB, Bailly F, et al. Hepatitis B virus replication is blocked by a 2-hydroxyisoquinoline-1,3(2H,4H)-dione (HID) inhibitor of the viral ribonuclease H activity. Antiviral Res. 2014;108:48–55. Epub 2014/05/27. doi: 10.1016/j.antiviral.2014.05.007 S0166-3542(14)00136-3 [pii]. ; PubMed Central PMCID: PMC4101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu G, Lomonosova E, Cheng X, Moran EA, Meyers MJ, Le Grice SF, et al. Hydroxylated tropolones inhibit hepatitis B virus replication by blocking viral ribonuclease H activity. Antimicrob Agents Chemother. 2015;59(2):1070–9. Epub 2014/12/03. doi: 10.1128/AAC.04617-14 [pii]. ; PubMed Central PMCID: PMC4335860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11(9):3413–20. Epub 1992/09/01. ; PubMed Central PMCID: PMC556876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin L, Hu J. Inhibition of hepadnavirus reverse transcriptase-epsilon RNA interaction by porphyrin compounds. J Virol. 2008;82(5):2305–12. Epub 2007/12/21. doi: JVI.02147-07 [pii] doi: 10.1128/JVI.02147-07 ; PubMed Central PMCID: PMC2258913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang YX, Wen YM, Nassal M. Carbonyl J acid derivatives block protein priming of hepadnaviral P protein and DNA-dependent DNA synthesis activity of hepadnaviral nucleocapsids. J Virol. 2012;86(18):10079–92. Epub 2012/07/13. doi: 10.1128/JVI.00816-12 JVI.00816-12 [pii]. ; PubMed Central PMCID: PMC3446557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe N, Iwamura T, Shinoda T, Fujita T. Regulation of NFKB1 proteins by the candidate oncoprotein BCL-3: generation of NF-kappaB homodimers from the cytoplasmic pool of p50-p105 and nuclear translocation. EMBO J. 1997;16(12):3609–20. Epub 1997/06/01. doi: 10.1093/emboj/16.12.3609 ; PubMed Central PMCID: PMC1169985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao WL, Ikeda S, Tsukamoto Y, Shindo K, Otakaki Y, Qin M, et al. Establishment of a human hepatocellular cell line capable of maintaining long-term replication of hepatitis B virus. Int Immunol. 2017. Epub 2017/03/25. doi: 10.1093/intimm/dxx0123066062 [pii]. . [DOI] [PubMed] [Google Scholar]
- 27.Ogura N, Watashi K, Noguchi T, Wakita T. Formation of covalently closed circular DNA in Hep38.7-Tet cells, a tetracycline inducible hepatitis B virus expression cell line. Biochem Biophys Res Commun. 2014;452(3):315–21. Epub 2014/08/26. doi: 10.1016/j.bbrc.2014.08.029 S0006-291X(14)01444-2 [pii]. . [DOI] [PubMed] [Google Scholar]
- 28.Taguchi R, Hatayama K, Takahashi T, Hayashi T, Sato Y, Sato D, et al. Structure-activity relations of rosmarinic acid derivatives for the amyloid beta aggregation inhibition and antioxidant properties. Eur J Med Chem. 2017;138:1066–75. Epub 2017/08/02. doi: S0223-5234(17)30548-2 [pii] doi: 10.1016/j.ejmech.2017.07.026 . [DOI] [PubMed] [Google Scholar]
- 29.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29(4):428–40. Epub 2008/02/05. doi: 10.1016/j.molcel.2007.11.028 S1097-2765(07)00859-3 [pii]. . [DOI] [PubMed] [Google Scholar]
- 30.Sato S, Li K, Kameyama T, Hayashi T, Ishida Y, Murakami S, et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity. 2015;42(1):123–32. Epub 2015/01/06. doi: 10.1016/j.immuni.2014.12.016 S1074-7613(14)00483-X [pii]. . [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Nie H, Mao R, Mitra B, Cai D, Yan R, et al. Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS Pathog. 2017;13(4):e1006296 Epub 2017/04/12. doi: 10.1371/journal.ppat.1006296 PPATHOGENS-D-16-02833 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Z, Sun G, Guo W, Huang Y, Sun W, Zhao F, et al. Inhibition of hepatitis B virus replication by quercetin in human hepatoma cell lines. Virol Sin. 2015;30(4):261–8. Epub 2015/08/14. doi: 10.1007/s12250-015-3584-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–7. Epub 2004/06/23. doi: 10.1038/ni1087 [pii]. . [DOI] [PubMed] [Google Scholar]
- 34.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243(1):91–8. Epub 2011/09/03. doi: 10.1111/j.1600-065X.2011.01052.x . [DOI] [PubMed] [Google Scholar]
- 35.Leong CR, Funami K, Oshiumi H, Mengao D, Takaki H, Matsumoto M, et al. Interferon-stimulated gene of 20 kDa protein (ISG20) degrades RNA of Hepatitis B virus to impede the replication of HBV in vitro and in vivo. Oncotarget. 2016. Epub 2016/09/15. doi: 10.18632/oncotarget.11907 11907 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) A Western blot analysis for Pol pulled-down by ε-biotin in the presence of a chemical mix containing 10 kinds of compounds. The final concentration of each compound was 30 μM. A total of 3,965 compounds were screened. (B) Candidate mixes were divided into single compounds, and analyzed as in (A). The final concentration of the compounds was 30 μM. Arrows: 3×FLAG-Pol.
(TIF)
The RIG-I helicase assay was conducted in the presence of the indicated compounds. #: monomeric ssRNA, ##: annealed dsRNA, (n.s.): non-specific band.
(TIF)
PXB cells were infected with HBV, and treated with Rosmarinic acid at indicated concentrations. On day 12, cells were subjected to WST-1 cell proliferation assay. Data are from one representative of at least two independent experiments; means and S.D. of duplicate experiments are shown.
(TIF)
PXB cells were infected with HBV, and treated with 30 μM Quercetin. Extracellular HBV DNA, intracellular HBV 3.5 kb RNA, and SHBs were measured as in Fig 4A–4C. Data are from one representative of at least three independent experiments; means and S.D. of duplicate experiments are shown (* p < 0.05).
(TIF)
Uncropped and unadjusted gels and Western blots.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.