Abstract
Wang comments that the diffraction data for the structure of the A139T mutant of translocator protein TSPO from Rhodobacter sphaeroides should be used to 1.65 instead of 1.8 angstroms and that the density interpreted as porphyrin and monoolein is better fitted as polyethylene glycol. Although different practices of data processing exist, in this case they do not substantially influence the final map. Additional data are presented supporting the fit of a porphyrin and monooleins.
We appreciate Wang’s Comment (1) on our structure of the A139T mutant of translocator protein TSPO from Rhodobacter sphaeroides (RsTSPO) [Protein Data Bank (PDB) accession 4UC1)] (2) and the opportunity to reemphasize the basis for our interpretation of the electron density. Wang claims that a better structure of RsTSPO-A139T could be obtained if 1.65 Å, instead of 1.8 Å, were used as the high-resolution cut-off for the diffraction data and if Se, instead of S, were used throughout the refinement. His Comment also questions the interpretation of portions of the electron density as monoolein molecules, despite the fact that this membrane protein structure was obtained within a lipidic cubic phase (LCP) environment with 60% monoolein. Wang argues that reanalysis of our data shows that polyethylene glycol (PEG) fits the electron density better for both the monooleins and the ring-shaped density that we interpret as a porphyrin-like molecule. Although we welcome reanalyses of our data, we disagree strongly with several key points raised by Wang.
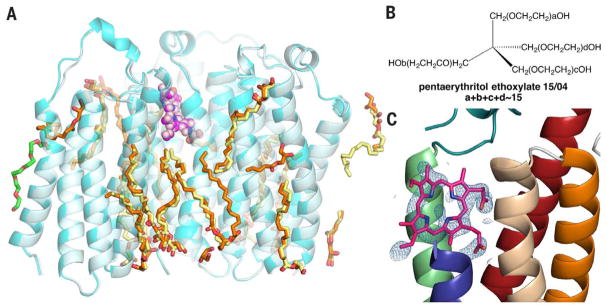
Different practices of data processing and refinement exist, but they rarely substantially influence the final map obtained. In our case, the decision to treat Se as S during refinement was based on the following: (i) the phase for this structure was solved by multiple isomorphous replacement with anomalous scattering with Hg and (Ta6Br12)Br2 derivatives before obtaining the 1.8-Å data set and (ii) the data set was not collected at the Se absorption peak (noted in PDB) and does not reliably result in extra density on the S atoms. Because no additional information would be gained by refining the structure as an anomalous data set, all methionine was treated as S-containing. In fact, we obtained a structure of RsTSPO-A139T at 2.4-Å resolution with an unlabeled protein (PDB 5DUO). Our results confirm that there are no substantial changes in the protein, the ring-shaped density, and most of the observed monoolein molecules (Fig. 1A). Importantly, this crystal was grown in LCP with monoolein, but with a different precipitating agent, pentaerythritol ethoxylate 15/04 (Fig. 1B). This bulkier, tetrahedral-shaped molecule could not substitute for the linear PEG 400 nor occupy any sites of electron density that Wang suggests as PEG 400 molecules. Specifically, it could not fit into the region that we have suggested might be a porphyrin binding site. Yet, in the overlay of the two structures, the positions of various putative monooleins are essentially identical, although a few more are resolved in the 1.8- Å structure (Fig. 1A). In both structures, the extra ring-shaped density (2Fobs – Fcalc) interpreted as a porphyrin is very similar (Fig. 1C) (2).
Fig. 1. Structure of RsTSPO-A139Tobtained in pentaerythritol ethoxylate 15/04 is very similar to that obtained in PEG 400.
(A) Structure of RsTSPO-A139T crystallized in PEG 400 (cyan) and pentaerythritol ethoxylate 15/04 (light cyan) show identical features of the porphyrin-like molecules (magenta and light magenta spheres) and most monooleins (orange and yellow sticks). A PEG molecule (green) could be fitted into the 1.8 Å structure but not in the 2.4 Å structure. One asymmetric unit containing three monomers is shown. (B) Structure of pentaerythritol ethoxylate 15/04. (C) Ring-shaped density (2Fobs – Fcalc) and the fitting of a porphyrin-like molecule in the 2.4-Å structure obtained with pentaerythritol ethoxylate 15/04.
Identification of the ring-shaped density was also supported by biochemical data. As shown in the ultraviolet-visible spectrum in the supplementary materials of our Report (fig. S8A) (2), the absence of most of the Soret band [as also seen in (3)] and the presence of alpha bands in the 500-to 600-nm region is consistent with an oxidized form of porphyrin, as we suggested. Therefore, it was not expected that the modeled PpIX would fit the density very well. In our Report we stated, “A partially oxidized porphyrin is suggested by the spectrum (fig. S8A) that shows absence of a Soret band” (2). Wang also questions the non-planar nature of the fitted porphyrin, but there are many examples in the literature and structural databases with distorted and “ruffled” porphyrins [e.g., PDB 3QGT (4), PDB 4GPH (5), and PDB 3EEE (6)]. Distorted porphyrins are often found in proteins whose function is to degrade heme, where the distortion is thought to contribute to the mechanism. It has been proposed that one of TSPO’s functions is to facilitate porphyrin breakdown (3), and as such, a distorted porphyrin in TSPO might be expected. Nevertheless, the high affinity binding of protoporphyrin IX to RsTSPO would still be observed by fluorescence quenching experiments because the oxidized porphyrin is present at less than 20% in the crystal structure (only one monomer out of three contains porphyrin, at <60% occupancy).
Wang’s Comment states that at 32% in the crystallization medium, PEG 400 is the likely non-protein ligand within the crystal. However, Wang neglected to consider how LCP crystallization works. Within the lipidic cubic phase, the 60% (v/v) monoolein roughly corresponds to a concentration of 1.6 to 1.7 M, whereas the PEG 400 can only be as high as 0.35 M. Given that monooleins are critical for the stability of the LCP and the resulting membrane protein crystals, it is highly unlikely that the observed electron densities are correctly interpreted as PEG 400 molecules. Wang further contends that the ligand library density-fitting (LLDF) scores support his argument. However, the LLDF scores are known not to be suitable criteria for surface-bound ligands, such as lipids (7). A quick survey of membrane protein crystal structures determined in LCP with monoolein shows that most designated monooleins have an LLDF higher than 2, demonstrating that this criterion does not work well for lipidic molecules on the surface of the protein. (Structures surveyed include 4QND, 4N6H, 4JKV, 3VW7, 4EIY, 4E1S, 3S8F, 3V5U, 2RH1, 3ZE3, and 4RYR.) On the other hand, we analyzed the re-refined structure of 4UC1 modeled with PEG provided by Wang using the PDB validation server. The result shows that most of the PEGs that Wang fitted into the transmembrane region in the place of monoolein have a LLDF score higher than 2 (as high as 206), which undermines his own argument.
However, we agree that raw data instead of merged data deposited in PDB provide more accurate information on data quality. The chosen resolution cut-off of the data at 1.8 Å was supported by the statistics of the raw data. As shown in Table 1, data at 1.65 Å have very high R factors and very low I/σ and CC(1/2)—all indications of high noise—and therefore were excluded in the refinement. Wang has recommended (8) that data should be cut at I/σ around 1, which is ~1.8 Å in this data set, the same as the resolution we reported (2). The decision to limit our data set to 1.8 Å was also supported by the merging statistics for the raw data, which clearly revealed significant anisotropy in the highest shells (1.78 to 1.69 Å). This is quite evident in the analysis of the data by merging and scaling with the program Aimless (9); data within the resolution shell of 1.69 to 1.79 Å, the CC(1/2) for two of the principal directions drops precipitously to under 0.3, and the mean I/σ values have dropped well below 1. In addition, Xtriage from Phenix (10) shows that the data in the highest shells are very weak; less than 23% of the reflections have an I/σ > 2, and less than 2.6% have an I/σ > 3. Given that the data in the highest shells display a clear anisotropic distribution, there could be a significant anisotropic contribution to any added “detail” in the resulting electron density maps. This could potentially lead to misleading refinement results and interpretation. Thus, limiting our working data to 1.8 Å is a best-case compromise that adds as much useful data as possible at high resolution but excludes the weak anisotropic data.
Table 1. Statistics of x-ray data of RsTSPO-A139T.
Diffraction data are processed with XDS package (11, 12). Statistics of the unmerged data are shown. I/σ, mean of intensity/sigma (intensity) of unique reflections (after merging symmetry-related observations); CC(1/2), percentage of correlation between intensities from random half data sets (correlation significance at the 0.1% level is marked by an asterisk); Anomal corr, percentage of correlation between random half-sets of anomalous intensity differences; Sig ano, mean anomalous difference in units of its estimated standard deviation (|F(+)-F(−)|/σ); Nano, number of unique reflections used to calculate Anomal corr and Sig ano.
| Resolution limit | Number of reflections
|
Completeness of data (%) |
R factor
(%) |
Compared | I/σ | R meas (%) | CC (1/2) | Anomal corr | Sig ano | Nano | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Unique | Possible | Observed | Expected | |||||||||
| 2.21 | 29059 | 7599 | 7609 | 99.90 | 20.90 | 21.00 | 29059 | 7.13 | 24.30 | 96.6* | −1 | 0.782 | 4772 |
|
| |||||||||||||
| 2.02 | 31920 | 8379 | 8391 | 99.90 | 39.10 | 39.30 | 31920 | 4.03 | 45.50 | 89.6* | 0 | 0.751 | 5117 |
|
| |||||||||||||
| 1.87 | 34512 | 9135 | 9145 | 99.90 | 81.80 | 80.90 | 34509 | 1.81 | 95.50 | 62.2* | 0 | 0.717 | 5422 |
|
| |||||||||||||
| 1.75 | 36929 | 9797 | 9807 | 99.90 | 151.50 | 156.30 | 36922 | 0.88 | 176.90 | 29.1* | 0 | 0.646 | 5707 |
|
| |||||||||||||
| 1.65 | 38230 | 10289 | 10420 | 98.70 | 272.90 | 293.00 | 38149 | 0.44 | 319.20 | 12.5* | 0 | 0.589 | 5780 |
|
| |||||||||||||
| Total | 244407 | 64547 | 64747 | 99.70 | 10.40 | 10.40 | 244313 | 9.03 | 12.10 | 99.8* | −1 | 0.731 | 39945 |
In conclusion, we believe that the new structure (5DUO) in pentaerithritol ethoxylate, along with chemical considerations and the statistics of the data, confirm our original conclusion that the best interpretation of the ring-shaped density is a porphyrin-type molecule and that the densities associated with the hydrophobic region of the protein are monooleins.
References
- 1.Wang J. Science. 2015;350:519. doi: 10.1126/science.aab1432. [DOI] [PubMed] [Google Scholar]
- 2.Li F, Liu J, Zheng Y, Garavito RM, Ferguson-Miller S. Science. 2015;347:555–558. doi: 10.1126/science.1260590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Y, et al. Science. 2015;347:551–555. doi: 10.1126/science.aaa1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takayama SJ, Ukpabi G, Murphy ME, Mauk AG. Proc Natl Acad Sci USA. 2011;108:13071–13076. doi: 10.1073/pnas.1101459108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unno M, Ardèvol A, Rovira C, Ikeda-Saito M. J Biol Chem. 2013;288:34443–34458. doi: 10.1074/jbc.M113.486936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olea C, Boon EM, Pellicena P, Kuriyan J, Marletta MA. ACS Chem Biol. 2008;3:703–710. doi: 10.1021/cb800185h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Protein Data Bank. User guide to the wwPDB X-ray validation reports. www.wwpdb.org/validation/ValidationPDFNotes.html.
- 8.Wang J. Protein Sci. 2015;24:661–669. doi: 10.1002/pro.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans P. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 10.Adams PD, et al. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabsch W. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabsch W. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]