SUMMARY
The neuromodulator dopamine (DA) plays a key role in motor control, motivated behaviors, and higher-order cognitive processes. Dissecting how these DA neural networks tune the activity of local neural circuits to regulate behavior requires tools for manipulating small groups of DA neurons. To address this need, we assembled a genetic toolkit that allows for an exquisite level of control over the DA neural network in Drosophila. To further refine targeting of specific DA neurons, we also created reagents that allow for the conversion of any existing GAL4 line into Split GAL4 or GAL80 lines. We demonstrated how this toolkit can be used with recently developed computational methods to rapidly generate additional reagents for manipulating small subsets or individual DA neurons. Finally, we used the toolkit to reveal a dynamic interaction between a small subset of DA neurons and rearing conditions in a social space behavioral assay.
In Brief
The rapid analysis of how dopaminergic circuits regulate behavior is limited by the genetic tools available to target and manipulate small numbers of these neurons. Xie et al. present genetic tools in Drosophila that allow rational targeting of sparse dopaminergic neuronal subsets and selective knockdown of dopamine signaling.

INTRODUCTION
Unraveling the circuit mechanisms underlying behavior is a principal goal of current neuroscience research. Beyond the anatomical characterization of neurons and their connectivity, understanding the full repertoire of animal behavior requires delineating how neuromodulators convey information about the environment and internal states by tuning the activity of neural circuits (Bargmann, 2012; Marder, 2012). Dopamine (DA) is a conserved neuromodulator that is important for a variety of behavioral processes, including attention, sleep/wake, motor control, motivation, reward processing, and learning/memory in fruit flies and mammals (Nieoullon, 2002; van Swinderen and Andretic, 2011; Lebestky et al., 2009; Liu et al., 2012a; Wisor et al., 2001; Kume et al., 2005; Riemensperger et al., 2011; Calabresi et al., 2014; Wise, 2004; Berridge and Robinson, 1998; Westbrook and Braver, 2016; Schwaerzel et al., 2003; Waddell, 2013). Thus, there is intense interest in dissecting the DA neural circuits that regulate behavior.
In both Drosophila and mammals, there are relatively few DA neurons compared to the total number of brain neurons (<0.5% in flies and mice) (Mao and Davis, 2009; Chiang et al., 2011; Herculano-Houzel, 2007; Björklund and Dunnett, 2007), but these neurons have extensive projections and differ in their functional targets. For example, in Drosophila, small subsets or individual DA neurons that regulate arousal, aggression, courtship learning, aversive odor memory, sucrose feeding behavior, and protein hunger have been identified (Liu et al., 2012a, 2017; Ueno et al., 2012; Alekseyenko et al., 2013; Keleman et al., 2012; Marella et al., 2012; Aso et al., 2010). These observations underscore the importance of finely dissecting DA neuromodulatory networks, but a key rate-limiting factor in the analysis of DA neural circuits is the relative dearth of genetic tools for convenient targeting and manipulation of small subsets of DA neurons.
To address this issue, we present here a set of genetic tools designed to facilitate the functional dissection of DA neural networks in Drosophila. We have generated and characterized a series of GAL4, GAL80, Split GAL4, Killer Zipper, homology-assisted CRISPR knockin (HACK), flippase (FLP), and microRNA transgenic lines for this purpose. Together with information derived from the annotation of these lines and driver screens that we have conducted, these reagents should, in combination, provide a high degree of control over DA neural circuits in fruit flies and assist in elucidating the function of DA in behavior.
RESULTS
Generation of Transgenic Lines for Intersectional Analyses of DA Circuits
The DA network in the adult central brain of Drosophila, defined by immunoreactivity to tyrosine hydroxylase (TH), contains ~125 neurons per hemisphere and is organized into a number of anatomically defined clusters, including several anterior groups (PAM, PAL, T1, and Sb) and 6 major posterior groups (PPL1, PPL2ab, PPL2c, PPM1, PPM2, PPM3), as well as a few other variably labeled cells (Figures 1A–1D) (Mao and Davis, 2009; Nässel and Elekes, 1992; White et al., 2010; Hartenstein et al., 2017). To generate transgenic lines that can be used to label specific DA neurons in an intersectional manner, we chose 10 “founder” enhancer sequences that collectively drive diverse expression patterns in these cells (Figures 1E–1N and S1). These enhancer sequences were derived from a combination of lines generated by our laboratory and from the FlyLight GAL4 collection (Liu et al., 2012a; Jenett et al., 2012) and were used to create GAL4, GAL80, and Split GAL4 lines (Brand and Perrimon 1993; Pfeiffer et al., 2010; Lee and Luo, 1999; Luan et al., 2006). For all GAL4 driver lines, we characterized the number of DA neurons labeled in each cluster (Table S1) and, as discussed below, we annotated the individual DA cells within each cluster for Split GAL4 combinations. As shown in Figure S2, these GAL4 lines exhibited no or sparse expression in DA neurons in the ventral nerve cord (VNC). We did not include the PAM neurons in our analyses because most if not all PAM neurons project to various subdomains of the mushroom bodies (MBs) (Liu et al., 2012b), and Aso et al. (2014) systematically identified intersectional drivers that sparsely label DA neurons innervating the MBs. However, a number of our Split GAL4 combinations do exhibit labeling in these cells, as noted in Table S2.
Figure 1. Expression Patterns of the Founder DA GAL4 Driver Lines.
(A–D) Whole-mount brain immunostaining of wild-type flies showing TH+ neuron cell bodies (green) with nc82 neuropil counterstaining (blue). Maximal intensity projections are shown for image stacks for anterior (A) and posterior (C) thirds of the brains.
(B and D) Schematics demonstrating DA neuron clusters in the anterior (B) and posterior (D) regions, as defined by Nässel and Elekes (1992).
(E–N) Whole-mount brain immunostaining of TH-C-GAL4 (E), TH-D-GAL4 (F), TH-F-GAL4 (G), Ddc-R60F07-GAL4 (H), Ddc-R61H03-GAL4 (I), DAT-B-GAL4 (J), DAT-R55C10-GAL4 (K), Vmat-R76F01-GAL4 (L), Vmat-R76F02-GAL4 (M), and Vmat-R76F05-GAL4 (N) lines driving expression of 10XUAS-IVS-Syn21-GFP-p10 with anti-TH (magenta), anti-GFP (green), and anti-nc82 (blue).
Maximal intensity projections of anterior (subscript 1) and posterior (subscript 2) stacks are shown. DA neurons in a given cluster are indicated by dashed ellipses. Scale bars, 100 μm.
For intersectional analyses, GAL80 inhibits the GAL4 transactivator, allowing for “not” logic gating, whereas the Split GAL4 system requires the presence of both GAL4 activation (AD) and DNA-binding domains (DBDs) for driving expression, thus enabling “and” logic gating. We generated GAL80 transgenes under control of the 10 founder enhancer sequences and found that these transgenes largely recapitulated the expression patterns observed with the GAL4 lines (Figure 2; Table S1). We next used a similar approach to generate Split GAL4 AD and DBD transgenic lines. To broadly characterize their expression patterns in DA neurons, we first used each line, in combination with TH-ZpGAL4DBD or TH-p65ADZp animals generated by Aso et al. (2014), to drive expression of 10XUAS-GFP (Table S3).
Figure 2. Restriction of DA Expression Using GAL80 Transgenes.
(A and B) Schematics of anterior (A) and posterior (B) DA neuron clusters as shown in Figure 1.
(C–L) Whole-mount brain immunostaining of TH-GAL4 >10XUAS-IVS-Syn21-GFP-p10 with TH-C-GAL80 (C), TH-D-GAL80 (D), TH-F-GAL80 (E), Ddc-R60F07-GAL80 (F), Ddc-R61H03-GAL80 (G), DAT-B-GAL80 (H), DAT-R55C10-GAL80 (I), Vmat-R76F01-GAL80 (J), Vmat-R76F02-GAL80 (K), or Vmat-R76F05-GAL80 (L) using anti-TH (magenta) and anti-GFP (green). Anterior and posterior third image stacks are shown.
Arrowheads and dashed ellipses denote individual or groups of DA neurons in which GFP expression is suppressed by GAL80. Scale bar, 100 μm.
Because these Split GAL4 lines could yield useful sparse labeling in DA cells, we next annotated the precise DA neurons labeled by all 90 potential combinations. To do this, we performed immunostaining for all Split GAL4 combinations and registered all images to the Janelia standard fly brain (www.virtualflybrain.org). We then annotated TH-GAL4+ single neuron images from the FlyCircuit database (Chiang et al., 2011) (Table S4), merged the FlyCircuit images with each Split GAL4 confocal stack, and manually identified the DA neurons labeled by each Split GAL4 combination. As shown in Figures 3A–3D and Figure S3, these Split GAL4 transgenes allow for in some cases highly restricted expression in DA neurons. Despite this, these Split GAL4 combinations labeled on average approximately seven non-PAM TH+ cells, necessitating additional methods for restricting expression further.
Figure 3. Annotation of DA Neurons Labeled in Split GAL4 Combinations.
(A) Matrix summarizing individual DA neurons labeled by different Split GAL4 combinations. For a given DBD-AD pair, the combination yielding the more restricted expression pattern is shown. In some cases, both are shown, if both combinations of a DBD-AD pair provide useful patterns.
(B–D) Whole-mount brain immunostaining of DAT-B-DBD, Vmat-R76F02-AD (B), TH-C-DBD, Vmat-R76F02-AD (C), and DAT-R55C10-DBD, Vmat-R76F02-AD
(D) lines driving expression of 10XUAS-IVS-Syn21-GFP-p10 with anti-TH (magenta), anti-GFP (green), and anti-nc82 (blue).
Arrowheads denote individual DA neurons. Scale bar, 100 μm.
Killer Zipper and HACK Refine and Improve Fidelity of Expression Patterns
To further restrict the DA neurons labeled by our Split GAL4 combinations, we used the recently developed Killer Zipper (KZip+) method, given that GAL80 does not effectively inhibit Split GAL4 activity. KZip+ acts as a dominant-negative repressor of Split GAL4 and allows for the combination of “not” with “and” logic gating (Dolan et al., 2017). We thus generated transgenic lines expressing KZip+ under control of the 10 founder enhancers. As shown in Table S5 and Figures 4A–4C, these KZip+ lines repressed expression in specific DA clusters, which is largely consistent with results for the founder-derived GAL4 and GAL80 lines seen in Table S1. These KZip+ lines, particularly those with relatively broad expression patterns in DA neurons such as TH-C-KZip+ and TH-D-KZip+ should be useful in further restricting expression in our Split GAL4 lines.
Figure 4. Use of KZip+ and HACK to Refine DA Neuron Expression Patterns.
(A–C) Whole-mount brain immunostaining of TH-ZpGAL4DBD, TH-p65ADZp>10XUAS-IVS-myr::GFP without (A) or with TH-C-KZip+ (B) or TH-D-KZip+ (C) using … Anterior (subscript 1) and posterior (subscript 2) stacks are shown.
(D) Schematic of HACK-mediated conversion of GAL4 effectors to GAL4-DBD, GAL4-AD, or GAL80. Gene conversion of the T2A-effector-loxP-Pax-RFP-loxP cassette occurs between a target GAL4 and the HACK donor transgene, when double-strand DNA breaks are created at the target GAL4, bringing the desired effector under control of the promoter/enhancer driving expression in the original target GAL4 line.
(E and F) Whole-mountbrainimmunostaining of Vmat-R76F05-AD, TH-D-DBD>10XUAS-IVS-Syn21-GFP-p10 (E) or Vmat-R76F05-AD, TH-D-DBDG4HACK>10XUAS-IVS-Syn21-GFP-p10 (F) with anti-TH (magenta) and anti-GFP (green), demonstrating removal of KC expression.
(G and H) Whole-mount brain immunostaining of 201y-GAL4>UAS-mCD8::GFP without (G) or with (H) OK107-GAL80G4HACK.
Asterisks indicate KCs. Scale bars, 100 μm.
We found that 13 of 90 of the Split GAL4 combinations also exhibited weak expression in Kenyon cells (KCs), even though in the majority of these cases this was not observed in the corresponding founder GAL4 lines. To address the possibility that the integration site contributes to this KC expression, we regenerated all DBD lines using an independent attP site (86Fb); however, KC expression was still observed in these 13 Split GAL4 combinations (data not shown). Thus, to increase the fidelity of the expression patterns of the AD and DBD transgenes, we wanted to directly replace the GAL4 in the original founder GAL4 lines with AD or DBD. We chose to use the HACK method (Lin and Potter, 2016) and created a series of AD and DBD donor transgenes for this approach that were inserted at random genomic locations or at the attP2 landing site (Figures 4D and S4).
To demonstrate the utility of these lines, we replaced the GAL4 in the TH-D transgenic line with DBD. As shown in Figures 4E and 4F, use of this line in combination with VMAT-R76F05-AD allowed for expression in DA neurons, without KC labeling. To broaden the utility of the HACK toolkit and to provide another means for suppressing unwanted KC expression, we next generated GAL80 HACK donor lines and used these reagents to replace the GAL4 in the OK107-GAL4 transgenic line with GAL80. As shown in Figures 4G and 4H, OK107-GAL80G4HACK efficiently suppressed KC labeling in the 201Y-GAL4 driver line. The development of these HACK AD, DBD, and GAL80 reagents will facilitate the analysis of not only DA neural circuits but also any neural circuits labeled by a GAL4 driver. Together, the KZip+ and HACK lines described here enable the refinement of DA neuron expression patterns from transgenic driver lines.
An Independent Strategy for Labeling Sparse Populations of DA Neurons
To develop an independent method for targeting small numbers of DA neurons, we additionally generated a transgenic line bearing FLP recombinase (Golic and Lindquist, 1989) under control of the original ~11-kb TH enhancer sequence (TH-FLP) (Friggi-Grelin et al., 2003). This FLP line, when used in combination with non-DA-related GAL4 drivers and an effector transgene under control of an FLP recognition target (FRT)-stop-FRT cassette, allows for highly restricted intersectional expression in DA neurons. As shown in Figure 5A, TH-FLP expresses FLP in the majority of the DA neurons labeled by TH-GAL4. To access additional DA neurons not covered by TH-FLP, we also generated TH-C-FLP, which drives FLP expression in the TH-C-GAL4 expression pattern (Figure 5B). We conducted an FLP-based screen to identify FlyLight GAL4 drivers that could be used to label a particular DA neuron (Table S6). To do this, we first used FlyCircuit single DA neuron images (Table S4) as query images for NBLAST (Costa et al., 2016) to choose the GAL4 drivers. We then used these GAL4 drivers in combination with TH-FLP and/or TH-C-FLP to drive expression of UAS-FRT-stop-FRT-CD8::GFP and confirmed cell identity by manual annotation. This approach led to highly restricted expression in DA neurons in some cases, and collectively we were able to determine GAL4/FLP combinations that allowed for genetic access to 29 of 38 classes (76%) of non-PAM DA neurons (Figures 5C–5F; Table S6).
Figure 5. TH-FLP Transgenes for Restricted DA Neuron Expression.

(A and B) Whole-mount brain immunostaining of TH-FLP, TH-GAL4>UAS-FRT-stop-FRT-CD8::GFP (A) or TH-C-FLP, TH-GAL4>UAS-FRT-stop-FRT-CD8::GFP (B) with anti-TH (magenta), anti-GFP (green), and anti-nc82 (blue). Anterior and posterior stacks are shown, and dashed ellipses indicate DA neuron clusters as indicated.
(C) Matrix summarizing individual DA neurons labeled by different TH-FLP-based intersectional combinations. Because these expression patterns were sufficiently sparse, annotation of individual DA neurons was possible without brain registration.
(D–F) Whole-mount brain immunostaining of TH-FLP>UAS-FRT-stop-FRT-CD8::GFP in combination with R36A10-GAL4 (D), R91E12-GAL4 (E), or R92G05-GAL4 (F) with anti-TH (magenta), anti-GFP (green), and anti-nc82 (blue).
Labeled DA neurons are indicated by arrowheads. Scale bars, 100 μm.
Intersectional approaches with FLP require the use of an FRT-stop-FRT cassette, limiting the choices of effector transgenes used. To address this limitation, the HACK Split GAL4 donor lines generated above can be used to convert the FlyLight GAL4 lines from the FLP screen to GAL4-AD or GAL4-DBD lines. Because the FlyLight GAL4 transgenes often are integrated into the attP2 site (Jenett et al., 2012), use of our HACK AD or DBD trans-genes inserted into this same site provides high-efficiency gene conversion (Lin and Potter, 2016). Thus, the information derived from our FLP-based screen, combined with our HACK reagents, should permit convenient access to drivers that provide sparse labeling of non-PAM DA neurons.
Genetic Tools for Circuit-Specific Manipulation of DA Signaling
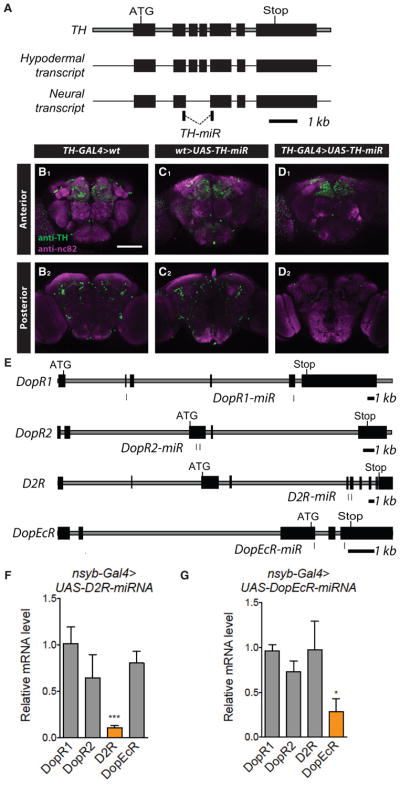
In addition to tools that allow for sparse labeling of DA neurons, we sought to generate reagents that would enable selective manipulation of DA signaling. To do so, we developed genetically targetable microRNA constructs that could be used to knock down expression of TH or DA receptors. One challenge in directly targeting TH in Drosophila DA neurons is that doing so typically affects the non-neuronal isoform, which is essential for cuticular sclerotization (Wright, 1987; Birman et al., 1994). Indeed, the first TH microRNA construct we generated (UAS-TH-miR-1), which targets an exon in both neuronal and non-neuronal isoforms, induced substantial lethality when expressed using TH-GAL4 (Figure S5). The TH locus undergoes alternative exon splicing to generate the neuronal isoform, which lacks exons 3 and 4. Thus, to selectively knock down the neural TH isoform, we generated two different miR constructs (UAS-TH-miR-2 and UAS-TH-miR-G), where the hairpin loops bridged exons 2 and 5 (Figure 6A). Expression of these miR transgenes in TH+ cells resulted in improved survival and cuticle pigmentation, particularly in the case of UAS-TH-miR-G, while still inducing widespread and substantial knockdown of TH in the brain (Figures S5 and 6B–6D)
Figure 6. miR Transgenes for Manipulation of DA Signaling.
(A) Schematic of the TH genomic locus, demonstrating exon structure of hypodermal and neural transcripts. Location of hairpin sequences for neural-specific TH-miRs, spanning two neural-specific exons is indicated.
(B–D) Whole-mount brain immunostaining of TH-GAL4>WT (wild-type) (B), WT>UAS-TH-miR-G (C), and TH-GAL4>UAS-TH-miR-G (D) using anti-TH (green) and anti-nc82 (magenta). Anterior and posterior third image stacks are shown. Scale bar, 100 μm.
(E) Schematic of the genomic loci for the DopR1, DopR2, D2R, and DopEcR genes. Location of hairpin sequences for miRs for each gene are shown. Each DA receptor miR transgene contains two 22mers that each form a hairpin loop targeting distinct sequences of the receptor.
(F and G) Relative mRNA level for DopR1, DopR2, D2R, and DopEcR in nsyb-GAL4>UAS-D2R-miR (F) or nsyb-Gal4>UAS-DopEcR-miR (G) compared to nsyb-Gal4>WT female flies (n = 3 trials). Statistical comparisons for a given gene were performed using t tests to the value in nsyb-GAL4>WT flies, which was normalized to 1.0. Because miRs act at both transcriptional and post-transcriptional levels, knockdown of the targeted protein is likely greater than the changes detected here.
*p < 0.05; ***p < 0.001.
Data are represented as means ± SEMs.
Another approach for inhibiting DA signaling would be to knock down the downstream DA receptors. Thus, we generated UAS transgenic lines that bear microRNA constructs, targeting each of the 4 DA receptors in Drosophila (DopR1, DopR2, D2R, and DopEcR) (Feng et al., 1996; Han et al., 1996; Hearn et al., 2002; Srivastava et al., 2005) (Figure 6E). To confirm the efficacy and specificity of these tools, we performed qPCR from the brains of flies expressing each miR under the control of nsyb-GAL4. We recently described and characterized two of these lines (for DopR1 and DopR2) (Liu et al., 2017) and obtained similar results for the transgenic lines targeting DopR2 and DopEcR (Figures 6F and 6G). These tools should enhance the ability of researchers to manipulate DA signaling in specific circuits.
Dissection of the Dopaminergic Regulation of Social Space
To demonstrate the utility of these reagents, we sought to dissect the dopaminergic circuitry underlying a specific behavior. Drosophila exhibit a variety of social behaviors (Anderson, 2016), including a tendency to aggregate into groups (Simon et al., 2012; Burg et al., 2013). A simple social space assay to assess this phenotype has been described. In this assay, a group of flies are allowed to settle in a two-dimensional arena, and the distances between the flies are measured. It is interesting that this behavior is dependent on rearing conditions; in other words, social-enriched flies tend to cluster together, whereas socially isolated flies tend to exhibit increased social space between individuals (Simon et al., 2012) (Figures 7A–7C). This social space behavior recently was shown to be regulated by dopaminergic signaling (Fernandez et al., 2017); we asked whether we could quickly identify the specific DA neurons involved.
Figure 7. DA Neurons that Regulate Social Space Behavior.
(A and B) Representative digitized images for social space behavior for wild-type (WT) iso31 male flies under socially enriched (A) or isolated (B) conditions.
(C) Average number of flies within 1 cm of a given fly for control iso31 under socially enriched (n = 4 trials) versus isolated conditions (n = 5 trials).
(D) Average number of flies within 1 cm of a given fly for WT>UAS-TH-miR-2 (n = 4 trials), TH-GAL4>WT (n = 4 trials), TH-GAL4>UAS-TH-miR-2 (n = 4 trials), TH-C-GAL4>UAS-TH-miR-2 (n = 6 trials), TH-D-GAL4>UAS-TH-miR-2 (n = 5 trials), TH-D-GAL4>WT (n = 5 trials), or TH-C-GAL4>WT (n = 6 trials) under socially enriched conditions.
(E) Average number of flies within 1 cm of a given fly for WT>UAS-TH-miR-2 (n = 4 trials), TH-C-GAL4>WT (n = 3 trials), TH-C-GAL4>UAS-TH-miR-2 (n = 4 trials), TH-D-GAL4>UAS-TH-miR-2 (n = 3 trials), or TH-D-GAL4>WT (n = 4 trials) under isolated conditions.
(F) Average number of flies within 1 cm of a given fly for WT>UAS-TH-miR-2 (n = 4 trials), TH-C-DBD, TH-D-AD>UAS-TH-miR-2 (n = 4 trials), TH-C-DBD, TH-D-AD>WT (n = 4 trials) under socially enriched conditions.
(G) Average number of flies within 1 cm of a given fly for TH-C-DBD, TH-D-AD>WT (n = 3 trials), TH-C-DBD, TH-D-AD>UAS-TH-miR-2 (n = 3 trials), or WT>UAS-TH-miR-2 (n = 4 trials) under isolated conditions.
*p < 0.05; **p < 0.01; ***p < 0.001. Data are represented as means ± SEMs.
We drove expression of UAS-TH-miR-2 with either TH-C-GAL4 or TH-D-GAL4 to inhibit DA signaling in broad subsets of DA neurons and assayed the effects on social space behavior. Knockdown of DA production using either driver appeared to reverse the relation between social rearing and social space; isolated TH-depleted flies behaved like enriched control flies, whereas the opposite results were obtained under conditions of social enrichment (Figures 7D, 7E, and S6). We next used the Split GAL4 combination (TH-D-DBD/TH-C-AD), which specifically labels two PPM2 WED and 2 VLP DA neurons, and obtained results similar to the broad TH-C-GAL4 and TH-D-GAL4 drivers (Figures 7F, 7G, S6, and S7A), suggesting that these four neurons play a key role in social space behavior. To ensure that the phenotypes observed result from the manipulation of neuronal (and not peripheral) DA, we next assessed TH-C-GAL4, elav-GAL80>UAS-TH-miR-2 and found no significant change in social space behavior compared to controls (Figure S7B). To further examine the specificity of this effect, we performed knockdown of TH using drivers that label other groups of DA neurons and found no significant effects on social space behavior under socially enriched conditions (Figures S7C and S7D). These findings underscore the utility of our genetic reagents in quickly isolating small subsets of DA neurons mediating a specific behavior.
DISCUSSION
The tools generated here are designed to permit a refined analysis of dopaminergic signaling in behavior and neural circuit function. We estimate that the transgenic GAL4, GAL80, Split GAL4, and KZip+ lines that we have generated can be combined to yield >900 unique patterns of DA neuron expression. In addition, the computational and FLP-based screening approaches developed here provide access to additional expression patterns in DA neurons and can be used to identify drivers that are suitable for targeting minimal subsets of DA neurons. We also generated transgenic microRNA lines that directly inhibit DA signaling; this complementary approach should allow for not only physiological manipulation of DA neuron activity but also enhanced specificity in targeting only DA-expressing neurons labeled by a GAL4 driver or suppressing release of DA but not of co-transmitters. Finally, we developed additional HACK reagents that allow for conversion of GAL4 to GAL4-DBD, GAL4-AD, and GAL80. These HACK lines should not only facilitate analysis of DA circuits but also be broadly useful for investigators interested in neural circuit analyses or intersectional expression in other cell types in Drosophila.
Using these tools, we quickly identified four DA neurons that regulate social space behavior. Our data suggest that social space preference is not regulated by these DA neurons on their own, but rather by the interplay of DA signaling with social rearing conditions, suggesting a multicomponent circuit mechanism. We note that two of these four neurons are the DA-WED neurons we previously demonstrated to be critical for protein hunger (Liu et al., 2017), and previous work in Drosophila has identified mechanisms connecting social aggregation and feeding (Lin et al., 2015).
Computational methods are being used increasingly to anatomically map neurons in the brains of a variety of animals ranging from flies to humans (Oh et al., 2014; Takemura et al., 2013; Glasser et al., 2016) to define “connectomes.” Although the connectome for the 302 neurons in Caenorhabditis elegans has been available for >30 years (White et al., 1986), generating “circuit diagrams” for animals with a greater number of neurons and brain complexity is substantially more challenging, necessitating the use of these computational approaches. In Drosophila, software facilitating the mining of the large publicly available GAL4 driver image datasets has been developed recently (Costa et al., 2016; Panser et al., 2016). In this work, we took advantage of this approach to quickly identify drivers that potentially label a specific DA neuron. It is worth emphasizing that the use of our HACK Split GAL4 reagents with the GAL4 driver lines characterized in our FLP-based screen allows for relatively convenient access to driver combinations targeting individual or small subsets of DA neurons. From a broader perspective, following the complete annotation of all of the neurons in the Drosophila central brain, these computational methods, coupled with available GAL4 libraries and evolving intersectional tools such as these HACK transgenic lines, should make possible genetic access to any individual neuron in Drosophila.
During the past decade, interest in imaging neurons, their activity, and their connectivity at the whole brain level has been growing (Oh et al., 2014; Chung et al., 2013; Renier et al., 2014; Ahrens et al., 2012; Mann et al., 2017; Economo et al., 2016; Plaza et al., 2014); however, these efforts must be complemented by methods for understanding the function and interactions of neurons at the level of individual cells. The importance of analysis at the individual neuron level is particularly relevant for neuromodulatory neurons, such as DA cells, which can act individually or in small groups to broadly influence neural circuit function. The tools introduced here are designed to facilitate these types of investigations for DA neurons, and the integration of these “local” analyses with “global” approaches for understanding neural circuit function should expedite the development of a comprehensive understanding of the mechanisms underlying cognition and behavior.
EXPERIMENTAL PROCEDURES
Fly Strains
TH-C-GAL4, TH-D-GAL4, TH-F-GAL4, TH-C-FLP, UAS-TH-miR-2, UAS-DopR1-miR, UAS-DopR2-miR, and UAS-FRT-stop-FRT-CD8::GFP were described previously (Liu et al., 2012a, 2017; Potter et al., 2010). DAT-R55C10-GAL4 (no. 39108), Ddc-R60F07-GAL4 (no. 45358), Ddc-R61H03-GAL4 (no. 39280), Vmat-R76F01-GAL4 (no. 39934), Vmat-R76F02-GAL4 (no. 39935), Vmat-R76F05-GAL4 (no. 41305), and P{10XUAS-IVS-myr::GFP} su(Hw)attP1 (no. 32200), and GAL4 driver lines used in the FLP screen (see Table S6) were obtained from the Bloomington Drosophila Stock Center. TH-GAL4 was obtained from S. Birman. elav-Gal80 was obtained from Y.-N. Jan. pJFRC81-10XUAS-IVS-Syn21-GFP-p10, TH-ZpGAL4DBD, and TH-p65ADZp were obtained from G. Rubin. All fly lines used in this study were either generated in the iso31 background or were backcrossed Rfour times into this background.
Molecular Biology
GAL4, GAL80, and Split GAL4 Constructs
To identify founder enhancer sequences that would provide diverse expression patterns in DA neurons, we performed “promoter-bashing” experiments for genes expressed in these cells—TH (Liu et al., 2012a), DAT (data not shown), and DDC (data not shown)—and examined all FlyLight GAL4 lines for TH, DAT, DDC, and VMAT. The DAT-B-GAL4 construct was generated by PCR amplifying genomic sequence from the first intron of the DAT gene using the primers 5′-TTT GCG GCC GCT CAA ACT AAG ATC GCG CAT ACG C-3′ and 5′-CCT TAA TTA ACC ACC AAA CTG TAA GAG TTG TAC-3′ and subcloning this enhancer element into the InSite vector pBMPGAL4LWL (Gohl et al., 2011). Enhancer elements for C-, D-, and F-GAL80 were subcloned into a modified pCasper4 vector that included a TATA box and the codon-optimized GAL80 derived from pBPGAL80Uw-6 (no. 26236). All of the other constructs for GAL80 and Split GAL4 lines were generated by Gateway cloning (Thermo Fisher Scientific) into pBPGAL80Uw-6 (no. 26236), pBPp65ADZpUw (no. 26234), or pBPZpGAL4DBDUw (no. 26233) vectors, respectively. Genomic regions were amplified by PCR using the following primers: TH-C: 5′-CAC CGC GAG ATG TTC GCC ATC AAG-3′ and 5′-GGA TGC AAT CTT CCA AGG C-3′; TH-D: 5′-CAC CGC GTA AGT ATC TAT CAT C-3′ and 5′-TGA CCA TGT TCC TTG CAG AGA-3′; TH-F: 5′-CAC CGA CGA TTC CCT GGA AGA TTG C-3′ and 5′-GAC AGC TTC TCG ATT TCT TCG-3′; and DAT-B: 5′-CAC CTC AAA CTA AGA TCG CGC ATA CGC-3′ and 5′-TCT ACC GAA CTA TCC AAG CC-3′. For enhancer sequences derived from G. Rubin GAL4 lines (DAT-R55C10, Ddc-R60F07, Ddc-R61H03, Vmat-R76F01, Vmat-R76F02, and Vmat-R76F05), the primers used were as described previously (Pfeiffer et al., 2008).
KZip+ Constructs
We first digested pBPZpGAL4DBDUw with KpnI and HindIII and replaced the GAL4 DBD with the KZip+ open reading frame (ORF) from the CCAP-IVS-Syn21-KZip+-p10 plasmid (Dolan et al., 2017) by using the In-Fusion cloning system (Clontech) and the following primers: 5′-GAC GCA TAC CAA ACG GTA CCA GAT CTA AAA GGT AGG TTC AAC CAC-3′ and 5′-CGG TAT CGA TAA GCT TGC CGT TAA CTC GAA TCG C-3′. We then used Gateway cloning to generate KZip+ constructs for all 10 founder enhancer elements.
HACK Split GAL4 and GAL80 Constructs
The HACK system (Lin and Potter, 2016) was used to convert existing GAL4 transgenic lines into Split GAL4 and GAL80 lines. pHACK-GAL4>AD and pHACK-GAL4>DBD were generated by replacing the QF2 sequence of pHACK-GAL4>QF2 (Addgene no. 80275) with GAL4-AD or GAL4-DBD sequences from pBPp65ADZpUw or pBPZpGAL4DBDUw, respectively. The GAL4-AD or GAL4-DBD cassettes were PCR amplified with the primers AD: 5′-CCC CGG GCC CCC TAG GAT GGA TAA AGC GGA ATT AAT TCC CGA GCC TCC-3′ and 5′-TGT ATT CAA TGC TAG CTT TAC TTG CCG CCG CCC AG-3′ and DBD: 5′-CCC CGG GCC CCC TAG GAT GCT GGA GAT CCG CGC CGC-3′ and 5′-TGT ATT CAA TGC TAG CTT TAC GAT ACC GTC AGT TGC CGT TGA CCC-3′, and were then inserted into the NheI/AvrII-digested pHACK-GAL4>QF2 plasmid using In-Fusion cloning. Note that this approach leaves 36 aa of the original QF2 sequence following the stop codon, which is out of frame with the GAL4-AD or GAL4-DBD sequence. Thus, to generate pHACK-GAL4>GAL80, the pHACK-GAL4>QF2 plasmid was first modified to allow simple replacement of the QF2-hsp70 cassette with a target cassette of choice following SphI/AvrII digestion (pHACK-GAL4>QF2-newHA1, Addgene no. 80275). A GAL80-SV40 cassette was PCR amplified from pQUAST-GAL80 (Addgene no. 46137) using the primers 5′-CCC CGG GCC CCC TAG GAT GGA CTA CAA CAA GAG ATC TTC-3′ and 5′-TAT ACG AAG TTA TGC ATG CGA TCC AGA CAT GAT AAG ATA CAT T-3′ and was inserted into the SphI/AvrII-digested pHACK-GAL4>QF2-newHA1 plasmid by In-Fusion cloning. Genomic insertion sites of the transgenes for all of the HACK donor lines were identified using the splinkerette method (Potter and Luo, 2010) (Figure S4).
FLP Constructs
The TH-FLP construct was generated by replacing the GAL4 coding sequence in pCasper-TH-GAL4 (gift from S. Birman) (Friggi-Grelin et al., 2003) with codon-optimized FLP. The FLP sequence was PCR amplified with primers 5′-CGG GAT CCA TGA GCC AGT TCG ACA TCC TG-3′ and 5′-GGA CTA GTT CAG ATC CGC CTG TTG ATG TA-3′ and subcloned into pCasper-TH-GAL4 using BamHI and SpeI. Generation of the TH-C-FLP construct was described previously (Liu et al., 2017).
microRNA Constructs
In general, microRNA (miR) constructs were generated as previously described (Chen et al., 2007). For UAS-TH-miR-1, two 22mers in exon 2 (5′-TGG TCA AGC AGA CCA AAC AAA C-3′) and exon 5 (5′-AAT CTG ATG GCC GAC AAT AAC T-3′), respectively, were used to create the two hairpin loops. The construct for UAS-TH-miR-2 was described previously (Liu et al., 2017). For UAS-TH-miR-G, two 22mers bridging the second and third coding exons of the neural transcript of the TH gene (5′-CGC AGC AAG GCA AAT GAT TAC G-3′ and 5′-GGC AAA TGA TTA CGG TCT CAC C-3′) were used to create the two hairpin loops. The UAS-DopR1-miR and UAS-DopR2-miR constructs were described previously (Liu et al., 2017). To generate UAS-D2R-miR, two 22mers within the third (5′-TCG TTT GTG ATT TCT ATA TAG C-3′) and fourth (5′-TCT ACA ACG CCG ACT TTA TAC T-3′) coding exons were used to create the two hairpin loops. For UAS-DopEcR-miR, two 22mers in the first (5′-CGT CCT GTC CAA CC T CCT CAT TAT C-3′) and third (5′-GCG CCA CCT TGG CGA TAT TAG T-3′) coding exons were used to create the two hairpin loops. miR sequences were synthesized in vitro (GeneArt) and then subcloned into pUAST using EcoRI and NotI.
Transgenic Animals
Transgenic animals were generated by standard techniques (Rainbow Transgenic Flies), as follows:
GAL4, GAL80, and Split GAL4 lines: DAT-B-GAL4 was integrated into the attP2 site using the PhiC31 system. TH-C-GAL80, TH-D-GAL80, and TH-F-GAL80 were generated by random P-element-mediated insertion. All of the other GAL80 lines were generated by PhiC31-mediated integration into JK22C sites. GAL4-DBD and GAL4-AD lines were inserted into attP2 and attP40, respectively, using the PhiC31 integrase system.
Killer Zipper lines: KZip+ transgenic lines for the 10 founder enhancer sequences were integrated into JK22C sites using the PhiC31 system.
HACK-generated lines: We used the HACK system as described by Lin and Potter (2016) to convert TH-D-GAL4 to TH-D-GAL4-DBDG4HACK. OK107-GAL4 was converted to OK107-GAL80G4HACK using the 76D5 GAL80 HACK donor line. Putative HACK lines were confirmed by PCR and by immunostaining.
FLP recombinase lines: The TH-FLP transgenic line was generated via random P-element-mediated integration. We screened 13 lines by crossing to TH-GAL4 and UAS-FRT-stop-FRT-CD8::GFP and performing immunostaining with anti-GFP and anti-TH to select the specific transgenic line with the broadest coverage in TH+ cells. The TH-C-FLP transgenic line was described previously (Liu et al., 2017).
microRNA lines: All miR lines were generated via random P-element-mediated insertion. Neuronally targeted TH miR transgenic lines were screened for viability and general health, cuticle pigmentation, and efficacy in TH knockdown, as assessed by immunostaining (Figures 6 and S5). For the DA receptor miR lines, the efficacy and specificity of knockdown was assessed by qPCR (Figure 6).
Immunostaining
Immunostaining of whole-mount brains or VNCs was performed as follows: brains or VNCs of 3- to 7-day-old flies were dissected in PBS, fixed in 4% paraformaldehyde (PFA) for 20 min, and subjected to chicken anti-GFP (1:1000, In-vitrogen), rabbit anti-TH (1:1000, Millipore), rat anti-CD8 (1:200, Invitrogen), mouse anti-nc82 (1:25, Developmental Studies Hybridoma Bank), or all four at 4°C for 18–40 hr, followed by incubation with fluorescent Alexa 488 anti-chicken (1:1000, Invitrogen), Alexa 488 anti-rat (1:200, Invitrogen), Alexa 568 anti-rabbit (1:1000, Invitrogen), Alexa 647 anti-mouse (1:1000, Invitrogen), or Cy3 anti-mouse (1:200, Jackson Immunoresearch) secondary antibodies for ~16 hr at 4°C. Brains or VNCs were mounted in Vectashield (Vector Laboratories) or SlowFade Gold using SS8X9-SecureSeal spacers (both Thermo Fisher Scientific). Images were obtained on an LSM510 or LSM700 confocal microscope (Zeiss) with 1-μm- or 3-μm-thick sections under 10x or 25x magnification. MultiColor FlpOut (MCFO) analysis of select driver combinations was performed as described previously (Nern et al., 2015) using rat anti-FLAG (1:200, Novus Biologicals) and rabbit anti-HA (1:300, Cell Signaling Technology) antibodies.
Brain Registration and Neuron Annotation
We used custom scripts and Jefferis laboratory scripts (https://github.com/jefferis/AnalysisSuite) in conjunction with LSM2NRRD (https://github.com/Robbie1977/lsm2nrrd), Computational Morphometry Toolkit (CMTK) (http://nitrc.org/projects/cmtk), and Fiji/ImageJ (Schindelin et al., 2012) to implement a brain registration pipeline to analyze confocal stack images for the Split GAL4 combinations. Registration of collected brain confocal stack images to the Janelia adult female Drosophila template brain (JFRC2/JFRC2010 derived by Virtual Fly Brain from Jenett et al., 2012) was performed using morphometric warping of the nc82 staining with CMTK and methods developed by Cachero et al., 2010.
Single neuron confocal stack images from FlyCircuit (Chiang et al., 2011) for TH-GAL4 were used to annotate our confocal stack images and were obtained from Virtual Fly Brain (Milyaev et al., 2012) and then aligned to the JFRC2010 template. For each Split GAL4 combination (Figures 3 and S3), we chose a brain confocal stack with a representative expression pattern, superimposed each FlyCircuit TH-GAL4+ neuron image, and then manually determined the identity of each labeled neuron.
Registered Split GAL4 images are hosted at Virtual Fly Brain (www.virtualflybrain.org), and flattened confocal images and raw .lsm files are publicly available at www.markwulab.net.
FLP Screen
To identify GAL4 drivers potentially labeling each of the non-PAM DA neurons, we used NBLAST (Costa et al., 2016) with the relevant single neuron confocal stack from FlyCircuit as a query image set. Using this approach, we identified ~200 GAL4 lines, which were then examined by performing whole-mount brain immunostaining of these GAL4 lines crossed to TH-FLP, TH-C-FLP, or both driving expression of UAS-FRT-stop-FRT-mCD8::GFP using anti-GFP and anti-TH antibodies, as described above. In cases in which DA neuron projections could not be visualized clearly, we performed a modified protocol to increase the signal, in which anti-CD8 and anti-GFP antibodies were used simultaneously on brains from flies bearing two copies of UAS-FRT-stop-FRT-CD8::GFP.
Social Space Behavior
Social space behavioral assays were performed largely as described in Simon et al. (2012). Briefly, flies were grown in bottles, and 1- to 2-day-old males were collected and kept in vials (40 per vial) for 2 days. For the socially enriched condition, flies were transferred into vials in groups of 40 for 2 more days, whereas for the isolated condition, individual flies were transferred to a single vial and maintained for 2 more days. For testing, flies were transferred to a triangular testing chamber (width and height, 15 cm), composed of glass plates with spacers held in a vertical position. After 20–40 min, when flies stopped moving, a digital image was taken using a camera. Images were imported into Fiji, and the location of flies was manually annotated by marking the thoraco-abdominal junction. The number of flies within 1 cm of a given fly was calculated using a custom Microsoft Excel macro. Experiments were conducted between ZT5 and ZT8, at 23°–25°C and ~60% relative humidity.
Statistical Analysis
Statistical analyses were performed with Prism 5 (GraphPad). For comparisons of two groups of normally distributed data, Student’s t tests were performed. For multiple comparisons, one-way ANOVAs followed by Tukey’s post hoc test were performed.
Supplementary Material
Highlights.
More than 40 transgenic lines allow for restricted expression in dopamine (DA) neurons
TH-based FLP recombinase permits isolation of DA neurons from any GAL4 driver
microRNA transgenic lines enable selective manipulation of DA signaling
HACK reagents can be used to replace any GAL4 with GAL80 or Split GAL4
Acknowledgments
We thank G. Rubin, Y.-N. Jan, and the Bloomington Drosophila Stock Center for fly stocks and S. Birman and Addgene for DNA plasmids. We thank S. Chin for pilot studies of the GAL80 lines, E. Marr for splinkerette mapping of HACK GAL80 donor lines, and J. Machamer for assistance with social space analyses. T.X. was supported by a China Scholarship Council Fellowship. M.C.W.H. was supported by NIH training grant T32HL110952. This work was supported by NIH grants R21NS088521 and R01NS079584 (to M.N.W.).
Footnotes
Supplemental Information includes supplemental references, seven figures, and six tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.068.
AUTHOR CONTRIBUTIONS
T.X. and M.C.W.H. performed the majority of experiments and data analysis. Q.L. performed the annotation of DA neurons and experiments related to miR lines. W.H. mapped the HACK donor lines and performed immunostaining. C.-C.L., D.T., and C.J.P. developed and provided reagents related to the HACK system. H.L. and B.H.W. developed and provided reagents related to the KZip+ system. T.X., M.C.W.H., and M.N.W. wrote the manuscript with input from all of the authors.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Ahrens MB, Li JM, Orger MB, Robson DN, Schier AF, Engert F, Portugues R. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature. 2012;485:471–477. doi: 10.1038/nature11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko OV, Chan YB, Li R, Kravitz EA. Single dopaminergic neurons that modulate aggression in Drosophila. Proc Natl Acad Sci USA. 2013;110:6151–6156. doi: 10.1073/pnas.1303446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ. Circuit modules linking internal states and social behaviour in flies and mice. Nat Rev Neurosci. 2016;17:692–704. doi: 10.1038/nrn.2016.125. [DOI] [PubMed] [Google Scholar]
- Aso Y, Siwanowicz I, Bräcker L, Ito K, Kitamoto T, Tanimoto H. Specific dopaminergic neurons for the formation of labile aversive memory. Curr Biol. 2010;20:1445–1451. doi: 10.1016/j.cub.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, Rubin GM. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife. 2014;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. Beyond the connectome: how neuromodulators shape neural circuits. BioEssays. 2012;34:458–465. doi: 10.1002/bies.201100185. [DOI] [PubMed] [Google Scholar]
- Berridge KC, Robinson TE. What is the role of dopamine in reward: hedonic impact, reward learning, or incentive salience? Brain Res Brain Res Rev. 1998;28:309–369. doi: 10.1016/s0165-0173(98)00019-8. [DOI] [PubMed] [Google Scholar]
- Birman S, Morgan B, Anzivino M, Hirsh J. A novel and major isoform of tyrosine hydroxylase in Drosophila is generated by alternative RNA processing. J Biol Chem. 1994;269:26559–26567. [PubMed] [Google Scholar]
- Björklund A, Dunnett SB. Dopamine neuron systems in the brain: an update. Trends Neurosci. 2007;30:194–202. doi: 10.1016/j.tins.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Burg ED, Langan ST, Nash HA. Drosophila social clustering is disrupted by anesthetics and in narrow abdomen ion channel mutants. Genes Brain Behav. 2013;12:338–347. doi: 10.1111/gbb.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachero S, Ostrovsky AD, Yu JY, Dickson BJ, Jefferis GS. Sexual dimorphism in the fly brain. Curr Biol. 2010;20:1589–1601. doi: 10.1016/j.cub.2010.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabresi P, Picconi B, Tozzi A, Ghiglieri V, Di Filippo M. Direct and indirect pathways of basal ganglia: a critical reappraisal. Nat Neurosci. 2014;17:1022–1030. doi: 10.1038/nn.3743. [DOI] [PubMed] [Google Scholar]
- Chen CH, Huang H, Ward CM, Su JT, Schaeffer LV, Guo M, Hay BA. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, Shih CT, Wu JJ, Wang GT, Chen YC, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Manton JD, Ostrovsky AD, Prohaska S, Jefferis GS. NBLAST: rapid, sensitive comparison of neuronal structure and construction of neuron family databases. Neuron. 2016;91:293–311. doi: 10.1016/j.neuron.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan MJ, Luan H, Shropshire WC, Sutcliffe B, Cocanougher B, Scott RL, Frechter S, Zlatic M, Jefferis GSXE, White BH. Facilitating neuron-specific genetic manipulations in Drosophila melanogaster using a Split GAL4 repressor. Genetics. 2017;206:775–784. doi: 10.1534/genetics.116.199687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economo MN, Clack NG, Lavis LD, Gerfen CR, Svoboda K, Myers EW, Chandrashekar J. A platform for brain-wide imaging and reconstruction of individual neurons. eLife. 2016;5:e10566. doi: 10.7554/eLife.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Hannan F, Reale V, Hon YY, Kousky CT, Evans PD, Hall LM. Cloning and functional characterization of a novel dopamine receptor from Drosophila melanogaster. J Neurosci. 1996;16:3925–3933. doi: 10.1523/JNEUROSCI.16-12-03925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez RW, Akinleye AA, Nurilov M, Feliciano O, Lollar M, Aijuri RR, O’Donnell JM, Simon AF. Modulation of social space by dopamine in Drosophila melanogaster, but no effect on the avoidance of the Drosophila stress odorant. Biol Lett. 2017;13:20170369. doi: 10.1098/rsbl.2017.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Smith SM, Marcus DS, Andersson JL, Auerbach EJ, Behrens TE, Coalson TS, Harms MP, Jenkinson M, Moeller S, et al. The Human Connectome Project’s neuroimaging approach. Nat Neurosci. 2016;19:1175–1187. doi: 10.1038/nn.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin CC, Potter CJ, Clandinin TR. A versatile in vivo system for directed dissection of gene expression patterns. Nat Methods. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Han KA, Millar NS, Grotewiel MS, Davis RL. DAMB, a novel dopamine receptor expressed specifically in Drosophila mushroom bodies. Neuron. 1996;16:1127–1135. doi: 10.1016/s0896-6273(00)80139-7. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Cruz L, Lovick JK, Guo M. Developmental analysis of the dopamine-containing neurons of the Drosophila brain. J Comp Neurol. 2017;525:363–379. doi: 10.1002/cne.24069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn MG, Ren Y, McBride EW, Reveillaud I, Beinborn M, Kopin AS. A Drosophila dopamine 2-like receptor: molecular characterization and identification of multiple alternatively spliced variants. Proc Natl Acad Sci USA. 2002;99:14554–14559. doi: 10.1073/pnas.202498299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. Encephalization, neuronal excess, and neuronal index in rodents. Anat Rec (Hoboken) 2007;290:1280–1287. doi: 10.1002/ar.20598. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleman K, Vrontou E, Krüttner S, Yu JY, Kurtovic-Kozaric A, Dickson BJ. Dopamine neurons modulate pheromone responses in Drosophila courtship learning. Nature. 2012;489:145–149. doi: 10.1038/nature11345. [DOI] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T, Chang JS, Dankert H, Zelnik L, Kim YC, Han KA, Wolf FW, Perona P, Anderson DJ. Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron. 2009;64:522–536. doi: 10.1016/j.neuron.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lin CC, Potter CJ. Editing transgenic DNA components by inducible gene replacement in Drosophila melanogaster. Genetics. 2016;203:1613–1628. doi: 10.1534/genetics.116.191783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Prokop-Prigge KA, Preti G, Potter CJ. Food odors trigger Drosophila males to deposit a pheromone that guides aggregation and female oviposition decisions. eLife. 2015;4:e08688. doi: 10.7554/eLife.08688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol. 2012a;22:2114–2123. doi: 10.1016/j.cub.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Plaçais PY, Yamagata N, Pfeiffer BD, Aso Y, Friedrich AB, Siwanowicz I, Rubin GM, Preat T, Tanimoto H. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature. 2012b;488:512–516. doi: 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- Liu Q, Tabuchi M, Liu S, Kodama L, Horiuchi W, Daniels J, Chiu L, Baldoni D, Wu MN. Branch-specific plasticity of a bifunctional dopamine circuit encodes protein hunger. Science. 2017;356:534–539. doi: 10.1126/science.aal3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Gallen CL, Clandinin TR. Whole-brain calcium imaging reveals an intrinsic functional network in Drosophila. Curr Biol. 2017;27:2389–2396. e4. doi: 10.1016/j.cub.2017.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Davis RL. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E. Neuromodulation of neuronal circuits: back to the future. Neuron. 2012;76:1–11. doi: 10.1016/j.neuron.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella S, Mann K, Scott K. Dopaminergic modulation of sucrose acceptance behavior in Drosophila. Neuron. 2012;73:941–950. doi: 10.1016/j.neuron.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milyaev N, Osumi-Sutherland D, Reeve S, Burton N, Baldock RA, Armstrong JD. The Virtual Fly Brain browser and query interface. Bioinformatics. 2012;28:411–415. doi: 10.1093/bioinformatics/btr677. [DOI] [PubMed] [Google Scholar]
- Nässel DR, Elekes K. Aminergic neurons in the brain of blowflies and Drosophila: dopamine- and tyrosine hydroxylase-immunoreactive neurons and their relationship with putative histaminergic neurons. Cell Tissue Res. 1992;267:147–167. doi: 10.1007/BF00318701. [DOI] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, Rubin GM. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci USA. 2015;112:E2967–E2976. doi: 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieoullon A. Dopamine and the regulation of cognition and attention. Prog Neurobiol. 2002;67:53–83. doi: 10.1016/s0301-0082(02)00011-4. [DOI] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panser K, Tirian L, Schulze F, Villalba S, Jefferis GSXE, Bühler K, Straw AD. Automatic segmentation of Drosophila neural compartments using GAL4 expression data reveals novel visual pathways. Curr Biol. 2016;26:1943–1954. doi: 10.1016/j.cub.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuro-anatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza SM, Scheffer LK, Chklovskii DB. Toward large-scale connectome reconstructions. Curr Opin Neurobiol. 2014;25:201–210. doi: 10.1016/j.conb.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Potter CJ, Luo L. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS One. 2010;5:e10168. doi: 10.1371/journal.pone.0010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Riemensperger T, Isabel G, Coulom H, Neuser K, Seugnet L, Kume K, Iché-Torres M, Cassar M, Strauss R, Preat T, et al. Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc Natl Acad Sci USA. 2011;108:834–839. doi: 10.1073/pnas.1010930108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AF, Chou MT, Salazar ED, Nicholson T, Saini N, Metchev S, Krantz DE. A simple assay to study social behavior in Drosophila: measurement of social space within a group. Genes Brain Behav. 2012;11:243–252. doi: 10.1111/j.1601-183X.2011.00740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava DP, Yu EJ, Kennedy K, Chatwin H, Reale V, Hamon M, Smith T, Evans PD. Rapid, nongenomic responses to ecdysteroids and catecholamines mediated by a novel Drosophila G-protein-coupled receptor. J Neurosci. 2005;25:6145–6155. doi: 10.1523/JNEUROSCI.1005-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, Kume K. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci. 2012;15:1516–1523. doi: 10.1038/nn.3238. [DOI] [PubMed] [Google Scholar]
- van Swinderen B, Andretic R. Dopamine in Drosophila: setting arousal thresholds in a miniature brain. Proc Biol Sci. 2011;278:906–913. doi: 10.1098/rspb.2010.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell S. Reinforcement signalling in Drosophila; dopamine does it all after all. Curr Opin Neurobiol. 2013;23:324–329. doi: 10.1016/j.conb.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook A, Braver TS. Dopamine does double duty in motivating cognitive effort. Neuron. 2016;89:695–710. doi: 10.1016/j.neuron.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- White KE, Humphrey DM, Hirth F. The dopaminergic system in the aging brain of Drosophila. Front Neurosci. 2010;4:205. doi: 10.3389/fnins.2010.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RA. Dopamine, learning and motivation. Nat Rev Neurosci. 2004;5:483–494. doi: 10.1038/nrn1406. [DOI] [PubMed] [Google Scholar]
- Wisor JP, Nishino S, Sora I, Uhl GH, Mignot E, Edgar DM. Dopaminergic role in stimulant-induced wakefulness. J Neurosci. 2001;21:1787–1794. doi: 10.1523/JNEUROSCI.21-05-01787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TR. The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv Genet. 1987;24:127–222. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.