Abstract
Intrinsically disordered proteins (IDPs) have roles in myriad biological processes and numerous human diseases. However, kinetic and amplitude information regarding their ground-state conformational fluctuations have remained elusive. We demonstrate using nuclear magnetic resonance (NMR)-based relaxation dispersion that the D2 domain of p27Kip1, a prototypical IDP, samples multiple discrete, rapidly exchanging conformational states. By combining NMR with mutagenesis and small angle X-ray scattering (SAXS), we show that these states involve aromatic residue clustering through long-range hydrophobic interactions. Theoretical studies have proposed that small molecules bind promiscuously to IDPs, causing expansion of their conformational landscapes. However, based on previous NMR-based screening results, we show here that compound binding only shifts the populations of states that existed within the ground-state of apo p27-D2 without changing the barriers between states. Our results provide atomic resolution insight into how a small molecule binds an IDP and emphasize the need to examine motions on the low microsecond timescale when probing these types of interactions.
Keywords: Intrinsically disordered protein, Nuclear Magnetic Resonance Spectroscopy, Relaxation Dispersion, small molecule interaction
SYNOPSIS TOC
INTRODUCTION
Intrinsically disordered proteins (IDPs) and disordered regions (IDRs) within proteins mediate myriad cellular functions1 and underlie numerous human diseases.1–3 While the functions of many are understood1,3, open questions remain regarding the dynamic features of IDPs and how they affect interactions with other macromolecules and small molecule ligands.4–6 IDPs are inherently flexible and are generally described as ensembles of disordered conformations. Nuclear Magnetic Resonance spectroscopy (NMR) has allowed characterization of the physical features of IDPs7 under near physiological conditions without chemical modification and at atomic resolution. NMR and other methods8,9 have revealed that some IDPs transiently populate secondary structures7,10,11 and experience long-range tertiary interactions12–14; fluctuations of these features define the conformational landscape of an IDP and, in turn, mediate function. However, the timescales at which these states interconvert and, thus, the features of the conformational energy landscape remain unknown. Single molecule fluorescence correlation spectroscopy15,16 and NMR-based studies have identified nanosecond fluctuations for the backbone of IDPs that have been attributed to main chain intersegmental motion.8,17
Conventional NMR-based relaxation techniques that monitor dipolar and chemical shift anisotropy relaxation mechanisms can report on motion of nuclei on timescales up to nanoseconds at atomic resolution.18 Analysis of conventional relaxation data typically invokes use of the model-free formalism.19 However, this is predicated on separation of the timescales for global molecular tumbling and local internal motions. Because local and global motions are indistinguishable for unfolded proteins, IDPs, or IDRs, spectral density mapping has been applied to characterize backbone fluctuations that occur on the picosecond to nanosecond timescale.17,20,21 More recently, several groups have performed relaxation experiments at a range of static magnetic field strengths21 and temperatures.8 For the IDP, Engrailed, a comprehensive relaxation dataset was analyzed using a novel model which interpreted the data using an array of global correlation times, between picoseconds and nanoseconds, that subsequently allowed determination of nucleus specific motional amplitudes for each residue.21 A more recent study of an IDP included temperature-dependent conventional relaxation data. Three distinct correlation times were used to describe the relaxation data coupled with an Arrhenius-type analysis which provided nucleus specific motional activation energies.8 Proton relaxometry, which provides bulk relaxation information about a protein, allows for a more complete sampling of the spectral density function that describes relaxation because a broad range of Larmor frequencies can be measured. This technique has recently been applied to several IDPs.22 The ability to measure the bulk proton relaxation rate across a broad range of magnetic fields (0.5 mT to 1.1 T) has allowed detection of nanosecond timescale correlated motions for those IDPs.22 However, the aforementioned studies have not characterized the kinetics associated with the transitions between discrete conformational states that are related to the formation of long-range interactions.
IDPs/IDRs have diverse regulatory roles23 and have associations with a wide variety of human diseases.24 Therefore, IDPs are now considered as therapeutic targets25; however, drug discovery against IDPs is challenging due to their highly dynamic nature. Remarkably, small molecules that modulate the activity of a variety of IDPs/IDRs have been identified26–32 and, in a few cases, NMR was used to map small molecule:IDP/IDR interactions.27,32 However, the mechanism(s) of these interactions and their influence on IDP/IDR conformational landscapes are unknown. Theoretical studies have led to several models for small molecule:IDP interactions; in one, termed “ligand clouds/protein clouds”, a small molecule binds many sites within an IDP that fluctuates between many distinct conformations5 while in a second, small molecule binding was proposed to increase the number of conformational states of an IDP4; however, experimental confirmation of these theoretical models remains unavailable. Here, we characterized the conformational landscape of a prototypical IDP, p27Kip1, in the absence and presence of a small molecule, to provide insight into the mechanism and conformational consequences of small molecule:IDP interactions.
p27Kip1 (p27) is largely disordered in isolation and regulates eukaryotic cell division33 by folding upon binding and inhibiting nuclear cyclin-dependent kinase (Cdk)/cyclin complexes.34 Two regions within the N-terminal kinase inhibitory domain (KID) of p27 termed sub-domains D1 and D2 specifically bind to cyclin A and Cdk2, respectively, within the Cdk2/cyclin A complex while sub-domain LH forms a kinked α-helix that links the two other sub-domains.35 The Cdk inhibitory activity and stability of p27 is modulated by phosphorylation of tyrosine residues within the KID and this regulatory mechanism is aberrantly activated in chronic myelogenous leukemia33 and breast cancer.36 In addition, also in breast cancer, aberrant phosphorylation of p27 on a threonine residue within the disordered C-terminal regulatory domain37 causes cytoplasmic mislocalization, which is associated with increased cell migration and metastasis.38–41 Furthermore, in sensory and non-sensory epithelial hearing cells, terminal differentiation is maintained by p27 expression, which enforces cell cycle exit.42,43 Genetic ablation of p27 in these cells allowed their proliferation and suggested an avenue toward hearing cell regeneration through inhibition of p27.44 Motivated by these various disease associations and potential therapeutic strategies involving p27, we recently discovered a small molecule inhibitor through an NMR-based fragment screen that bound specifically to the D2 sub-domain of p27 (p27-D2) and caused partial reactivation of Cdk2 kinase activity when titrated into the inhibited p27-D2/Cdk2/cyclin A complex.32 The availability of this inhibitor (termed SJ403 here) provided the opportunity to experimentally characterize the mechanism of small molecule:IDP interactions. Here, we used NMR high-power relaxation dispersion (RD), small angle X-ray scattering (SAXS) and mutagenesis to describe the conformational landscape of p27-D2 and how this is altered by binding to SJ403. Our results show that, in contrast to proposals from theoretical studies, in this case small molecule binding induces a population shift within the conformational landscape of p27-D2.
RESULTS
Large amplitude NMR RD detects rapid interconversion between multiple states on two distinct microsecond regime timescales for an IDP We utilized NMR relaxation dispersion methods to characterize conformational fluctuations within p27-D2 with atomic spatial resolution and high temporal resolution. NMR RD monitors the process of chemical exchange which causes nuclei to sample different conformational states and provides information on, i) the exchange lifetime ((τex = 1/kex); where kex is the rate of exchange) for the process, ii) the populations (pi) of the different states, and iii) the difference in structural environment between populated states [reported as the chemical shift difference between the states (Δω)]. The NMR chemical shift timescale defines the exchange regime; for the transient events studied here, they were anticipated to be in the fast regime (τexΔω)−1 > 1) which allows for the determination of τex and conformational amplitudes (Φex = pApbΔω2 for a two-state exchange process). Spin-lattice rotating-frame (R1ρ) RD spectroscopy monitors relaxation rates for individual nuclei as a function of increasing effective radiofrequency field strengths ( ; υRF and υoff are the amplitude of the radiofrequency field and offset between the applied radiofrequency and the frequency of the observed resonance in Hz, respectively). The effective radiofrequency field acts to spin-lock a queried magnetization. If chemical exchange occurs within the fast regime, increases in ωeff decrease departures of the magnetization from the effective field thereby reducing the R1ρ rate. Here, we utilized recently developed large amplitude spin-lock field strength R1ρ experiments applied to 1HN nuclei45 that use large effective refocusing fields (“high-power”; ωeff up to 2.2·105 rad s−1) to monitor exchange processes with τex values as low as ~4 µs (limiting τex ~ 1/ωeff) (Figure S2).45–48 The implementation of the 1HN-R1ρ experiment used here is also advantageous because an effective transverse relaxation rate (R2,eff) can be calculated. Therefore, the longitudinal component of R1ρ can be removed. We applied this methodology at temperatures from 274 to 290 K to characterize the conformational landscape of p27-D2 in isolation and when bound to SJ403.
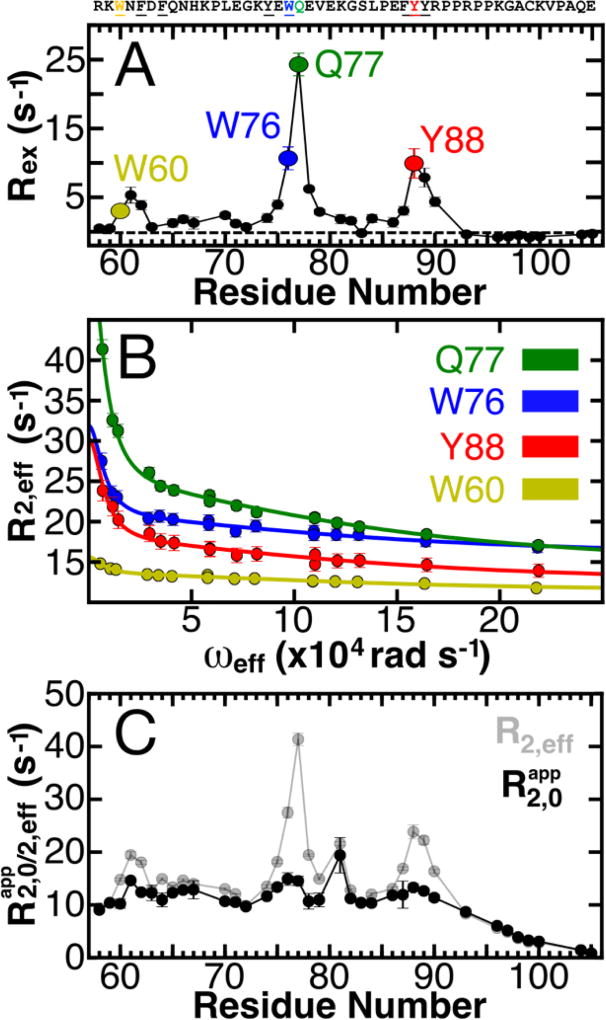
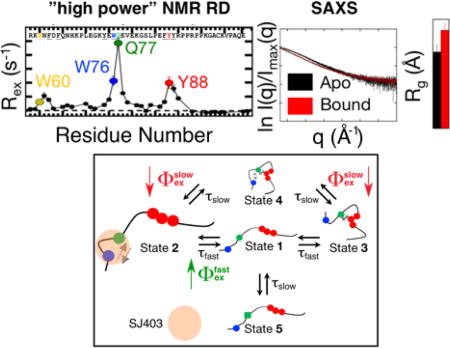
Significant contributions of conformational exchange were observed for many 1HN nuclei in wild-type p27-D2 at 274 K (Figure 1A). Of 36 well resolved 1HN resonances for p27-D2 (p27-D2 is comprised of 48 residues, in total; Figure S1), 22 resonances exhibited quantifiable conformational exchange (Figure 1A). Analysis of RD curves revealed that data for 11 of the 22 1HN nuclei, which exhibited large values of Rex ( ; on average ~8 s−1), could not be described by a standard two-state model but instead required fitting with a model that has two independent exchange lifetimes49,50 (Figure 1A, 1B and Figure S4–5; see the Supporting Information for details pertaining to the quantification of NMR RD). For these 11 residues, curve fitting indicated two processes whose kinetic parameters differ by one order of magnitude and are manifested in the RD data as a sum of two Lorentzian functions. This is manifested in our data, as follows. The slower kinetic process is refocused and no longer contributes to the observed RD at ωeff values greater than 5·104 rad s−1. In contrast, the fast kinetic process contributes to RD at ωeff values less than 5·104 rad s−1 and is only readily apparent at the higher ωeff values (Figure 1B). Therefore, the large amplitude 1HN-R1ρ experiment was necessary to resolve the two processes. Importantly, large Rex values were observed for several aromatic residues within three regions of p27-D2 (Figure 1A), including W60-F62 (termed the W60 region), E75-E78, which contains W76 and Q77 (the W76 region; note that Q77 exhibited the largest Rex value), and F87-Y88 (the Y88 region); many of these aromatic moieties interact with Cdk2 within the ternary p27/Cdk2/cyclin A complex35 (Figure S6). These RD results, we believe for the first time, define the kinetics for transitions between discrete conformational states for a region of an IDP (p27-D2).
Figure 1. Defining the conformational landscape of p27-D2.
(A) Large effective refocusing field strength (ωeff) and low temperature 1HN relaxation dispersion (RD) experiments reveal multi-state conformational exchange for the intrinsically disordered protein, p27-D2. Conformational exchange is sensed by many residues within p27-D2, which exhibit quantifiable exchange contributions ( ; the dashed line indicates a Rex value of zero). The greatest observed conformational exchange is observed for residues in p27-D2 within three aromatic clusters, including W60 (yellow), W76 (blue), and Y88 (red); the 1HN displaying the largest amplitude of motion is Q77 (green). (B) The observed RD monitors the change in the effective transverse relaxation rate (R2,eff) as a function of ωeff and reports that some residues undergo conformational exchange that has two kinetic phases. Solid lines correspond to global fits of all RD data to a thermodynamic model that describes nuclei which sense two separate kinetic phases. Data presented in A & B were collected at 274 K. (C) Plot of R2,eff measured at low ωeff (grey points) where a maximal contribution of microsecond exchange is detected as compared to the transverse relaxation rate due to the apparent intrinsic fast nanosecond exchange ( ; black points) across all residues. was determined from the fitted RD data and shows that large amplitude 1HN RD permits extensive sampling of Rex by using high ωeff which substantially quenches RD. The fast microsecond exchange detected in p27-D2 accounts for the elevated R2,eff values and once these are taken into account, a flat, featureless profile is observed as expected for a prototypical disordered protein.
To more completely define the conformational landscape of p27-D2 captured by the NMR RD data (Figure 1A & B), we performed temperature dependent 1HN-R1ρ RD experiments for apo p27-D2 from 274 to 290 K (Figures S4–5; Tables S2–3). All 1HN nuclei that displayed two kinetic phase exchange behavior over this temperature range were then globally fit using a model in which the states are separated by two different activation barriers defined by two different Arrhenius-type activation energies . The global fit yielded and values of 49.7 ± 10.5 kJ/mol (attempt frequency: 3.8·1014 ± 1.1·1014) and 46.6 ± 4.6 kJ/mol (attempt frequency: 6.0·1012 ± 3.0·1012), respectively (Figure 1A; see Supporting Information). Please note that over this narrow temperature range the attempt frequencies cannot be known precisely.51 Measuring RD at temperatures greater than 290 K was not possible because the exchange lifetimes for the fast process (which is characterized by large conformational amplitudes), becomes too rapid to be detected with the current experimental parameters. The slow process is characterized by small conformational amplitudes (Table S2), which limit detection at elevated temperatures.
The multi-temperature RD data indicates that p27-D2 undergoes transitions with two different exchange lifetimes; these kinetically accessible states constitute the “ground-state” of p27-D2 (Table S2). Although we describe these processes as being slow and fast, extrapolation based on the fitted Ea values to 310 K (physiological temperature) shows that the slower (τslow) and faster (τfast) processes would indeed be rapid, with exchange lifetimes of ~12 µs and ~620 ns, respectively. A fit to an Eyring-type equation, which may not be applicable for protein conformational transitions, was also used to analyze the multi-temperature 1HN RD data and yielded similar activation energy values (τ G‡) ( ; see the Supporting Information).
Two mechanisms contribute to the effective transverse relaxation rate (R2,eff); local conformational fluctuations on the low nanoseconds timescale which create intrinsic relaxation due to dipolar and chemical shift anisotropy mechanisms and conformational exchange (Rex), which is caused by the modulation of the isotropic chemical shift on a supra-ns timescale and manifests as an additional source of relaxation. These two contributions can be deconvoluted through analysis of RD data.49 Residues in IDPs, IDRs, and unfolded proteins often exhibit elevated R2,eff values8,12,13,52, which have been attributed to transient long-range tertiary contacts12,13 and transient secondary structure.8 However, the kinetic and thermodynamic parameters associated with these transient features have not been measured. The relaxation behavior of a disordered chain is dominated by segmental motions that allow individual 1HN nuclei to independently reorient and is influenced by the sequence and persistence length of the polypeptide chain.8,53 For a completely disordered polypeptide, the transverse relaxation profile is expected to be flat and featureless with a gradual decrease in relaxation rates near the termini. The RD data recorded with the lowest ωeff values (with maximal contributions of conformational exchange) revealed elevated R2,eff values for residues within several regions of the p27-D2 sequence, especially those containing aromatic residues (Figure 1C, grey points). In contrast, the apparent R2,0 values (Figure 1C, black points) for p27-D2, which were determined from the RD fit procedure, are relatively continuous with few extrema showing that only the relaxation effects of fast segmental motions are retained (as expected for a disordered polypeptide) and that the contribution of Rex is largely quenched. We note that, due to the manner in which the 1HN RD data is recorded47, is an approximate measure between the transverse proton relaxation rate and the longitudinal nitrogen relaxation rate. This is because of the constant relaxation period implemented after the spin-lock period which permits facile conversion of R1ρ to R2,eff (see the Supporting Information). However, cannot be directly compared to a conventional R2,0 relaxation rate. Our ability to sample the contributions of Rex for 1HN nuclei was enhanced through the use of large amplitude ωeff RD methods45–48, which overcame limitations associated with measurements of either 13C or 15N relaxation (Figure S2). Application of large amplitude ωeff 1HN RD methods in the future will allow detection of low microsecond motions within other IDPs, IDRs, and unfolded proteins (e.g., as has been proposed to exist in a recent report for the IDP Engrailed21) and unfolded proteins that exhibit elevated transverse relaxation rates.8,12,13,52 This approach can complement studies that utilize conventional relaxation methods that probe picosecond to nanosecond motions.
The ground-state fluctuations for p27-D2 involve differential hydrophobic clustering
We next sought to elucidate the physical basis of the observed exchange behavior for 1HN nuclei within p27-D2 (Figure 1). 1HN nuclei are exquisitely sensitive to ring current effects. Therefore, the observed RD may involve ring current effects that are due to transient shuffling of hydrophobic residues within clusters. This could possibly be enhanced by aromatic residue sidechain rotamer interconversion, which, due to clustering, may be slow enough to contribute to the observable RD. Given the large variance of Rex values for aromatic residues within p27-D2, the details of the physical processes that cause the RD for each 1HN are anticipated to be complex (Figure 1A). Therefore, with the current data, we cannot determine whether the motion that underlies the observed 1HN RD is driven by sidechain fluctuations that are imprinted on the backbone, by only backbone fluctuations, or a combination of the two. We performed mutagenesis of aromatic residues that exhibited large R2,eff and Rex values (Figure 1) and studied the residual motions through 1HN RD measurements at 274 K to gain deeper insight into the exchange processes experienced by p27-D2.
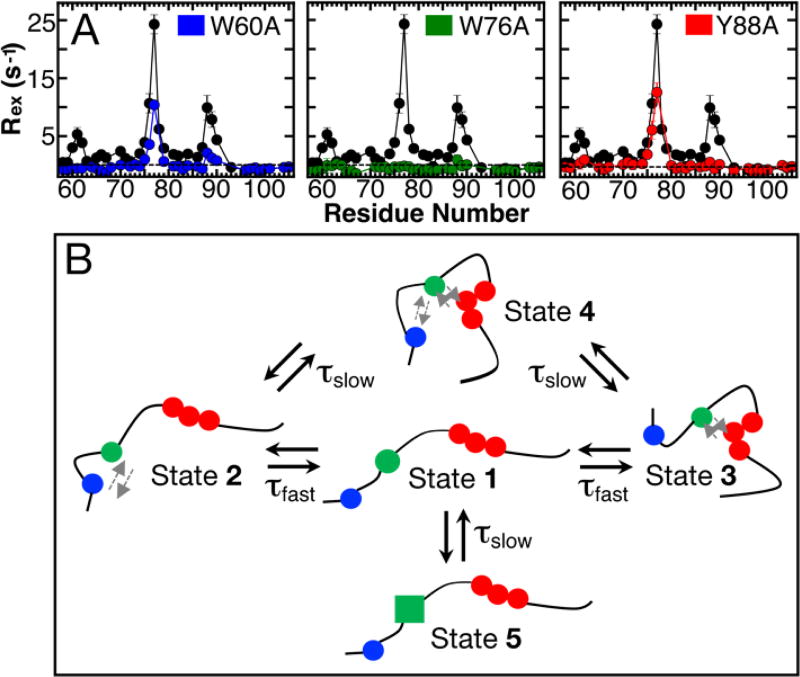
Analysis of differences in chemical shift values (1HN, 15N, and secondary 13Cα shifts; Figure S7) between all mutants and wild-type p27-D2 indicated no significant changes in average chemical shift values except near the mutation site, as expected. However, the magnitude of Rex did change significantly for all p27-D2 mutants as compared to the wild-type protein (Figure 2A), indicating changes in the conformational landscapes of the mutant p27-D2 constructs. Mutation of W60 to alanine (W60A) was associated with Rex values near zero for residues surrounding the mutation site while within the Y88 region the values were reduced by more than 50% and for W76 and surrounding residues the values were partially reduced (Figure 2A, blue circles). Similar reductions in Rex values for residues distal to the mutation site were observed upon mutation of Y88 to alanine but exchange in the W76 region persisted (Y88A; Figure 2A, red circles). Mutation of centrally positioned W76 (W76A), however, uniformly reduced Rex values to zero throughout the p27-D2 sequence and thus completely quenched the µs timescale motions observed in wild-type p27-D2 and the other two aromatic residue mutants (Figure 2A, green circles). In addition to the effects of mutations on Rex values for p27-D2 (Figure 2A), we also observed effects on values (Figure S9).
Figure 2. Mutagenesis of p27-D2 affect RD.
(A) Contributions of Rex for different point mutants of p27-D2 in which critical aromatic residues were substituted with alanine. Mutational analysis showed that W76 (green) abrogates all microsecond exchange in p27-D2 whereas the other mutants, W60A (blue) and Y88A (red), exhibit little exchange near the site of mutation and reduced exchange at the two non-mutated sites. Points in black correspond to wild-type p27-D2 data and the dashed black line is drawn at zero which indicates no detected exchange. Analysis of residual RD within the p27-D2 mutants can be found in Figure S8 and Table S4. (B) Schematic of the proposed model for observed microsecond p27-D2 dynamics. The three critical hydrophobic residues are colored in blue, green, and red and correspond to W60, W76 in the central region W76, and F87-Y89, respectively. The grey arrows indicate transient hydrophobic interactions.
The results above on the effects of mutations on residue-specific Rex values (Figure 2A) clearly indicated that the central region of p27-D2 (including residues W76 & Q77) is a nexus for interactions that create a complex conformational landscape. We developed a multi-state kinetic scheme to represent this landscape that is consistent with our experimental findings, as follows. First, we define State 1 (Figure 2B), which is poised for transitions that cause the observed RD. Next, we invoke transitions from State 1 to two states, States 2 and 3, through formation of interactions between the central region and either the W60 region or the Y88 region, respectively. These transitions, we propose, both exhibit the fast process kinetics (Table S4). We further invoke transitions of States 2 and 3 to State 4 through collective interactions of all three aromatic residue-containing regions of p27-D2 that are both connected by the slower kinetic process (Table S4). In this scheme, two similar fast and slow processes, respectively, contribute to the RD observed for p27-D2. The topology and kinetic features of States 1 – 4 in this scheme are consistent with the results of our analyses of RD data for the W60A and Y88A p27-D2 mutants—termed the “distal” aromatic residue mutants.
For example, these mutations completely abrogated exchange for residues near the mutated site and abrogated slow processes, but did not affect fast processes, for the non-mutated distal site (Figure S8 and Table S4). In particular, the conformational amplitudes for Y88 when W60 was mutated, and for W60 when Y88 was mutated, were the same (within error) as that for these residues in wild-type p27-D2 (Tables S1 and S4). These mutations did not affect values because the molecular populations associated with the slow transitions re-equilibrated amongst the remaining, unaltered states, giving values similar to those observed in wild-type p27-D2. In our scheme (Figure 2B), mutation of W60 eliminates States 2 and 4, leaving only transitions between States 1 and 3 to contribute to RD for residues in the W76 and Y88 regions. A similar scenario explains the effects of mutation of Y88 on the exchange behavior and RD for residues near W60 and W76. However, a four-state scheme does not account for the observation of both slow and fast exchange for residues in the central region of p27-D2 (W76 and Q77) in the two distal mutants (Table S4). To account for this, we invoked an additional slow process between State 1 and State 5, which we propose is a conformation in which interactions between W76 and either of the distal aromatic regions are less favorable than in State 1 (indicated by the green square in Figure. 2B, State 5). Analysis of RD data for W76 and Q77 in the W60A and Y88A mutants showed that the value of was unchanged while that for decreased by 50% (Table S4). The decrease by a factor of two for indicates that at least two slow processes exist in wild-type p27-D2. We note that W76 within the Y88A mutant showed statistical significance for fitting with the two exchange lifetime model (with independent fast and slow process) but the errors were larger than the fitted parameters. We attribute this to a larger measurement error for RD collected with the Y88A mutant (factor of 2 difference relative to wild-type p27-D2 and other mutants). However, Q77 in the same mutant exhibited statistically robust multi-state behavior, supporting the existence of State 5 in our conformational landscape scheme. Our scheme includes two fast processes and three slow processes that, respectively, exhibit similar kinetic parameters; this type of multi-pathway kinetic scheme has previously been observed for ubiquitin and GB3.51,54 Furthermore, our scheme is consistent with the observation that mutation of W76 to alanine completely abrogated RD in p27-D2. W76 participates in all of the inter-state transitions; therefore, the W76A mutation eliminates all possible structural transitions and consequently the associated NMR observable RD. We acknowledge that other kinetic schemes and interconversion pathways may be possible; however, to avoid over interpretation55, we argue that our model (Figure 2B) is the simplest that is consistent with our experimental data and describes the detected ground-state conformation transitions of p27-D2. Additional information about the proposed model is available in the Supporting Information. In summary, the conformational landscape of p27-D2 is comprised of minimally five states, many of which involve pairwise or ternary interactions between three aromatic residue-containing regions, that interconvert on either ~100s ns or ~10 µs (at 310 K) timescales. Although, the temperature range employed here was narrow, which can affect the validity of the Arrhenius extrapolation, the temperature dependence of 1HN and 15N chemical shifts for all residues follow a monotonic trend at temperatures up to 308 K (Figure S10). This indicates that the average ground-state behavior is preserved even with extrapolations derived from low temperature measurements. We propose that hydrophobic interactions, especially those involving aromatic amino acid sidechains, mediate the interactions captured in our model.
1HN RD was repeated on a wild-type sample containing 1 M urea (Figure S11) and revealed that Rex was undetectable for the W60 and Y88 regions of p27-D2, corroborating that the ground-state fluctuations of p27-D2 are due to transient hydrophobic interactions. The molar ratio of urea to p27-D2 was set to two for all experiments performed with urea. Urea is well known to disrupt hydrophobic interactions.56 It is important to note that the addition of urea to p27-D2 caused minimal chemical shift changes, consistent with negligible effects on the average local conformation34,57 and indicating that p27-D2 is similarly locally disordered in the absence and presence of urea (Figure S11).
Nonetheless, the motion that allows distal aromatic regions to form transient hydrophobic interactions was highly sensitive to the urea. This was evidenced by the absence of Rex, which yielded an R2,eff profile (Figure S11) that was as flat and featureless as the profile for urea-free p27-D2 (Figure 1C). Although we cannot provide detailed atomic models to explain the slower kinetic processes (Figure 2B; between States 2 and 4, 3 and 4, and States 1 and 5), we argue that these involve conformational rearrangements which modulate the favorability for different hydrophobic interactions.
NMR RD and SAXS reveal a population shift for p27-D2 when bound to a small molecule
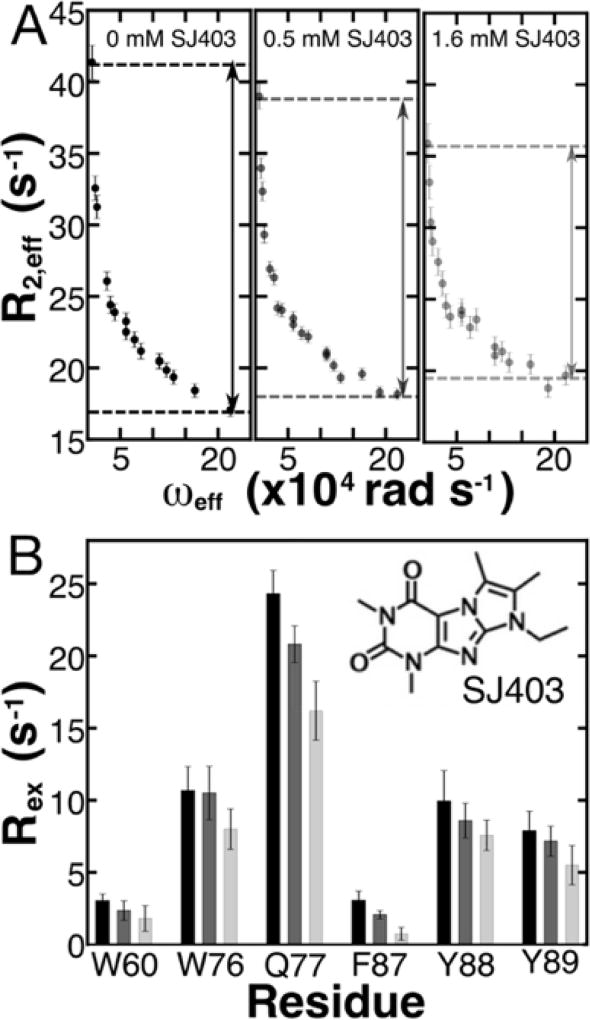
Several recent studies have identified small molecules that bind to IDPs26–32; however, the physical nature of these interactions is still unknown. We previously demonstrated using NMR that the small molecule, SJ403 (full name, SJ572403), binds specifically to aromatic residues within p27-D2.32 Based upon our findings from NMR RD experiments on aromatic residue dynamics discussed above, we hypothesized that the binding of SJ403 would alter the dynamic conformational landscape of p27-D2. We repeated 1HN RD experiments at 274 K in the presence of the small molecule and observed that Rex decreased with increasing concentration of SJ403 (Figure 3). For all residues that reported measureable RD (Figure 3 & S11; Table S1, 2, 5), Rex values decreased on average by approximately 50% from a [p27-D2]:[SJ403] ratio of 1:0 to 1:4. The decrease in the average value of Rex (Figure 3) was statistically significant with p-values of 1.2·10−3 and 7.16·10−14 when 500 µM and 1600 µM SJ403 were added, respectively, as all queried resonances showed decreases in Rex (Figure S12). To more fully understand the effects of SJ403 on the conformational landscape of p27-D2, we repeated the temperature dependent 1HN RD experiments and globally reanalyzed data for 9 1HN nuclei that displayed two separate kinetic phases (Figure S13 and Table S5) to quantify changes in exchange kinetics and their conformational amplitudes .
Figure 3. Binding of SJ403 to p27-D2 causes concentration-dependent reductions in Rex for 1HN nuclei.
(A) Increasing concentration of SJ572403 (SJ403) reveals a concentration-dependent decrease in the observable contribution of conformational exchange. The black, grey and light grey points correspond to 1HN RD performed with 0 µM, 500 µM and 1600 µM, respectively, with a constant p27-D2 concentration of 400 µM at 274 K. The example shown in A is for residue Q77. The dependence of relaxation on compound concentration is readily observed in the RD data, as highlighted by the dashed lines in the three panels. The value of Rex decreased by 14% and 32%, respectively, at 0.5 and 1.6 mM SJ403 for the example plotted above. See Figure S12 in the Supporting Information for RD curves for other residues of p27-D2. The double headed arrows are used to guide the reader to see the change in Rex as a function of SJ403 concentration. (B) All residues that displayed large Rex and multi-state exchange processes show a significant decrease in Rex due to titration of SJ403. The RD performed with 500 µM and 1600 µM were statistically relevant across all values with a p-value of 1.2·10−3 and 7.16·10−14, respectively. The chemical structure of SJ403 is shown as an inset in B.
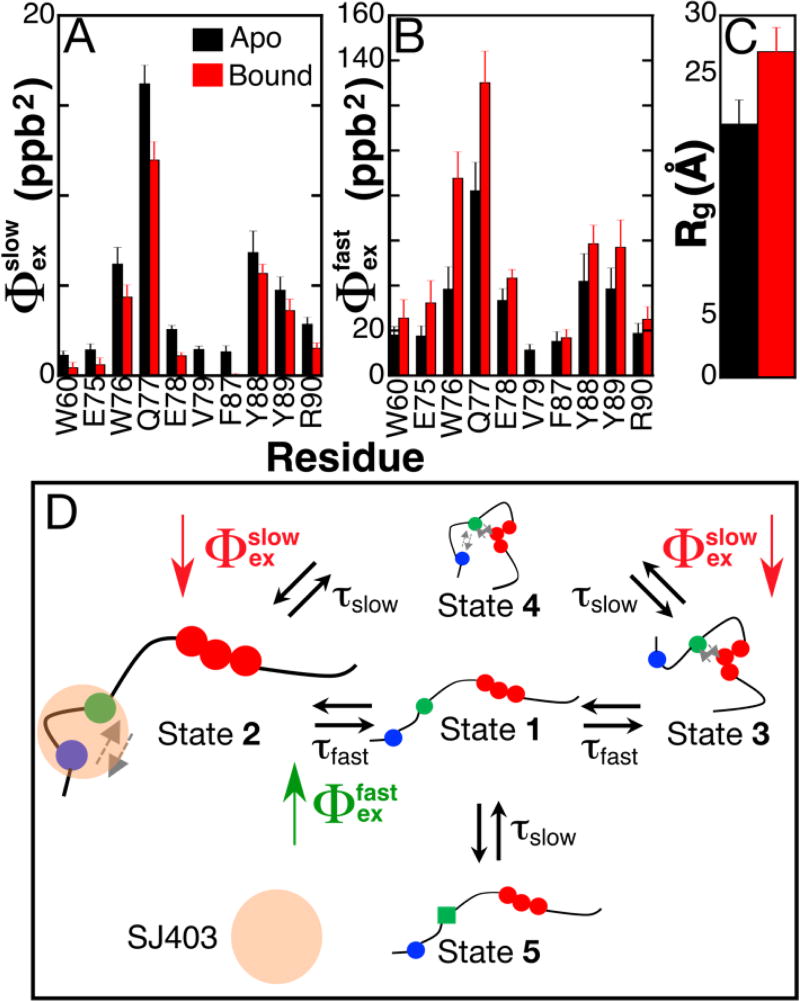
Data for all nuclei were globally fit to the same kinetic model with two distinct activation barriers as was done for apo p27-D2 (Table S5). From the global fits, and were found to be 48.5 ± 16.3 kJ/mol and 51.9 ± 10.3 kJ/mol and are similar within error to the values obtained in the absence of SJ403 (Table S2; 49.7 ± 10.5 kJ/mol and 46.6 ± 4.6 kJ/mol, respectively). Please note that the higher error in the activation energies for p27-D2 with SJ403 is due to the limit in the accessible temperature range for which 1HN RD could be performed. These results show that the binding of SJ403 did not alter the energy barriers associated with the conformational landscape of p27-D2. Since there were distinct decreases in Rex as a function of increasing concentrations of SJ403 (Figure 3), the decrease must be due to changes in the Φex values for p27-D2. Opposite changes were observed for and (Figure 4A & B); , with an average value in apo p27-D2 of 4.5 parts per billion squared (ppb2), decreased on average by 51 ± 29% while , with an average value in apo p27-D2 of 32 parts per billion squared (ppb2), increased by 61 ± 33% when SJ403 was added to p27-D2. In addition, RD was no longer observable for V79 and F87. These results suggest strongly that SJ403 caused a shift in the populations of the discrete states sampled by p27-D2. RD can be influenced by the on-/off-rates and the chemical shift changes induced by the binding of SJ403 to p27-D2. However, dissociation effects of SJ403 can be ruled out because the chemical shift perturbations induced by binding are too small and, with a dissociation constant of 2.2 ± 0.3 mM, any contribution to Rex would be undetectable by the current 1HN RD experiment (Figure S14).
Figure 4. A small molecule, SJ403, induces a shift in the populations of the ground-state for p27-D2.
(A & B). Comparison of conformational amplitudes for all residues that displayed multi-state exchange from fits of temperature dependent RD data for apo p27-D2 (black bars) and p27-D2 bound to SJ403 (red bars). The ratio of p27-D2 to SJ403 was 1:4. The conformational amplitudes associated with the slower kinetic phase decrease when SJ403 is bound as compared with those associated with the faster kinetic process , which increase for all residues. Note that for the slow process, the value of for V79 and F87 was zero. (C) Binding of SJ403 causes an increase in the Rg of p27-D2. Rg values were determined from a Guinier analysis of small angle X-ray scattering (SAXS) data. The increased Rg is also in agreement with the elevated Rg measured for p27-D2-Y88A mutant which is displaced due to its inability to engage in long-range hydrophobic interactions. The concentration of SJ403 during the SAXS measurement was 3 mM; therefore, based on the known dissociation constant, the bound population of p27-D2 was 55%. (D) The schematic illustrates populations changes associated with SJ403 binding to p27-D2; the population of State 2 increases while that of State 4 decreases. Population differences are represented by the relative sizes of the graphical icons for States 2 and 4; larger relative size indicates increased population. SJ403 preferentially binds to the W60 and W76 regions of p27-D2 increasing the population of State 2 with a concomitant decrease in that of State 4. This scenario is supported by the SAXS data which showed expansion of the Y88A p27-D2 mutant (Figure S15). See the Supporting Information for a further discussion of this conformational landscape model. The grey arrows indicate the hydrophobic interactions between residues.
In addition, if binding resulted in a detectable contribution to Rex, this would cause Rex values to increase, not decrease, as was observed (Figure 3). Because the chemical shift changes due to binding are too small to affect the measured RD, the chemical shift differences due to aromatic clustering which are formulated in correspondingly remain the same for both apo and SJ403-bound p27-D2. Therefore, changes in must be due to changes in the populations of the sampled states. We note that the large errors in Φex values determined in the presence versus absence of SJ403 stem from the smaller contribution of Rex to RD for p27-D2 when SJ403 was titrated (Figure 3), which limited the accessible temperature range over which 1HN RD was detectable.
Our NMR RD analyses showed clearly that the binding of SJ403 alters the conformational landscape of p27-D2. Since the features of this landscape arise through interactions of the distal and central aromatic residues, which influence the shape of p27-D2, we reasoned that, by altering aromatic residue interactions, the binding of SJ403 may alter the shape of p27-D2. Analysis using small angle X-ray scattering (SAXS)58,59 showed that the radius of gyration (Rg) for p27-D2 increased from 21 ± 2 Å to 27 ± 2 Å in the presence of SJ403 ([p27-D2]:[SJ403] ratio of 1:5; Figure 4C), consistent with our hypothesis. We next determined Rg values for the three aromatic residue mutants for which we measured RD (Figure 2A) to gain insight into how disrupting specific aromatic residue interactions affected the shape of p27-D2. Interestingly, p27-Y88A was significantly expanded in comparison with the other two mutants and wild-type p27-D2 (Figure S15), suggesting that the binding of SJ403 alters interactions between Y88 and the other two critical aromatic residues in p27-D2, W60 and W76. Our previous mutagenesis studies32 showed that SJ403 predominately interacts with W60 and W76, supporting a model in which SJ403 competes with Y88 for interactions with W60 and W76, resulting in displacement of Y88 and its neighboring residues. Because the changes in conformational amplitudes measured here must reflect changes in populations (Figure S14), the increase in and the decrease in suggests that the population of at least one state involved in a fast transition has increased and that this is accompanied by a decrease in the population of at least one state connected by a slow transition. Based on our RD data (Figure 4B), SAXS data (Figures 4C and S15) and previous results32, we assign these compound binding-induced changes to an increase in the population of State 2 (due to stabilization of the interaction between W60 and W76) and an accompanying decrease in the population of State 4 (due to destabilization of interactions between all three aromatic regions; Figure 4D). The reader is directed to the section entitled, “Explanation of the conformational landscape of p27-D2 and its interaction with SJ403”, in the Supporting Information for a detailed explanation of the models discussed in Figures 2B and 4D.
DISCUSSION AND CONCLUSION
Our analysis has shown that while two distinct kinetic phases emerged from the NMR RD data, mutagenesis of critical aromatic residues within p27-D2 revealed differential behavior that was manifested as altered conformational amplitudes (Figure S8 and Table S4) for nuclei that still showed quantifiable RD. To explain the residual RD observed for the distal and central aromatic regions in the various p27-D2 mutants, multiple states that share the same activation kinetics were necessary (Figure 2B) (see the Supporting Information). We emphasize that the mutagenesis data was essential for identifying multiple, kinetically similar conformational transitions in p27-D2. Even though the dissociation constant is 2.2 ± 0.3 mM32 for the p27-D2:SJ403 interaction, we observed measureable changes in values for p27-D2 and have determined that these are due to changes in populations of certain states and not to changes in their structural features. This behavior is in contrast to several models in which small molecules are proposed to interact with an ensemble of conformations and that small molecule:IDP interactions expand the conformational landscape of the IDP creating new conformations.4–6 We propose that the SJ403 small molecule binds preferentially to clusters of distal aromatic residues, including W60 and W76, and that these interactions lead to the displacement of Y88 that otherwise transiently engages the two tryptophan residues within these clusters (Figure 4). It is possible that different small molecules and IDPs utilize fundamentally different mechanisms when they interact. Importantly, we feel that the insights into the mechanism of small molecule:IDP interactions gained from our studies provide a potential avenue for the rational design of tighter binding compounds in the future. Given that the current small molecule (SJ403) was shown to displace p27-D2 from Cdk2/cyclin A and partially restore kinase activity32, we propose that improved molecules that further drive the population shift to the conformational state with W60/W76 clusters (State 2) hold promise for modulating p27 function in the cells.
In conclusion, we have demonstrated that a prototypical IDP, p27-D2, populates multiple, rapidly interconverting conformational states (Figures 1 and 2). Although ground-state conformational fluctuations of IDPs are thought to exhibit shallow energy wells with low activation energy barriers, we observed that the ground-state for p27-D2 exhibits energetic barriers that have the same magnitude as ground-state motional events observed in some folded proteins.51,54 Our experimental findings provide unique insights in terms of the structural and kinetic features of a multi-state conformational landscape into how a small molecule (SJ403) binds to an IDP (p27-D2). The NMR RD methods applied here can be used to define the conformational landscapes of myriad other IDPs in the future as a basis for understanding their diverse roles in biology and disease and for investigating their possible interactions with small molecules.
Supplementary Material
Acknowledgments
The SAXS work was conducted at the Advanced Light Source (ALS), at the SIBYLS beamline through its mail-in/hand-in program (sibyls.als.lbl.gov/htsaxs) which is funded by DOE/BER through IDAT and NIH/NCI’s SBDR grant (CA92584). This work was supported by the US National Institutes of Health (R01CA082491 to R.W.K and a National Cancer Institute Cancer Center Support Grant (P30CA21765) to St. Jude Children’s Research Hospital) and ALSAC. D.B. acknowledges support from the James Graham Brown Foundation and the National Center for Research Resources COBRE 1P30GM106396. J.Z. acknowledges support from the National Institutes of Health (1R01DC015010, 1R01DC015444, and 1R21DC013879) and the Office of Naval Research (N000141612315). L.I.I. was the recipient of the Garwood Foundation Fellowship from St. Jude Children’s Research Hospital.
Footnotes
ASSOCIATED CONTENT
Full details pertaining to sample preparation, experimental methods and analysis, extended discussion about the kinetic model and how it is changed by small molecule binding, p27-D2 mutant analyses, temperature dependent RD data, temperature dependence of chemical shifts, tables containing parameters extracted from individual and global fits to RD data for wild-type and mutant p27-D2, and SAXS data for wild-type, SJ403 bound, and mutant p27-D2.
The Supporting Information is available free of charge on the ACS Publications website.
References
- 1.Csizmok V, Follis AV, Kriwacki RW, Forman-Kay JD. Chem. Rev. 2016;116:6424–6462. doi: 10.1021/acs.chemrev.5b00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW. eLIFE. 2016;5:e13571. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dyson HJ, Wright PE. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 4.Heller GT, Pietro Sormanni, Vendruscolo M. Trends Biochem. Sci. 2015;40:491–496. doi: 10.1016/j.tibs.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Jin F, Yu C, Lai L, Liu Z. PLoS Comput. Biol. 2013;9(10):e1003249. doi: 10.1371/journal.pcbi.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunker AK, Uversky VN. Curr. Opin. in Pharm. 2010;10:782–788. doi: 10.1016/j.coph.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Jensen MR, Ruigrok RW, Blackledge M. Curr. Opin. Struct. Biol. 2013;23:426–435. doi: 10.1016/j.sbi.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Abyzov A, Salvi N, Schneider R, Maurin D, Ruigrok RWH, Jensen MR, Blackledge M. J. Am. Chem. Soc. 2016;138:6240–6251. doi: 10.1021/jacs.6b02424. [DOI] [PubMed] [Google Scholar]
- 9.Varadi M, Kosol S, Lebrun P, Valentini E, Blackledge M, Dunker AK, Felli IC, Forman-Kay JD, Kriwacki RW, Pierattelli R, Sussman J, Svergun DI, Uversky VN, Vendruscolo M, Wishart D, Wright PE, Tompa P. Nucl. Acids Res. 2013;42:326–335. doi: 10.1093/nar/gkt960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukrasch MD, Markwick P, Biernat J, Bergen von M, Bernadó P, Griesinger C, Mandelkow E, Zweckstetter M, Blackledge M. J. Am. Chem. Soc. 2007;129:5235–5243. doi: 10.1021/ja0690159. [DOI] [PubMed] [Google Scholar]
- 11.Jensen MR, Communie G, Ribeiro EA, Jr, Martinez N, Desfosses A, Salmon L, Mollica L, Gabel F, Jamin M, Longhi S, Ruigrok RWH, Blackledge M. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9839–9844. doi: 10.1073/pnas.1103270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wirmer J, Schlörb C, Klein-Seetharaman J, Hirano R, Ueda T, Imoto T, Schwalbe H. Angew. Chem. Int. Ed. 2004;43:5780–5785. doi: 10.1002/anie.200460907. [DOI] [PubMed] [Google Scholar]
- 13.Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H. Science. 2002;295:1719–1722. doi: 10.1126/science.1067680. [DOI] [PubMed] [Google Scholar]
- 14.Bernadó P, Bertoncini CW, Griesinger C, Zweckstetter M, Blackledge M. J. Am. Chem. Soc. 2005;127:17968–17969. doi: 10.1021/ja055538p. [DOI] [PubMed] [Google Scholar]
- 15.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. Proc Natl Acad Sci USA. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chattopadhyay K, Elson EL, Frieden C. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2385–2389. doi: 10.1073/pnas.0500127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modig K, Poulsen FM. J. Biomol. NMR. 2008;42:163–177. doi: 10.1007/s10858-008-9280-0. [DOI] [PubMed] [Google Scholar]
- 18.Palmer AG., III Chem. Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 19.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- 20.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. Biochemistry. 1997;36:2390–2402. doi: 10.1021/bi962548h. [DOI] [PubMed] [Google Scholar]
- 21.Khan SN, Charlier C, Augustyniak R, Salvi N, Déjean V, Bodenhausen G, Lequin O, Pelupessy P, Ferrage F. Biophys. J. 2015;109:988–999. doi: 10.1016/j.bpj.2015.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parigi G, Rezaei-Ghaleh N, Giachetti A, Becker S, Fernandez C, Blackledge M, Griesinger C, Zweckstetter M, Luchinat C. J. Am. Chem. Soc. 2014;136:16201–16209. doi: 10.1021/ja506820r. [DOI] [PubMed] [Google Scholar]
- 23.Forman-Kay JD, Mittag T. Struct./Fold. and Design. 2013;21:1492–1499. doi: 10.1016/j.str.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uversky VN, Oldfield CJ, Dunker AK. Annu. Rev. Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 25.Metallo SJ. Curr. Opin. in Chem. Biol. 2010;14:481–488. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erkizan HV, Kong Y, Merchant M, Schlottmann S, Barber-Rotenberg JS, Yuan L, Abaan OD, Chou T-H, Dakshanamurthy S, Brown ML, Üren A, Toretsky JA. Nat. Med. 2009;15:750–756. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishnan N, Koveal D, Miller DH, Bin Xue, Akshinthala SD, Kragelj J, Jensen MROB, Gauss C-M, Page R, Blackledge M, Muthuswamy SK, Peti W, Tonks NK. Nat. Chem. Biol. 2014;10:558–566. doi: 10.1038/nchembio.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin X, Giap C, Lazo JS, Prochownik EV. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 29.Tóth G, Gardai SJ, Zago W, Bertoncini CW, Cremades N, Roy SL, Tambe MA, Rochet J-C, Galvagnion C, Skibinski G, Finkbeiner S, Bova M, Regnstrom K, Chiou S-S, Johnston J, Callaway K, Anderson JP, Jobling MF, Buell AK, Yednock TA, Knowles TPJ, Vendruscolo M, Christodoulou J, Dobson CM, Schenk D, McConlogue L. PLoS One. 2014;9:e87133. doi: 10.1371/journal.pone.0087133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Convertino M, Vitalis A, Caflisch A. J. Biol. Chem. 2011;286:41578–41588. doi: 10.1074/jbc.M111.285957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu C, Niu X, Jin F, Liu Z, Jin C, Lai L. Sci. Rep. 2016;6:22298:1–11. doi: 10.1038/srep22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iconaru LI, Ban D, Bharatham K, Ramanathan A, Zhang W, Shelat AA, Zuo J, Kriwacki RW. Sci. Rep. 2015;5:15686. doi: 10.1038/srep15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimmler M, Wang Y, Mund T, Cilenšek Z, Keidel E-M, Waddell MB, Jäkel H, Kullmann M, Kriwacki RW, Hengst L. Cell. 2007;128:269–280. doi: 10.1016/j.cell.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 34.Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao L, Weiss S, Hengst L, Kriwacki RW. Nat. Struct. Mol. Biol. 2004;11:358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 35.Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletic NP. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 36.Chu I, Sun J, Arnaout A, Kahn H, Hanna W, Narod S, Sun P, Tan C-K, Hengst L, Slingerland J. Cell. 2007;128:281–294. doi: 10.1016/j.cell.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galea CA, Nourse A, Wang Y, Sivakolundu SG, Heller WT, Kriwacki RW. J. Mol. Biol. 2008;376:827–838. doi: 10.1016/j.jmb.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin I, Yakes FM, Rojo F, Shin N-Y, Bakin AV, Baselga J, Arteaga CL. Nat. Med. 2002;8:1145–1152. doi: 10.1038/nm759. [DOI] [PubMed] [Google Scholar]
- 39.Zhao H, Faltermeier CM, Mendelsohn L, Porter PL, Clurman BE, Roberts JM. Oncotarget. 2015;5:12704–12714. doi: 10.18632/oncotarget.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viglietto G, Motti ML, Bruni P, Melillo RM, D'Alessio A, Califano D, Vinci F, Chiappetta G, Tsichlis P, Bellacosa A, Fusco A, Santoro M. Nat. Med. 2002;8:1136–1144. doi: 10.1038/nm762. [DOI] [PubMed] [Google Scholar]
- 41.Liang J, Zubovitz J, Petrocelli T, Kotchetkov R, Connor MK, Han K, Lee J-H, Ciarallo S, Catzavelos C, Beniston R, Franssen E, Slingerland JM. Nat. Med. 2002;8:1153–1160. doi: 10.1038/nm761. [DOI] [PubMed] [Google Scholar]
- 42.Chen P, Segil N. Development. 1999;126:1581–1590. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- 43.White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Nature. 2006;441:984–987. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- 44.Walters BJ, Liu Z, Crabtree M, Coak E, Cox BC, Zuo J. J. Neuro. 2014;34:15751–15763. doi: 10.1523/JNEUROSCI.3200-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ban D, Gossert AD, Giller K, Becker S, Griesinger C, Lee D. J. Magn. Reson. 2012;221:1–4. doi: 10.1016/j.jmr.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Smith CA, Ban D, Pratihar S, Giller K, Schwiegk C, de Groot BL, Becker S, Griesinger C, Lee D. Angew. Chem. Int. Ed. Engl. 2015;54:207–210. doi: 10.1002/anie.201408890. [DOI] [PubMed] [Google Scholar]
- 47.Eichmüller C, Skrynnikov NR. J. Biomol. NMR. 2005;32:281–293. doi: 10.1007/s10858-005-0658-y. [DOI] [PubMed] [Google Scholar]
- 48.Smith CA, Ban D, Pratihar S, Giller K, Paulat M, Becker S, Griesinger C, Lee D, de Groot BL. Proc. Natl. Acad. Sci. U.S.A. 2016;113:3269–3274. doi: 10.1073/pnas.1519609113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer AG, Massi F. Chem. Rev. 2006;106:1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 50.Grey MJ, Wang C, Palmer AG. J. Am. Chem. Soc. 2003;125:14324–14335. doi: 10.1021/ja0367389. [DOI] [PubMed] [Google Scholar]
- 51.Pratihar S, Sabo TM, Ban D, Fenwick RB, Becker S, Salvatella X, Griesinger C, Lee D. Angew. Chem. 2016;128:9719–9722. doi: 10.1002/anie.201603501. [DOI] [PubMed] [Google Scholar]
- 52.Schwalbe H, Fiebig KM, Buck M, Jones JA, Grimshaw SB, Spencer A, Glaser SJ, Smith LJ, Dobson CM. Biochemistry. 1997:1–15. doi: 10.1021/bi970049q. [DOI] [PubMed] [Google Scholar]
- 53.Xue Y, Podkorytov IS, Rao DK, Benjamin N, Sun H, Skrynnikov NR. Prot. Sci. 2009;18:1401–1424. doi: 10.1002/pro.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ban D, Funk M, Gulich R, Egger D, Sabo TM, Walter KFA, Fenwick RB, Giller K, Pichierri F, de Groot BL, Lange OF, Grubmüller H, Salvatella X, Wolf M, Loidl A, Kree R, Becker S, Lakomek N-A, Lee D, Lunkenheimer P, Griesinger C. Angew. Chem. Int. Ed. Engl. 2011;50:11437–11440. doi: 10.1002/anie.201105086. [DOI] [PubMed] [Google Scholar]
- 55.Li P, Martins IRS, Rosen MK. J. Biomol. NMR. 2011;51:57–70. doi: 10.1007/s10858-011-9541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng W, Borgia A, Buholzer K, Grishaev A, Schuler B, Best RB. J. Am. Chem. Soc. 2016;138:11702–11713. doi: 10.1021/jacs.6b05443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sivakolundu SG, Bashford D, Kriwacki RW. J. Mol. Biol. 2005;353:1118–1128. doi: 10.1016/j.jmb.2005.08.074. [DOI] [PubMed] [Google Scholar]
- 58.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, II, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, Yang S-J, Scott JW, Dillard BD, Adams MWW, Tainer JA. Nat. Meth. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Classen S, Hura GL, Holton JM, Rambo RP, Rodic I, McGuire PJ, Dyer K, Hammel M, Meigs G, Frankel KA, Tainer JA. J. Appl. Cryst. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.