Abstract
Objectives
Although different methods have been established to detect epidermal growth factor receptor (EGFR) T790M mutation in circulating tumor DNA (ctDNA), a wide range of diagnostic accuracy values were reported in previous studies. The aim of this meta-analysis was to provide pooled diagnostic accuracy measures for droplet digital PCR (ddPCR) in the diagnosis of EGFR T790M mutation based on ctDNA.
Materials and methods
A systematic review and meta-analysis were carried out based on resources from Pubmed, Web of Science, Embase and Cochrane Library up to October 11, 2017. Data were extracted to assess the pooled sensitivity, specificity, positive likelihood ratio, negative likelihood ratio (NLR), diagnostic OR (DOR), and areas under the summary receiver-operating characteristic curve (SROC).
Results
Eleven of 311 studies identified have met the including criteria. The sensitivity and specificity of ddPCR for the detection of T790M mutation in ctDNA ranged from 0.0% to 100.0% and 63.2% to 100.0%, respectively. For the pooled analysis, ddPCR had a performance of 70.1% (95% CI, 62.7%–76.7%) sensitivity, 86.9 % (95% CI, 80.6%–91.7%) specificity, 3.67 (95% CI, 2.33–5.79) PLR, 0.41 (95% CI, 0.32–0.55) NLR, and 10.83 (95% CI, 5.86–20.03) DOR, with the area under the SROC curve being 0.82.
Conclusion
The ddPCR harbored a good performance for detection of EGFR T790M mutation in ctDNA.
Keywords: T790M, droplet digital PCR, circulating tumor DNA, lung cancer
Introduction
As reported, epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) could drastically increase the median survival time from 1 year to 3–5 years in EGFR-mutant nonsmall cell lung cancer (NSCLC) patients.1,2 However, despite of initial response on first-generation TKIs, patients finally develop resistance within 1–2 years and about 50%–65% of resistance is gained because of EGFR T790M mutation.3–6 Therefore, rebiopsy has been recommended to explore whether there is the T790M resistant mutation when disease has progressed. But, due to the invasiveness of biopsy procedures, sometimes the inaccessibility of tumor tissues, heterogeneity of the tumor or patients’ unwillingness, it has always become difficult and problematic to carry out rebiopsy in routine clinical work.7,8 Alternatively, circulating tumor DNA (ctDNA), which can be collected and extracted from peripheral blood, has been identified as clinically significant biomarker to help reveal the EGFR mutation status.9–11
Testing platforms of EGFR T790M mutation in ctDNA are numerous, including the real-time PCR [Cobas, Amplification Refractory Mutation System (ARMS)], digital platforms [droplet digital PCR (ddPCR), Beads, Emulsions, Amplification and Magnetic (BEAMing)], and next generation sequencing technologies, respectively.10,12,13 Regarding to PCR-based techniques, data revealed that ddPCR had superior sensitivity compared with Cobas and ARMS.14–18 Thress et al conducted a cross-platform comparison of these leading technologies and found the sensitivity of Cobas, ARMS, and ddPCR were 41%, 29%, and 71% in EGFR T790M mutation detection, respectively.18 Besides, using Cobas tissue test as the reference, Zhang et al found the positive percent agreement (PPA) of Cobas plasmas, Super-ARMS, and ddPCR were 42%, 49%, and 56% respectively. The sensitivity for plasma T790M detection slightly increased with ddPCR compared with Super-ARMS and Cobas plasma test.15
The ddPCR technology harbors detection limit of 0.01%–0.04% for EGFR mutation.19 Unlike EGFR sensitizing mutation, there is only a small fraction of mutant T790M alleles among plenty of wild-type alleles in clinical samples.20 Therefore, the design of ddPCR assures the partitioning of competing backgrounds of wild-type alleles by thousands of even millions of droplets, leading to decrease in their PCR inhibitory effects and improvements in detection sensitivity.21 However, the reported sensitivity and specificity of ddPCR for detection of EGFR T790M mutation in ctDNA varied. For example, Suzawa et al reported a sensitivity of 42.8%, whereas a high sensitivity of 100% was indicted by Yu et al.22,23 Similar discordance in specificity was also observed.24,25 Hence, the aim of the current study is to search related publications and then summarize data to provide pooled diagnostic accuracy values of ddPCR for detection of EGFR T790M mutation in ctDNA.
Material and methods
Literature search strategy
We comprehensively searched various online sources including PubMed, Web of Science, Embase and Cochrane Library up to October 11, 2017, using key words “digital PCR” and “T790M”. The language is limited to English and Chinese. After duplicates were removed, all searched results underwent title and abstract review and potentially eligible studies were reviewed through full texts. This analysis was carried out according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.26
Inclusion and exclusion criteria
Searched studies were assessed by two reviewers independently and disagreements were solved by discussion with the third person until consensus was reached. Those that satisfied the following inclusion criteria were selected for final analysis: 1) enrolled the NSCLC patients treated with EGFR-TKI therapy; 2) studied diagnostic accuracy of ddPCR for detection of EGFR T790M mutation based on ctDNA; 3) taking biopsy samples as reference method; and 4) reported necessary data for calculating pooled index. Studies were excluded if they were 1) not published in full-text, such as meeting abstracts; 2) not original article, like case report or review; 3) unrelated to research topics; and 4) duplicate publications.
Data extraction and quality assessment
Eligible studies were checked again by the two reviewers and a consensus was reached prior to further process. All necessary data for calculating pooled index were extracted and two-by-two tables were reconstructed in each study for the true-positive (TP), false-negative (FN), true-negative (TN), and false-positive (FP) values. Other relevant information, including the name of the first author, year of publication, country of origin, sample size, basic characteristics of studied population, tumor histology, clinical stage, source of biopsy samples and related detection methods for EGFR T790M mutation, and the manufacturer of ddPCR platform used in the study, were extracted from the included studies.
The quality of all included studies were assessed by the revised Quality Assessment of Diagnostic Accuracy Studies (QUADAS)-2 tool from four aspects, namely the methods of patients selection, the conduction and interpretation of index tests, reference standard, and flow and timing regarding to the index tests and reference standard. After being assessed, each study quality was categorized as being low, high, or unclear regarding to risk of bias, and applicability concerns, respectively. The same two reviewers carried out the assessment and disagreement was solved by consensus.27
Statistical analysis
Several pooled values were calculated in this meta-analysis, including sensitivity, specificity, the positive likelihood ratio (PLR), the negative likelihood ratio (NLR), and diagnostic OR (DOR). PLR equals to sensitivity/(1−specitity), and larger values mean high possibility of true positivity when the index test result is positive. Similarly, NLR is calculated by (1−sensitivity)/specificity, and the smaller the value, the higher the true negativity possibility of index test results. The DOR of a test is the ratio of the odds of positivity in disease relative to the odds of positivity in the nondiseased. Its value ranges from 0 to infinity, with higher values indicating better discriminatory test performance.28 Besides, PPA and negative percent agreement (NPA) were pooled, and summary receiver-operating characteristic curve (SROC) was drawn.
The Cochrane’s Q and the I2 statistic were used to analyze heterogeneity. For the Q statistic, P-value <0.10 was considered statistically significant for heterogeneity.
As for the I2 statistic, which is the percentage of the observed variability between studies due to heterogeneity rather than chance, its value >50% represented that heterogeneity existed in pooled values. In the current meta-analysis heterogeneity existed, and all statistics were calculated and combined using the random effects model with 95% CI. To shed light on heterogeneity among studies, subgroup analyses were conducted.
In addition, the publication bias was assessed by Deek’s funnel plot asymmetry test.29 The professional statistical software Meta-Disc 1.4 (Ramón y Cajal Hospital in Madrid, Spain) program was used for pooled analysis. Publication bias was performed using STATA 12.0 (STATA Corporation, College Station, TX, USA).
Results
Study selection
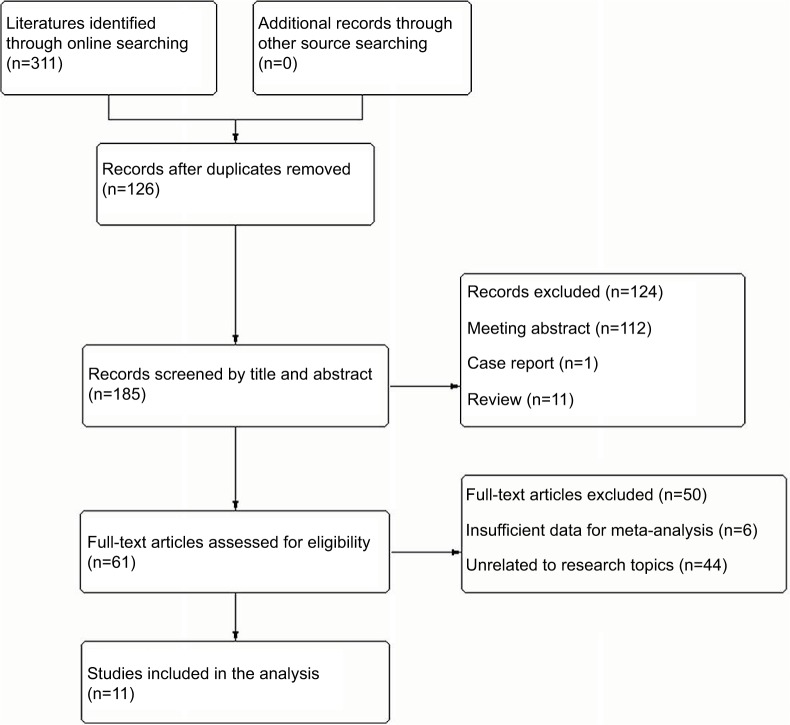
According to PRISMA guidelines, the process of study selection was shown in Figure 1. Total 311 literatures were initially identified through online searching, and there were no additional records being found through other source searching. After 126 duplicates were removed, 185 studies were screened by title and abstract for potential eligibility of the meta-analysis. Then, 124 of them were excluded because of the publication format being meeting abstracts (n=112), reviews (n=11), and case (n=1). Finally, 61 studies were identified and reviewed through the whole text, with 11 being satisfied to our inclusion criteria. The other 50 studies were excluded because of insufficient data for meta-analysis (n=6) or unrelated to research topics (n=8).
Figure 1.
Flow chart of the included studies in this meta-analysis.
Characteristics of included studies
The characteristics of the included 11 studies in this meta-analysis were summarized in Table 1. As a whole, there were 872 advanced NSCLC patients involved in these studies, most of whom were Asians. Among all the patients, EGFR-TKIs were used as a targeted therapy for disease control. To evaluate ddPCR performance of EGFR T790M mutation detection in ctDNA, 298 patients (337 blood samples) who have underwent both T790M mutation test in tumor tissue or malignant fluid and plasma ctDNA, were identified. In 7 studies (63.6%), all studied patients have already developed EGFR-TKI resistance when ddPCR tests were carried out.
Table 1.
Baseline characteristics of included studies
| Study | Country | Ethnicity | Sample size | Age (years) | Male (%) | Smoker (%) | Histology | Clinical stage | Time for T790M detection |
|---|---|---|---|---|---|---|---|---|---|
| Ishii et al36 | Japan | Japanese | 18 | 50–81 | 11.1 | 5.6 | Adenocarcinoma | Recurrent | Resistance developed |
| Thress et al18 | UK | Hesperian | 38 | NA | NA | NA | NSCLC | M0/M1a/M1b | Resistance developed |
| Wei et al42 | China | Chinese | 50a | 45–68 | 62.0 | 76.0 | Adenocarcinoma | IV | No resistance |
| Wang et al17 | China | Chinese | 108 | NA | 50.9 | 34.3 | NSCLC | IIIB/IV | Resistance developed |
| Takahama et al43 | Japan | Japanese | 260 | 36–90 | 30.0 | 27.3 | NSCLC | IIIB/IV/inoperable/recurrent | Resistance developed |
| Seki et al44 | Japan | Japanese | 35 | 47–74 | 40.0 | 31.4 | Adenocarcinoma | IV/recurrent | Resistance developed |
| Zheng et al24 | China | Chinese | 117 | NA | 39.3 | 24.8 | NSCLC | IIIB/IV/recurrent | Resistance developed |
| Sacher et al25 | USA | Hesperian | 180 | NA | 38.0 | NA | NSCLC | IIIB/IV/recurrent | Resistance developed |
| Yu et al22 | China | Chinese | 22 | 35–74 | 45.5 | NA | NSCLC | IIIB/IV | Resistance developed/no resistance |
| Suzawa et al23 | Japan | Japanese | 24a | 39–84 | 29.2 | 37.5 | NSCLC | NA | NA |
| Xu et al45 | China | Chinese | 20 | 37–76 | 50 | 30 | NSCLC | I–IV | NA |
Note:
Healthy volunteers in original study were not included here.
Abbreviations: NSCLC, nonsmall cell lung cancer; NA, not available.
Table 2 described the included sample size for 2×2 table, the samples were used for reference test and index text, as well as the time interval between tissue and plasma sampling. Most tissues were taken from primary sites. Almost all of the time interval were not specific, except for one study that told a median time of 12.5 months.
Table 2.
The included sample for 2×2 table analysis
| Study | Sample size | Included size for 2×2 table | Sample for reference test | Reference test | Sample for index test | Index test | Time intervalc |
|---|---|---|---|---|---|---|---|
| Ishii et al36 | 18 | 18 | Primary/MF | ddPCR | Plasma ctDNA | ddPCR (Bio-Rad) | Not specific both following progression |
| Thress et al18 | 38 | 23 | Primary | Cobas test | Plasma ctDNA | ddPCR (Bio-Rad) | Not specific both following progression |
| Wei et al42 | 50 | 50 | Primary | NA | Plasma ctDNA | ddPCR (Bio-Rad) | Not specific both at baseline |
| Wang et al17 | 108 | 16 | Primary | ARMS | Plasma ctDNA | ddPCR (AmoyDx) | Not specific both following progression |
| Takahama et al43 | 260 | 41 | Primary/MF | ARMS/Cobas | Plasma ctDNA | ddPCR (Bio-Rad) | Not specific both following progression |
| Seki et al44 | 35 | 10 | Primary | ARMS | Plasma ctDNA | Picoliter-ddPCR (RainDance) | Not specific both following progression |
| Zheng et al24 | 117 | 25 | Primary/MF | ARMS/ddPCR | Plasma ctDNA | PNA-TaqMan-ddPCR (Bio-Rad) | Not specific both following progression |
| Sacher et al25 | 180 | 54 | Primary/metastasis/MF | PCR/NGS | Plasma ctDNA | ddPCR (Bio-Rad) | Not specific |
| Yu et al22 | 22 | 20 | Primary/metastasis | ARMS | Plasma ctDNA | Picoliter-ddPCR (RainDance) | Not specific |
| Suzawa et al23 | 24 | 59a | Primary | PNA-LNA PCR | Plasma ctDNA | PNA-LNA-ddPCR (Bio-Rad) | Median 12.5 months both following progression |
| Xu et al45 | 20 | 21b | Primary/metastasis/MF | ARMS/NGS | Plasma ctDNA | ddPCR (Bio-Rad) | Not specific |
Notes:
59 plasma samples were withdrawn from 24 patients for analysis.
Total 20 patients, 17 were enrolled to compare EGFR mutation profiles between tissue and plasma, but 21 cases were analyzed.
Time interval between tissue and blood sampling used for T790M analysis.
Abbreviations: MF, malignant fluid; ddPCR, droplet digital polymerase chain reaction; ARMS, Amplification Refractory Mutation System; NGS, Next-Generation Sequencing; ctDNA, circulating tumor DNA; PNA, peptide nucleic acids; LNA, locked nucleic acids; NA, not available.
According to tissue or malignant fluid detection results, there were 165 T790M-mutation-positive patients (177 blood samples) and 133 T790M-mutation-negative patients (156 blood samples). The TP/FP/FN/TN values, sensitivity, and specificity of ddPCR test in each study were summarized in Table 3.
Table 3.
The ddPCR performance of each included study
| Study | Included size | Reference method
|
ddPCR for ctDNA
|
Sensitivity (%) | Specificity (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| T790M + | T790M – | Ref +/ddPCR + | Ref–/ddPCR + | Ref +/ddPCR – | Ref–/ddPCR – | ||||
| Ishii et al36 | 18 | 11 | 7 | 9 | 1 | 2 | 6 | 81.8% | 85.7% |
| Thress et al18 | 23 | 17 | 6 | 12 | 1 | 5 | 5 | 70.6% | 83.3% |
| Wei et al42 | 50 | 25 | 25 | 19 | 3 | 6 | 22 | 76.0% | 88.0% |
| Wang et al17 | 16 | 9 | 7 | 6 | 2 | 3 | 5 | 66.7% | 71.4% |
| Takahama et al43 | 41 | 31 | 10 | 20 | 3 | 11 | 7 | 64.5% | 70.0% |
| Seki et al44 | 10 | 7 | 3 | 5 | 0 | 2 | 3 | 71.4% | 100.0% |
| Zheng et al24 | 25 | 16 | 9 | 13 | 0 | 3 | 9 | 81.3% | 100.0% |
| Sacher et a 25 | 54 | 35 | 19 | 27 | 7 | 8 | 12 | 77.1% | 63.2% |
| Yu et al22 | 20 | 4 | 16 | 4 | 1 | 0 | 15 | 100.0% | 93.8% |
| Suzawa et al23 | 59 | 21 | 38 | 9 | 1 | 12 | 37 | 42.8% | 97.3% |
| Xu et al45 | 21 | 1 | 20 | 0 | 2 | 1 | 18 | 0.0% | 90.0% |
Abbreviations: ddPCR, droplet digital polymerase chain reaction; ctDNA, circulation tumor DNA; Ref, reference.
Overall accuracy of the ddPCR test
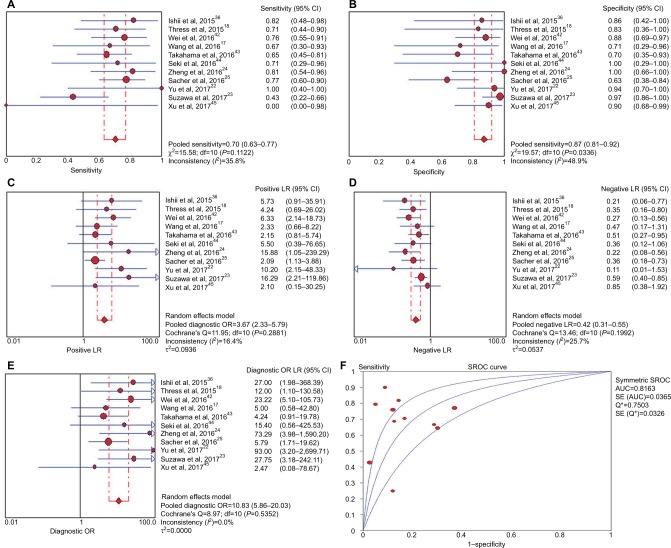
For all studies, the sensitivity and specificity of ddPCR for the detection of T790M mutation in ctDNA ranged from 0.0% to 100.0% and 63.2% to 100.0%, respectively. The concordance between plasma and tissue test was 81.2%, with the PPA being 71.2% and NPA being 90.0%. When it came to the pooled analysis, ddPCR test had a performance of 70.1% (95% CI, 62.7%–76.7%) sensitivity, 86.9 % (95% CI, 80.6% –91.7%) specificity, 3.67 (95% CI, 2.33–5.79) PLR, 0.41 (95% CI, 0.32–0.55) NLR, and 10.83 (95% CI, 5.86–20.03) DOR in diagnosis of EGFR T790M mutation based on ctDNA in NSCLC patients. In addition, the area under the SROC curves (AUC) was 0.82 (Figure 2). Because significant heterogeneity exists regarding to pooled specificity (P=0.03, I2=48.9%), the random-effect model was used for the whole analyses.
Figure 2.
Meta-analysis of (A) sensitivity, (B) specificity, (C) positive likelihood ratio, (D) negative likelihood ratio, (E) diagnostic odds ratio, (F) and SROC curve for droplet digital PCR for diagnosing epidermal growth factor receptor T790M mutation in circulating tumor DNA.
Abbreviations: LR, likelihood ratio; SROC, summary receiver-operating characteristic; AUC, area under the ROC curve; PCR, polymerase chain reaction..
Subgroup analysis
As specificity heterogeneity existed, we performed subgroup analyses regarding to ethnicity, tumor histology, and index test (Table 4). The index test was the most possible cause of specificity heterogeneity, with I2 changing from 48.9% to the maximum 8.1%. The picoliter-ddPCR and designed ddPCR assay presented higher specificity, 94.7% (95% CI, 74.0 %–99.9%) and 97.9% (95% CI, 88.7%–99.9%), respectively. In ethnicity and tumor histology subgroup analyses, specificity heterogeneity was still present.
Table 4.
Subgroup analysis of ddPCR performance
| Subgroup | Number of studies | Sensitivity % (95% CI) | Specificity % (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC | I2 b | P-valuec |
|---|---|---|---|---|---|---|---|---|---|
| Ethnicity | |||||||||
| Hesperian | 2 | 75.0 (61.1–86.0) | 68.0 (46.5–85.1) | 2.25 (1.26–4.03) | 0.36 (0.21–0.61) | 6.73 (2.27–19.96) | NA | 0.0% | 0.34 |
| Japanese | 4 | 61.4 (49.0–72.8) | 91.4 (81.0–97.1) | 4.27 (1.66–10.95) | 0.52 (0.38–0.70) | 10.48 (3.59–30.56) | 0.78 | 56.3% | 0.08* |
| Chinese | 5 | 76.4 (63.0–86.8) | 89.6 (80.6–95.4) | 5.12 (2.60–10.09) | 0.37 (0.19–0.70) | 17.13 (5.51–53.24) | 0.90 | 3.9% | 0.39 |
| Tumor histology | |||||||||
| Adenocarcinoma | 3 | 76.7 (61.4–88.2) | 88.6 (73.3–96.8) | 6.10 (2.53–14.69) | 0.28 (0.16–0.48) | 22.70 (6.71–76.86) | 0.92 | 0.0% | 0.67 |
| NSCLC | 8 | 67.9 (59.3–75.7) | 86.4 (79.1–91.9) | 3.35 (1.90–5.91) | 0.47 (0.34–0.63) | 8.46 (4.13–17.34) | 0.79 | 62.5% | 0.01* |
| Index test | |||||||||
| Picoliter-ddPCR (RainDance) | 2 | 81.8 (48.2–97.7) | 94.7 (74.0–99.9) | 8.70 (2.28–33.19) | 0.29 (0.09–0.95) | 37.35 (3.51–397.16) | NA | 0.0% | 0.55 |
| ddPCR (Bio-Rad) | 7 | 72.1 (63.5–79.6) | 79.8 (70.2–87.4) | 2.72 (1.80–4.11) | 0.41 (0.30–0.57) | 8.11 (4.09–16.03) | 0.80 | 8.1 % | 0.37 |
| Othersa | 2 | 59.5 (42.1–75.2) | 97.9 (88.7–99.9) | 16.14 (3.23–80.57) | 0.39 (0.15–1.05) | 38.28 (6.51–225.05) | NA | 0.0 % | 0.51 |
| Pool | 11 | 70.1 (62.7–76.7) | 86.9 (80.6–91.7) | 3.67 (2.33–5.79) | 0.41 (0.32–0.55) | 10.83 (5.86–20.03) | 0.82 | 48.9% | 0.03* |
Notes:
Others including technologies of PNA-TaqMan-ddPCR (Bio-Rad) and PNA-LNA-ddPCR (Bio-Rad).
The I2 for specificity in each subgroup.
Heterogeneity test for specificity in each subgroup.
Statistically significant for specificity heterogeneity
Abbreviations: ddPCR, droplet digital polymerase chain reaction; PLR, positive likelihood ratio; NLR, negative likelihood ratio; DOR, diagnosis odd ratio; AUC, area under the ROC curve; NA, not available.
Quality assessment
Based on QUADAS-2 tool, the details of quality assessment were listed in Table S1. As for risk of bias, the patient selection assessments were shown in column 2, index test assessments in column 3, reference standard assessments in column 4 and the flow and timing assessments in column 5. Except for one study, almost all patients were randomly enrolled without inappropriate exclusions. The results of ddPCR test were mostly interpreted independently, and reference methods were likely to correctly classify T790M mutation condition. However, regarding to the flow and timing, two studies showed the potential risk due to different detection strategies being used for T790M mutation in biopsy samples, and in addition one study showed long-time interval between tissue and plasma sampling. The applicability was high for most studies.
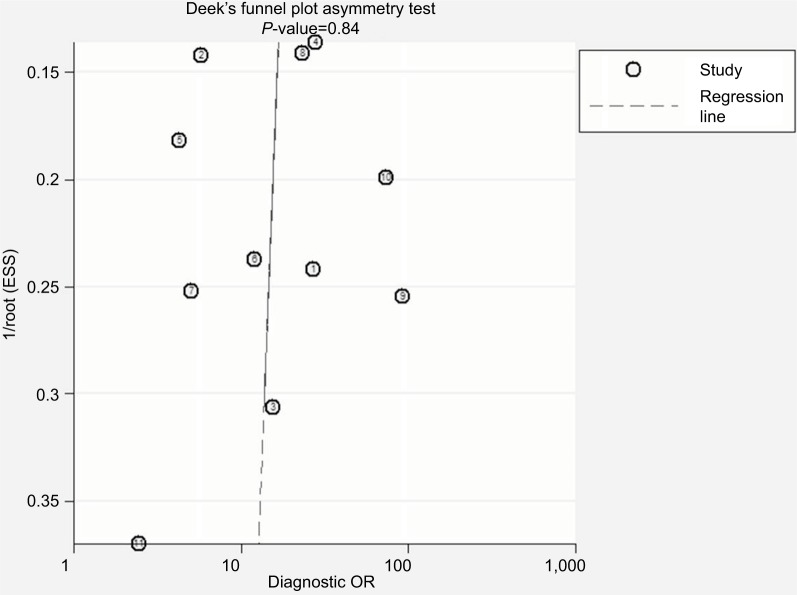
Publication bias
Deek’s funnel plot asymmetry test (P=0.84) did not show argument for publication bias (Figure 3).
Figure 3.
Assessment of publication bias by Deek’s funnel plot asymmetry test.
Abbreviation: ESS, effective sample size.
Discussion
Since the best detection method for EGFR T790M-resistant mutation is still in debate, a meta-analysis was conducted to evaluate the diagnostic performance of ddPCR. When testing in plasma ctDNA with tissue or malignant fluid test as reference method, ddPCR harbored a high sensitivity of 70.1%, a specificity of 86.9 %, and a DOR of 10.83. In stratified analysis, the picoliter-ddPCR performed well with the sensitivity, specificity, and DOR being 81.8%, 94.7%, and 37.35% respectively.
Compared to Cobas and ARMS, the pooled results confirmed that ddPCR performed a high sensitivity but relatively low specificity. Two previous meta-analyses, which studied the diagnostic value of ctDNA for the detection of EGFR mutation status in NSCLC, found that the sensitivity and specificity of ARMS were 54.9% (0.41.9%–0.67.2%) and 52.5% (35.7%–0.68.8%), 97.5% (93.7%–99.1%), and 94.7% (86.0%–98.1%), respectively.30,31 Therefore, ARMS has exhibited a low sensitivity but a definitely higher specificity. As for Cobas, in the ongoing ASTRIS study, the detection rate of T790M mutation in ctDNA was much lower than that in tissues, 27.5%–62.8%.32 That could be related to its lower sensitivity. Besides, Jenkins et al evaluated the performance of Cobas plasma test from patients screened for the AURA extension and AURA2 Phase II studies, finding the concordance between the Cobas plasma and the tissue tests for detection of T790M mutation was only 65.4%.33 However, the concordance of ddPCR was 81.2% in the current study.
For the T790M mutation detection in ctDNA, digital PCR platforms generally outperform the nondigital platforms.16,18 The familiar digital platforms include ddPCR, BEAMing, and Quant Studio 3D dPCR (QS3D dPCR), with a detection limit of around 0.01%–0.04%, 0.01%, and 0.1%, respectively.19 For the same samples, Thress et al reported the sensitivity and specificity for T790M mutation detection were 71% and 83%, respectively, with the ddPCR, and 71% and 67%, respectively, with BEAMing.18 Besides, the study by Karlovich et al also revealed that the sensitivity and specificity of BEAMing was 73.3% and 50.0%.34 Hence, the ddPCR and BEAMing performed similar sensitivity but BEAMing showed a lower specificity. As for QS3D dPCR, Gu et al studied 39 samples, finding its sensitivity, specificity, and concordance for T790M mutation detection in ctDNA was 100.0%, 97.3%, and 97.44%, respectively.35
The overall concordance rate of T790M testing between the paired tumor tissues and plasma was 81.2% in the pooled analysis, which was similar to the previous studies.24,36 The PPA and NPA were 71.2% and 90.0%, respectively. Therefore, ddPCR can be a practical and alternative method for T790M mutation detection in clinics. Due to its noninvasiveness and short turnaround time, serial monitoring of EGFR mutation status using ddPCR has been carried out in recent clinics. Related studies found the detection of T790M mutation in plasma ctDNA was around 2–12 months earlier than the clinical manifestation of disease progression.12,24,37 For example, Zheng et al found almost half of the T790M ctDNA-positive patients were identified at a median time of 2.2 months prior to clinically progressive disease.24 Oxnard et al revealed that the detection of resistance mutations was up to 3.5 months before radiographic progression.37 When resistance developed, the AURA3 phase 3 trial confirmed that osimertinib is superior to standard platinum–pemetrexed chemotherapy regarding to median progression-free survival (10.1 months vs. 4.4 months), objective response rate (71% vs. 31%) as well as the quality of life.38 Nevertheless, more reliable data need to be demonstrated in future trials.
On the other hand, Soria et al recently identified that the median progression-free survival was significantly longer with osimertinib as first line than with standard EGFR-TKIs (18.9 months vs. 10.2 months).39 Therefore, there is the prediction that the detection of T790M mutation will be less important.40 However, US Food and Drug Administration (FDA) has not approved the indications of osimertinib using as first-line treatment yet, and the data from Soria’s study have not shown overall survival benefits. Thus, ddPCR can be of great use clinically in a long time. Except for T790M, ddPCR is also being used to detect other EGFR aberrations in ctDNA from lung cancer patients. When compared with tissue samples, the sensitivity, and specificity are 62.5%–90.9% and 88.9%–100% for Exon 19 del, 74.0%–88.9% and 96.6%–100% for L858R, respectively.18,22,25,41
In spite of the thoroughly search of related articles as well as careful selection and evaluation, there were several limitations of our meta-analysis. First, included cases for analysis were limited and most publications were retrospective studies. Second, different biopsy materials and detection strategies were used regarding to the reference method. And, there could be FN even for the mutation detection in biopsy samples due to tumor heterogeneity or incorrect operation.42 Third, the time between index tests and reference tests were not provided in most studies. In one study, the median time interval between plasma collection and rebiopsy test was 12.5 months.23 That meant the discrepancy between the two results could be caused by real genomic change of tumor instead of technological capability. Last but not the least, specificity heterogeneity exited due to index tests. To sum up, the limitations above should be considered when interpreting our results.
In conclusion, we found ddPCR harbored a performance of 70.1% sensitivity and 86.9 % specificity for T790M mutation detection in ctDNA. Compared to ARMS and Cobas plasma test, it showed a higher sensitivity. Besides, though the ddPCR and BEAMing performed similar sensitivity, ddPCR showed a higher specificity. In addition to the utilization of third-generation TKIs as first-line treatment for EGFR mutant lung cancers, ddPCR could still play an important role in detecting T790M resistant mutation.
Supplimentary materials
Table S1.
Quality assessment of included studies according to QUADAS-2 tool
| Study | Risk of bias
|
Applicability concerns
|
|||||
|---|---|---|---|---|---|---|---|
| Patient selection | Indexed test | Reference methods | Flow and timing | Patient selection | Indexed test | Reference methods | |
| Ishii et al1 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Thress et al2 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Wei et al3 | □ | ☺ | ? | ☺ | □ | ☺ | ? |
| Wang et al4 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Takahama et al5 | ☺ | ☺ | ☺ | □ | ☺ | ☺ | ☺ |
| Seki et al6 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Zheng et al7 | ☺ | ☺ | ☺ | □ | ☺ | ☺ | ☺ |
| Sacher et al8 | ☺ | ☺ | ? | ☺ | ☺ | ☺ | ? |
| Yu et al9 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Suzawa et al10 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Xu et al11 | ☺ | ☺ | ☺ | □ | ☺ | ☺ | ☺ |
Notes:☺=low risk; □=high risk; ?=unclear risk.
Abbreviation: QUADAS-2, Quality Assessment of Diagnostic Accuracy Studies-2.
References
- 1.Ishii H, Azuma K, Sakai K, et al. Digital PCR analysis of plasma cellfree DNA for non-invasive detection of drug resistance mechanisms in EGFR mutant NSCLC: correlation with paired tumor samples. Oncotarget. 2015;6(31):30850–30858. doi: 10.18632/oncotarget.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90(3):509–515. doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Wei Z, Shah N, Deng C, Xiao X, Zhong T, Li X. Circulating DNA addresses cancer monitoring in non small cell lung cancer patients for detection and capturing the dynamic changes of the disease. Springerplus. 2016;5:531. doi: 10.1186/s40064-016-2141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W, Song Z, Zhang Y. A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance. Cancer Med. 2017;6(1):154–162. doi: 10.1002/cam4.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahama T, Sakai K, Takeda M, et al. Detection of the T790M mutation of EGFR in plasma of advanced non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan oncology group 8014LTR study) Oncotarget. 2016;7(36):58492–58499. doi: 10.18632/oncotarget.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seki Y, Fujiwara Y, Kohno T, et al. Picoliter-droplet digital polymerase chain reaction-based analysis of cell-free plasma dna to assess EGFR mutations in lung adenocarcinoma that confer resistance to tyrosinekinase inhibitors. Oncologist. 2016;21(2):156–164. doi: 10.1634/theoncologist.2015-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6:20913. doi: 10.1038/srep20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2(8):1014–1022. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Q, Huang F, Zhang M, et al. Multiplex picoliter-droplet digital PCR for quantitative assessment of EGFR mutations in circulating cell-free DNA derived from advanced non-small cell lung cancer patients. Mol Med Rep. 2017;16(2):1157–1166. doi: 10.3892/mmr.2017.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzawa K, Yamamoto H, Ohashi K, et al. Optimal method for quantitative detection of plasma EGFR T790M mutation using droplet digital PCR system. Oncol Rep. 2017;37(5):3100–3106. doi: 10.3892/or.2017.5567. [DOI] [PubMed] [Google Scholar]
- 11.Xu T, Kang X, You X, et al. Cross-platform comparison of four leading technologies for detecting EGFR mutations in circulating tumor dna from non-small cell lung carcinoma patient plasma. Theranostics. 2017;7(6):1437–1446. doi: 10.7150/thno.16558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Acknowledgments
This work was supported by the Transformation Projects of Sci-Tech Achievements of Sichuan Province (2016CZYD0001), and the Sci-Tech Support Program of Science and Technology Department of Sichuan Province (2016SZ0073).
Footnotes
Author contributions
PT and WL were involved in the study design, whereas RZ, BC, and XT carried out data collection, data analysis, and drafting of the manuscript. YW, CW, and JJ participated in the interpretation of study results and critical revision of the manuscript. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16(2):141–151. doi: 10.1016/S1470-2045(14)71173-8. [DOI] [PubMed] [Google Scholar]
- 2.Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J Clin Oncol. 2011;29(21):2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 3.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toyooka S, Kiura K, Mitsudomi T. EGFR mutation and response of lung cancer to gefitinib. N Engl J Med. 2005;352(20):2136. doi: 10.1056/NEJM200505193522019. [DOI] [PubMed] [Google Scholar]
- 5.Ji W, Choi CM, Rho JK, et al. Mechanisms of acquired resistance to EGFR-tyrosine kinase inhibitor in Korean patients with lung cancer. BMC Cancer. 2013;13:606. doi: 10.1186/1471-2407-13-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479–485. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber B, Meldgaard P, Hager H, et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer. 2014;14:294. doi: 10.1186/1471-2407-14-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang T, Ren S, Zhou C. Role of circulating-tumor DNA analysis in non-small cell lung cancer. Lung Cancer. 2015;90(2):128–134. doi: 10.1016/j.lungcan.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Bernabe R, Hickson N, Wallace A, Blackhall FH. What do we need to make circulating tumour DNA (ctDNA) a routine diagnostic test in lung cancer? Eur J Cancer. 2017;81:66–73. doi: 10.1016/j.ejca.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 11.Francis G, Stein S. Circulating cell-free tumour DNA in the management of cancer. Int J Mol Sci. 2015;16(6):14122–14142. doi: 10.3390/ijms160614122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang Z, Cheng Y, Chen Y, et al. EGFR T790M ctDNA testing platforms and their role as companion diagnostics: correlation with clinical outcomes to EGFR-TKIs. Cancer Lett. 2017;403:186–194. doi: 10.1016/j.canlet.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Lin CC, Huang WL, Wei F, Su WC, Wong DT. Emerging platforms using liquid biopsy to detect EGFR mutations in lung cancer. Expert Rev Mol Diagn. 2015;15(11):1427–1440. doi: 10.1586/14737159.2015.1094379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li R, Ye SB, He Y, et al. Detection of epidermal growth factor receptor gene mutations in different types of non-small cell lung cancer by droplet digital PCR and amplification refractory mutation system. Zhonghua Bing Li Xue Za Zhi. 2017;46(11):764–768. doi: 10.3760/cma.j.issn.0529-5807.2017.11.006. Chinese [with English abstract]. [DOI] [PubMed] [Google Scholar]
- 15.Zhou C, Wang M, Cheng Y, et al. Detection of EGFR T790M in Asia-Pacific patients (pts) with EGFR mutation-positive advanced non-small cell lung cancer (NSCLC): circulating tumour (ct) DNA analysis across 3 platforms. Ann Oncol. 2017;28(Suppl 5):v460–v496. [Google Scholar]
- 16.Zhu L, Zhang S, Xun Y, et al. Comparison of the Amplification Refractory Mutation System, Super Amplification Refractory Mutation System, and Droplet Digital PCR for T790M mutation detection in non-small cell lung cancer after failure of tyrosine kinase inhibitor treatment. Pathol Oncol Res. 2017 Sep 3; doi: 10.1007/s12253-017-0286-3. Epub. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Song Z, Zhang Y. A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance. Cancer Med. 2017;6(1):154–162. doi: 10.1002/cam4.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90(3):509–515. doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Kasahara N, Kenmotsu H, Serizawa M, et al. Plasma epidermal growth factor receptor mutation testing with a chip-based digital PCR system in patients with advanced non-small cell lung cancer. Lung Cancer. 2017;106:138–144. doi: 10.1016/j.lungcan.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66(16):7854–7858. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 21.Zhu G, Ye X, Dong Z, et al. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J Mol Diagn. 2015;17(3):265–272. doi: 10.1016/j.jmoldx.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Yu Q, Huang F, Zhang M, et al. Multiplex picoliter-droplet digital PCR for quantitative assessment of EGFR mutations in circulating cell-free DNA derived from advanced non-small cell lung cancer patients. Mol Med Rep. 2017;16(2):1157–1166. doi: 10.3892/mmr.2017.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzawa K, Yamamoto H, Ohashi K, et al. Optimal method for quantitative detection of plasma EGFR T790M mutation using droplet digital PCR system. Oncol Rep. 2017;37(5):3100–3106. doi: 10.3892/or.2017.5567. [DOI] [PubMed] [Google Scholar]
- 24.Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6:20913. doi: 10.1038/srep20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2(8):1014–1022. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 28.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PMM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56(11):1129–1135. doi: 10.1016/s0895-4356(03)00177-x. [DOI] [PubMed] [Google Scholar]
- 29.van Enst WA, Ochodo E, Scholten RJ, Hooft L, Leeflang MM. Investigation of publication bias in meta-analyses of diagnostic test accuracy: a meta-epidemiological study. BMC Med Res Methodol. 2014;14:70. doi: 10.1186/1471-2288-14-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu M, Wang J, Xu Y, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2015;24(1):206–212. doi: 10.1158/1055-9965.EPI-14-0895. [DOI] [PubMed] [Google Scholar]
- 31.Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep. 2014;4:6269. doi: 10.1038/srep06269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marinis FD, Cho BC, Kim DW, Kim SW, Hochmair MJ, Metro G. ASTRIS: A real world treatment study of osimertinib in patients (pts) with EGFR T790M positive non-small cell lung cancer (NSCLC) J Clin Oncol. 2017;35(15 Suppl):9036–9036. [Google Scholar]
- 33.Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA Analysis for detection of the EGFR T790M Mutation in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2017;12(7):1061–1070. doi: 10.1016/j.jtho.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR Mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO-1686) Clin Cancer Res. 2016;22(10):2386–2395. doi: 10.1158/1078-0432.CCR-15-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu J, Zang W, Liu B, et al. Evaluation of digital PCR for detecting low-level EGFR mutations in advanced lung adenocarcinoma patients: a cross-platform comparison study. Oncotarget. 2017;8(40):67810–67820. doi: 10.18632/oncotarget.18866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishii H, Azuma K, Sakai K, et al. Digital PCR analysis of plasma cell-free DNA for non-invasive detection of drug resistance mechanisms in EGFR mutant NSCLC: correlation with paired tumor samples. Oncotarget. 2015;6(31):30850–30858. doi: 10.18632/oncotarget.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-Mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20(6):1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376(7):629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-Mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378(2):113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 40.Popat S. Osimertinib as first-line treatment in EGFR-Mutated non-small-cell lung cancer. N Engl J Med. 2018;378(2):192–193. doi: 10.1056/NEJMe1714580. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Chang N, Yang G, et al. A comparison of ARMS-Plus and droplet digital PCR for detecting EGFR activating mutations in plasma. Oncotarget. 2017;8(67):112014–112023. doi: 10.18632/oncotarget.22997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei Z, Shah N, Deng C, Xiao X, Zhong T, Li X. Circulating DNA addresses cancer monitoring in non small cell lung cancer patients for detection and capturing the dynamic changes of the disease. Springer-plus. 2016;5:531. doi: 10.1186/s40064-016-2141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahama T, Sakai K, Takeda M, et al. Detection of the T790M mutation of EGFR in plasma of advanced non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan oncology group 8014LTR study) Oncotarget. 2016;7(36):58492–58499. doi: 10.18632/oncotarget.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seki Y, Fujiwara Y, Kohno T, et al. Picoliter-droplet digital polymerase chain reaction-based analysis of cell-free plasma dna to assess EGFR mutations in lung adenocarcinoma that confer resistance to tyrosine-kinase inhibitors. Oncologist. 2016;21(2):156–164. doi: 10.1634/theoncologist.2015-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu T, Kang X, You X, et al. Cross-platform comparison of four leading technologies for detecting EGFR mutations in circulating tumor dna from non-small cell lung carcinoma patient plasma. Theranostics. 2017;7(6):1437–1446. doi: 10.7150/thno.16558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Quality assessment of included studies according to QUADAS-2 tool
| Study | Risk of bias
|
Applicability concerns
|
|||||
|---|---|---|---|---|---|---|---|
| Patient selection | Indexed test | Reference methods | Flow and timing | Patient selection | Indexed test | Reference methods | |
| Ishii et al1 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Thress et al2 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Wei et al3 | □ | ☺ | ? | ☺ | □ | ☺ | ? |
| Wang et al4 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Takahama et al5 | ☺ | ☺ | ☺ | □ | ☺ | ☺ | ☺ |
| Seki et al6 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Zheng et al7 | ☺ | ☺ | ☺ | □ | ☺ | ☺ | ☺ |
| Sacher et al8 | ☺ | ☺ | ? | ☺ | ☺ | ☺ | ? |
| Yu et al9 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Suzawa et al10 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Xu et al11 | ☺ | ☺ | ☺ | □ | ☺ | ☺ | ☺ |
Notes:☺=low risk; □=high risk; ?=unclear risk.
Abbreviation: QUADAS-2, Quality Assessment of Diagnostic Accuracy Studies-2.