Abstract
Objective
This study was to investigate the expression and clinical significance of RRBP1 in esophageal carcinoma.
Materials and methods
RRBP1 expression was detected in 120 esophageal carcinoma and matched adjacent normal tissues, and the relationship of RRBP1 with clinicopathological characteristics and prognosis was analyzed.
Results
RRBP1 was highly expressed in esophageal carcinoma tissues compared with matched adjacent normal tissues (P<0.05). Moreover, RRBP1 expression was associated with T stage, lymph node metastasis, and TNM stage in esophageal carcinoma (P<0.05). Survival analysis revealed that RRBP1, T stage, lymph node metastasis, and TNM stage were significantly associated with patients’ prognosis.
Conclusion
RRBP1 is highly expressed in esophageal carcinoma and can serve as a potential biomarker to predict patients’ prognosis.
Keywords: RRBP1, prognosis, esophageal carcinoma, survival analysis
Introduction
Esophageal carcinoma is one of the most common malignant tumors in China, which accounts for the sixth most common cause of cancer-related death in the world.1,2 Surgical resection is the main treatment for esophageal cancer patients; however, the 5-year survival rate of esophageal cancer patients after surgery is still less than 25%.3,4 Currently, the incidence of esophageal carcinoma is still increasing in China.1,5,6 The early diagnosis of esophageal carcinoma is a tough challenge.7,8 Thus, it would be meaningful to explore novel molecular biomarkers associated with the early diagnosis and prognosis of esophageal cancer.
RRBP1 is an endoplasmic reticulum membrane protein, which plays a critical role in the transportation and secretion of nascent proteins.9 Recently, RRBP1 over-expression has been frequently observed in lung cancer, breast cancer, and colorectal cancer.10–12 Moreover, RRBP1 correlates with shorter survival and can serve as a valuable prognostic factor in Her-2-positive breast cancer patients.13 RRBP1 over-expression contributes to the progression of colorectal cancer and is useful for predicting patients’ prognosis.14 Thus, this evidence suggests that RRBP1 may be a key oncogene involved in tumor formation and progression. However, the expression and clinical significance of RRBP1 have never been reported in esophageal carcinoma.
In this study, we detected the expression of RRBP1 in 120 cases of esophageal carcinoma and matched adjacent normal tissues, and analyzed the correlation between RRBP1 expression and clinicopathological features. Moreover, whether RRBP1 could be a potential prognostic biomarker in patients with esophageal carcinoma was further assessed.
Materials and methods
Patients and samples
One hundred and twenty esophageal carcinoma (without chemotherapy and radiotherapy before surgery) specimens were collected from patients presenting to Cangzhou Central Hospital during 2010–2014. Matched adjacent normal tissues were collected 3 cm from esophageal carcinoma tissue. Patients included 57 males and 63 females with a mean age of 58 years (range, 32–74 years). Clinical pathological characteristics including age, gender, history of smoking, tumor location, T stage, lymph node metastasis, and TNM stage were obtained from hospital records. Follow-up time was from the day of surgery. No patient was lost during follow-up and the follow-up duration ranged from 1 to 65 months (mean, 38.1 months). All the samples were diagnosed as squamous cell carcinoma. The pathological diagnosis was confirmed by two pathologists in Cangzhou Central Hospital.
Quantitative real-time polymerase chain reaction (q-RT-PCR)
All tissues were frozen in liquid nitrogen. RNA was extracted by RNAisoTM PLUS (Thermo Fisher Scientific, Waltham, MA, USA) and reverse transcribed into cDNA by cDNA Synthesis Kit (TaKaRa Corp, Dalian, China). Quantitative analysis of RRBP1 was performed using 7500 SYBR Green Fast Real-Time PCR System (Thermo Fisher Scientific,). The reaction conditions were 95°C for 10 min, followed 95° for 15 s for 40 cycles and 60°C for 60 s. The primer sequences of RRBP1 were 5′-TGAATCCTCCAAAGACCACA-3′ and 5′-CTTTCCCTCTCGCGTCTCT-3′. The primer sequences of GAPDH were 5′-CTGAACGGGAAGCTCACTGG-3′ and 5′-TGAGGTCCACCACCCTGTTG-3′. The experiments were repeated three times under the same conditions.
Western blot analysis
All tissues were frozen in liquid nitrogen. Proteins were extracted by protease inhibitors and quantified by the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). An amount of 50 μg per sample was resolved on 5% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. After blocking in 5% fat-free milk at room temperature, membranes were incubated with RRBP1 (Epitomics, Inc., Burlingame, CA, USA) (diluted 1:1000) and GAPDH (Zhongshan Corp, Beijing, China) (diluted 1:1000) antibodies overnight at 4°C. Then, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h. The signals were measured by enhanced chemiluminescence detection reagents.
Immunohistochemical (IHC) staining
Sections (2 μm thick) were deparaffinized with xylene and rehydrated in graded ethanol. Endogenous peroxidase was wiped off with 3% hydrogen peroxide and antigenicity was repaired by 0.01 mol/L sodium citrate buffer (pH 6.0). All sections were incubated with rabbit monoclonal RRBP1 antibody (Epitomics, Inc.) (diluted 1:200) at room temperature for 2 h. After incubation with secondary biotinylated antibody, sections were stained with diaminobenzidine (DAB) and hematoxylin.
The staining of RRBP1 was analyzed by semi-quantitative method. The staining intensity was scored as blank (0), weak (1), moderate (2), and strong (3). The percentage of positive cells was scored as <5% (0), ≥5% –<25% (1), 25% –50% (2), and >50% (3). The scores were calculated by multiplying these two values (ranging from 0 to 9). These scores (≥4) were defined as RRBP1 high-expression, and others were defined as RRBP1 low-expression (<4). All IHC scores were assessed by two pathologists independently without the clinical information.
Statistical analysis
All data were analyzed with SPSS software (version 19.0; IBM Corporation, Armonk, NY, USA). IHC results were analyzed by chi-square test. Survival analysis was performed by the Kaplan–Meier method and log-rank test. Multivariate analysis was assessed by Cox’s proportional hazards model. The comparison of two-sample mean was evaluated using independent samples t-test. P-value of <0.05 was defined as statistically significant.
Ethics statement
This study was approved by the Cangzhou Central Hospital Ethnics Committee. All patients signed informed consent and agreed to the use of their tissue samples in this study.
Results
RRBP1 is highly expressed in esophageal carcinoma
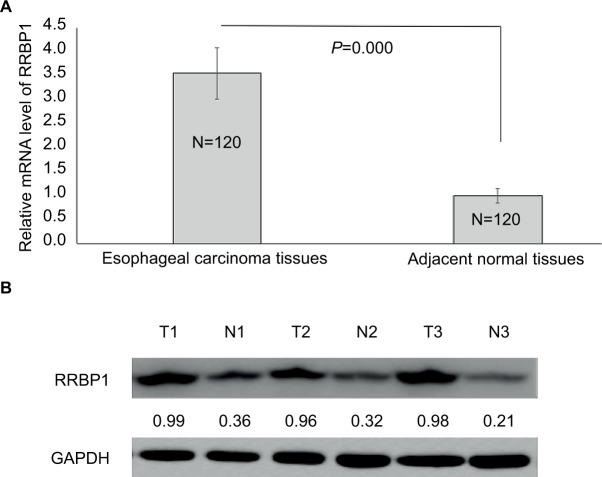
First, we detected the expression of RRBP1 in 120 esophageal carcinoma specimens and matched adjacent normal tissues by qRT-PCR and Western blot assays. qRT-PCR results indicated that RRBP1 mRNA level was significantly higher in esophageal carcinoma tissues compared with matched adjacent normal tissues (Figure 1A, P=0.000). Meanwhile, Western blot results revealed that RRBP1 protein was highly expressed in esophageal carcinoma tissues compared with matched adjacent normal tissues (Figure 1B, P=0.000). These data indicated that RRBP1 was highly expressed in esophageal carcinoma.
Figure 1.
RRBP1 expression.
Notes: The expression of RRBP was detected in esophageal carcinoma and matched adjacent normal tissues by qRT-PCR (A) and Western blot (B). T, esophageal carcinoma tissue; N, matched adjacent normal esophageal tissue.
RRBP1 expression correlates with clinical pathological characteristics in esophageal carcinoma
Subsequently, we detected the expression of RRBP1 in 120 esophageal carcinoma specimens and matched adjacent normal tissues by IHC. As shown in Figure 2, positive expression of RRBP1 was located in cell cytoplasm and easily observed in esophageal carcinoma tissues, but was hardly detected in normal esophageal tissues. The high-expression rates of RRBP1 in esophageal carcinoma and normal esophageal tissues were 59.2% and 11.7%, respectively, and the difference was statistically significant (Table 1, P=0.000). Moreover, RRBP1 expression was associated with T stage, lymph node metastasis, and TNM stage in esophageal carcinoma (Table 2, P<0.05), but was not associated with age, gender, history of smoking, and tumor location (Table 2, P>0.05).
Figure 2.
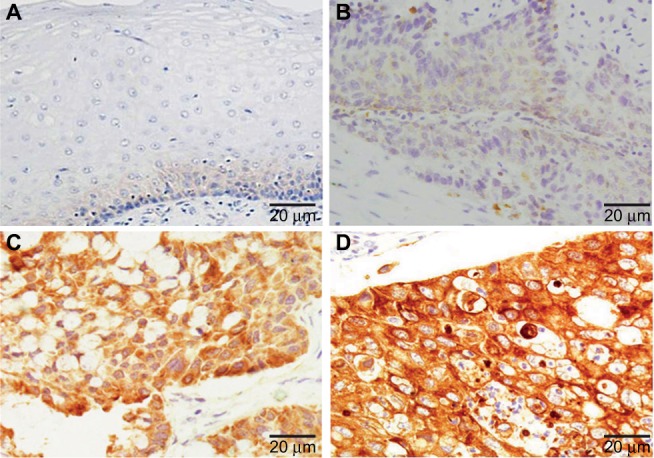
RRBP1 expression was detected in esophageal carcinoma and matched adjacent normal tissues by immunohistochemical staining.
Notes: (A) Adjacent normal tissues; (B) weak staining of RRBP1 in esophageal carcinoma; (C) moderate staining of RRBP1 in esophageal carcinoma; (D) strong staining of RRBP1 in esophageal carcinoma.
Table 1.
RRBP1 expression in esophageal carcinoma and normal esophageal tissues by immunohistochemical staining
| Types | N | RRBP1
|
P-value | |
|---|---|---|---|---|
| Low-expression (%) | High-expression (%) | |||
| Esophageal carcinoma tissues | 120 | 49 (40.8) | 71 (59.2) | 0.000 |
| Normal esophageal tissues | 120 | 106 (88.3) | 14 (11.7) | |
Table 2.
RRBP1 expression correlation with clinicopathological characteristics in esophageal carcinoma
| Clinicopathological characteristics | N | RRBP1
|
P-value | |
|---|---|---|---|---|
| Low-expression | High-expression | |||
| Age (years) | ||||
| ≤58 | 58 | 27 | 31 | 0.266 |
| >58 | 62 | 22 | 40 | |
| Gender | ||||
| Male | 57 | 24 | 33 | 0.714 |
| Female | 63 | 25 | 38 | |
| History of smoking | ||||
| Negative | 55 | 20 | 35 | 0.853 |
| Positive | 65 | 29 | 36 | |
| Tumor location | ||||
| Upper esophagus | 56 | 27 | 29 | 0.14 |
| Middle-lower esophagus | 64 | 22 | 42 | |
| T stage | ||||
| T1–T2 | 37 | 30 | 7 | 0.000 |
| T3–T4 | 83 | 19 | 64 | |
| Lymph node metastasis | ||||
| Negative | 83 | 42 | 41 | 0.001 |
| Positive | 37 | 7 | 30 | |
| TNM stages | ||||
| I–II | 37 | 30 | 7 | 0.000 |
| III–IV | 83 | 19 | 64 | |
High-expression of RRBP1 predicts an unfavorable survival rate in esophageal carcinoma patients
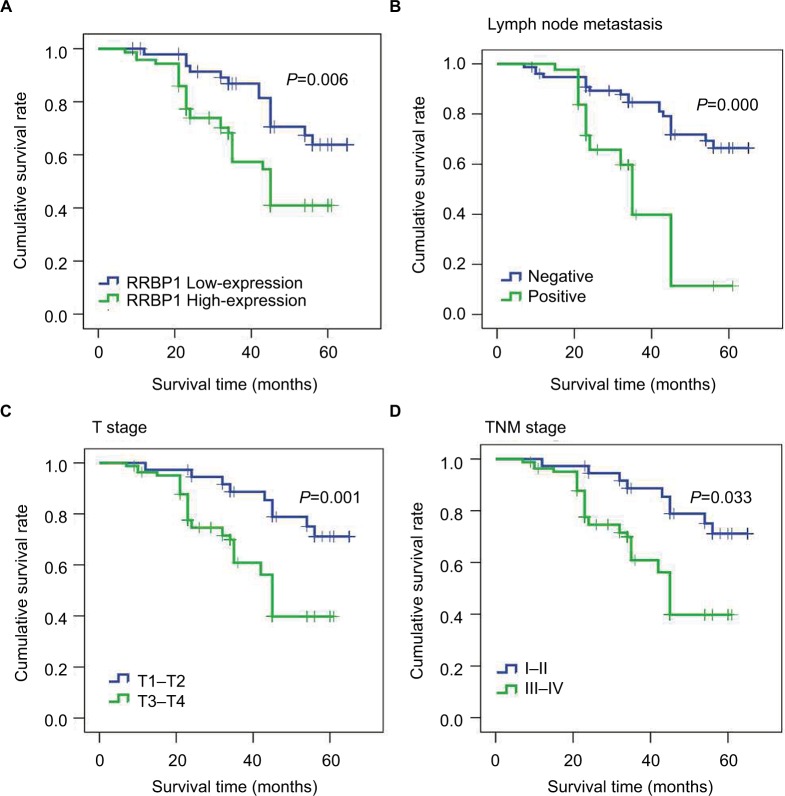
Then, we further analyzed the correlation between RRBP1 expression and patients’ survival by Kaplan–Meier method and Cox’s proportional hazards model. Kaplan–Meier analysis revealed that the median survival time of patients with RRBP1 high-expression was 43 months, which was significantly shorter compared with those with RRBP1 low-expression (56 months) (Table 3, Figure 3A, P=0.006). Moreover, T stage, lymph node metastasis, and TNM stage rather than age, gender, and history of smoking were confirmed to be associated with patients’ survival (Table 3, Figure 3B–D, P<0.05). Furthermore, multivariate Cox regression analysis showed RRBP1 high-expression was significantly associated with unfavorable survival rate in esophageal carcinoma. Except for age, gender, history of smoking and tumor location, T stage, lymph node metastasis and TNM stage were also confirmed to be correlated with patients’ survival (Table 4, P<0.05).
Table 3.
Patient survival: Kaplan–Meier survival analysis
| Variables | N | Survival time (months, 95% CI) | P-value |
|---|---|---|---|
| RRBP1 | |||
| Low-expression | 49 | 56 (51–60) | 0.006 |
| High-expression | 71 | 43 (39–48) | |
| Gender | |||
| Male | 57 | 51 (47–56) | 0.323 |
| Female | 63 | 47 (42–52) | |
| Age (years) | |||
| ≤58 | 58 | 50 (45–55) | 0.963 |
| >58 | 62 | 50 (45–54) | |
| History of smoking | |||
| Negative | 55 | 49 (44–55) | 0.845 |
| Positive | 65 | 50 (45–54) | |
| Tumor location | |||
| Upper esophagus | 56 | 50 (45–53) | 0.213 |
| Middle-lower esophagus | 64 | 49 (45–54) | |
| T stage | |||
| T1–T2 | 37 | 58 (53–62) | 0.001 |
| T3–T4 | 83 | 43 (39–48) | |
| Lymph node metastasis | |||
| Negative | 83 | 55 (51–59) | 0.000 |
| Positive | 37 | 36 (31–41) | |
| TNM stages | |||
| I–II | 37 | 58 (53–62) | 0.033 |
| III–IV | 83 | 43 (39–46) |
Figure 3.
Kaplan–Meier survival analysis.
Notes: Results indicated that RRBP1 expression (A), lymph node metastasis (B), T stage (C), and TNM (D) stage were associated with patients’ prognosis.
Table 4.
Patients’ survival evaluation by multivariate Cox regression analysis
| Variables | Hazard ratio | 95% CI | P-value |
|---|---|---|---|
| RRBP1 (high-expression vs low-expression) | 2.441 | 1.267–4.702 | 0.008 |
| Gender (male vs female) | 1.329 | 0.736–2.400 | 0.346 |
| Age (≤58 vs >58 years) | 0.994 | 0.516–1.916 | 0.987 |
| History of smoking (positive vs negative) | 0.963 | 0.498–1.862 | 0.912 |
| Tumor location (upper vs middle-lower) | 0.929 | 0.538–1.629 | 0.921 |
| T stage (T3–T4 vs T1–T2) | 3.054 | 1.453–6.421 | 0.003 |
| Lymph node metastasis (positive vs negative) | 4.024 | 2.180–7.424 | 0.000 |
| TNM stage (III–IV vs I–II) | 3.054 | 1.452–6.421 | 0.003 |
Discussion
RRBP1, an endoplasmic reticulum membrane protein, is mainly located on the endoplasmic reticulum membrane and plays an important role in the transportation and secretion of nascent proteins.9,11,15 Moreover, RRBP1 is crucial for the terminal differentiation of secretory tissues and the procollagen biosynthesis of secretory tissues.16–19 Recently, RRBP1 has been reported to be connected to the regulation of unfolded protein response signaling molecules and the accumulation of perinuclear autophagosomes of cancer cells.10,20,21 In addition, RRBP1 was confirmed as an oncogene highly expressed in lung cancer, breast cancer, and colorectal cancer.10–12 RRBP1 over-expression predicts unfavorable survival rates in colorectal cancer patients.14 However, the expression and clinical significance of RRBP1 have never been reported in esophageal carcinoma.
In this study, in order to investigate the clinical significance of RRBP1 in esophageal carcinoma, we detected the expression of RRBP1 in 120 cases of esophageal carcinoma and matched adjacent normal tissues by qRT-PCR, Western blot, and IHC assays. qRT-PCR and Western blot results both showed that RRBP1 was highly expressed in esophageal carcinoma tissues compared to matched adjacent normal tissues, suggesting that RRBP1 high-expression might contribute to the occurrence of esophageal carcinoma. Meanwhile, IHC results showed that RRBP1 high-expression was observed in 59.2% esophageal carcinoma, but only in 11.7% matched adjacent normal tissues. Thus, IHC results were consistent with qRT-PCR and Western blot results, which further supported that RRBP1 high-expression was correlated with the occurrence of esophageal carcinoma. In addition, our data revealed that RRBP1 expression was associated with T stage, lymph node metastasis, and TNM stage in esophageal carcinoma, which suggested that RRBP1 expression might be connected to the progression of esophageal carcinoma. Survival analysis showed that patients with RRBP1 high-expression presented shorter survival rates compared with those with RRBP1 low-expression, indicating that RRBP1 might serve as a prognostic biomarker in esophageal carcinoma. It is well-known that T stage, lymph node metastasis, and TNM stage are key factors associated with the progression of esophageal carcinoma and patients’ survival.22–28 In the present study, our data also indicated that T stage, lymph node metastasis, and TNM stage were independent prognostic factors in esophageal carcinoma. Thus, our data suggested that RRBP1 high-expression might contribute to the progression of esophageal carcinoma, which results in a poorer prognosis. In addition, Liang et al reported that RRBP1 was a valuable prognostic factor in Her-2-positive breast cancer patients.13 Pan et al reported that RRBP1 promoted the progression of colorectal cancer and predicted prognosis.14
Conclusion
This paper is the first to report that RRBP1 is an oncogene highly expressed in esophageal carcinoma. Additionally, our data indicate that RRBP1 may be connected with the occurrence and progression of esophageal carcinoma, and serve as an independent prognostic factor to predict patients’ prognosis. Of course, further investigations are needed to validate our findings.
Acknowledgments
Thanks to all patients who agreed to participate in this study.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Han T, Shu T, Dong S, et al. Chemokine-like factor-like MARVEL transmembrane domain-containing 3 expression is associated with a favorable prognosis in esophageal squamous cell carcinoma. Oncol Lett. 2017;13(5):2982–2988. doi: 10.3892/ol.2017.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Hu JM, Liu K, Liu JH, et al. CD163 as a marker of M2 macrophage, contribute to predict aggressiveness and prognosis of Kazakh esophageal squamous cell carcinoma. Oncotarget. 2017;8(13):21526–21538. doi: 10.18632/oncotarget.15630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349(23):2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 5.He Z, Li G, Tang L, Li Y. SIX1 overexpression predicts poor prognosis and induces radioresistance through AKT signaling in esophageal squamous cell carcinoma. Onco Targets Ther. 2017;10:1071–1079. doi: 10.2147/OTT.S125330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng S, Zhang X, Wang X, Li J. Downregulation of miR-138 predicts poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Biomark. 2017;20(1):49–54. doi: 10.3233/CBM-170079. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Wang W, Zhang R, et al. High expression of LAMP2 predicts poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Biomark. 2017;19(3):305–311. doi: 10.3233/CBM-160469. [DOI] [PubMed] [Google Scholar]
- 8.Zhan XH, Jiao JW, Zhang HF, et al. A three-gene signature from protein-protein interaction network of LOXL2- and actin-related proteins for esophageal squamous cell carcinoma prognosis. Cancer Med. 2017;6(7):1707–1719. doi: 10.1002/cam4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savitz AJ, Meyer DI. 180-kD ribosome receptor is essential for both ribosome binding and protein translocation. J Cell Biol. 1993;120(4):853–863. doi: 10.1083/jcb.120.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai HY, Yang YF, Wu AT, et al. Endoplasmic reticulum ribosome- binding protein 1 (RRBP1) overexpression is frequently found in lung cancer patients and alleviates intracellular stress-induced apoptosis through the enhancement of GRP78. Oncogene. 2013;32(41):4921–4931. doi: 10.1038/onc.2012.514. [DOI] [PubMed] [Google Scholar]
- 11.Telikicherla D, Marimuthu A, Kashyap MK, et al. Overexpression of ribosome binding protein 1 (RRBP1) in breast cancer. Clin Proteomics. 2012;9(1):7. doi: 10.1186/1559-0275-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krasnov GS, Oparina N, Khankin SL, et al. Colorectal cancer 2D- proteomics: identification of altered protein expression. Mol Biol (Mosk) 2009;43(2):348–356. doi: 10.1134/s0026893309020186. Russian. [DOI] [PubMed] [Google Scholar]
- 13.Liang X, Sun S, Zhang X, et al. Expression of ribosome-binding protein 1 correlates with shorter survival in Her-2 positive breast cancer. Cancer Sci. 2015;106(6):740–746. doi: 10.1111/cas.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Y, Cao F, Guo A, et al. Endoplasmic reticulum ribosome-binding protein 1, RRBP1, promotes progression of colorectal cancer and predicts an unfavourable prognosis. Br J Cancer. 2015;113(5):763–772. doi: 10.1038/bjc.2015.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao W, Li Q, Zhu R, Jin J. la autoantigen induces ribosome binding protein 1 (RRBP1) expression through internal ribosome entry site (IRES)-mediated translation during cellular stress condition. Int J Mol Sci. 2016;17(7) doi: 10.3390/ijms17071174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benyamini P, Webster P, Meyer DI. Knockdown of p180 eliminates the terminal differentiation of a secretory cell line. Mol Biol Cell. 2009;20(2):732–744. doi: 10.1091/mbc.E08-07-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa-Goto K, Tanaka K, Ueno T, et al. p180 is involved in the interaction between the endoplasmic reticulum and microtubules through a novel microtubule-binding and bundling domain. Mol Biol Cell. 2007;18(10):3741–3751. doi: 10.1091/mbc.E06-12-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbe L, Lundberg E, Oksvold P, et al. Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics. 2008;7(3):499–508. doi: 10.1074/mcp.M700325-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Olsen JV, Blagoev B, Gnad F, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 20.Cardoso CM, Groth-Pedersen L, Hoyer-Hansen M, et al. Depletion of kinesin 5B affects lysosomal distribution and stability and induces peri-nuclear accumulation of autophagosomes in cancer cells. PLoS One. 2009;4(2):e4424. doi: 10.1371/journal.pone.0004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diefenbach RJ, Diefenbach E, Douglas MW, Cunningham AL. The ribosome receptor, p180, interacts with kinesin heavy chain, KIF5B. Biochem Biophys Res Commun. 2004;319(3):987–992. doi: 10.1016/j.bbrc.2004.05.069. [DOI] [PubMed] [Google Scholar]
- 22.Chirieac LR, Swisher SG, Ajani JA, et al. Posttherapy pathologic stage predicts survival in patients with esophageal carcinoma receiving preoperative chemoradiation. Cancer. 2005;103(7):1347–1355. doi: 10.1002/cncr.20916. [DOI] [PubMed] [Google Scholar]
- 23.Chao YK, Chan SC, Liu YH, et al. Pretreatment T3–4 stage is an adverse prognostic factor in patients with esophageal squamous cell carcinoma who achieve pathological complete response following preoperative chemoradiotherapy. Ann Surg. 2009;249(3):392–396. doi: 10.1097/SLA.0b013e3181949e9f. [DOI] [PubMed] [Google Scholar]
- 24.Huang J, Hu W, Pang L, Chen J, Yang H. Value of positive lymph node ratio for predicting postoperative distant metastasis and prognosis in esophageal squamous cell carcinoma. Oncol Res Treat. 2015;38(9):424–428. doi: 10.1159/000439038. [DOI] [PubMed] [Google Scholar]
- 25.Akita H, Doki Y, Yano M, et al. Effects of neoadjuvant chemotherapy on primary tumor and lymph node metastasis in esophageal squamous cell carcinoma: additive association with prognosis. Dis Esophagus. 2009;22(4):291–297. doi: 10.1111/j.1442-2050.2008.00879.x. [DOI] [PubMed] [Google Scholar]
- 26.Conio M, Gostout CJ. Histopathologic findings predicting lymph node metastasis and prognosis of patients with superficial esophageal carcinoma. Analysis of 240 surgically resected tumors. Gastrointest Endosc. 2001;54(5):668–669. [PubMed] [Google Scholar]
- 27.Yu X, Zhang J, Zhong H, et al. Decreased tumor suppressor candidate 3 predicts poor prognosis of patients with esophageal squamous cell carcinoma. Int J Med Sci. 2016;13(12):963–969. doi: 10.7150/ijms.16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Yang T, Xu Z, Cao X. Upregulation of the long non-coding RNA BANCR correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Biomed Pharmacother. 2016;82:406–412. doi: 10.1016/j.biopha.2016.05.014. [DOI] [PubMed] [Google Scholar]