Abstract
Fatty acid esters of hydroxy fatty acids (FAHFAs) are a recently discovered class of endogenous lipids with anti-diabetic and anti-inflammatory activities. Interest in these lipids is due to their unique biological activity and the observation that insulin resistant people have lower palmitic acid esters of hydroxystearic acid (PAHSAs) levels, suggesting that a FAHFA deficiency may contribute to metabolic disease. Rigorous testing of this hypothesis will require the measurement of many clinical samples; however, current analytical workflows are too slow to enable samples to be analyzed quickly. Here, we describe the development of a significantly faster workflow to measure FAHFAs that optimizes the fractionation and chromatography of these lipids. We can measure FAHFAs in 30 minutes with this new protocol versus 90 minutes using the older protocol with comparable performance in regioisomer detection and quantitation. We also discovered through this optimization that oleic acid esters of hydroxystearic acids (OAHSAs), another family of FAHFAs, have a much lower background signal than PAHSAs, which makes them easier to measure. Our faster workflow was able to quantify changes in PAHSAs and OAHSAs in mouse tissues and human plasma, highlighting the potential of this protocol for basic and clinical applications.
Graphical Abstract

The adipose tissue Glut4 overexpressor (AG4OX) mouse is a valuable model for understanding the molecular pathways related to insulin sensitivity and metabolic disease1–5. Normally, obesity in rodents and humans is linked to the onset of insulin resistance and, eventually, the development of diabetes6–7. AG4OX mice are unique because they are obese, yet they remain insulin sensitive and are resistant to diabetes1–5. Gene expression studies followed by metabolic tracer studies revealed that AG4OX mice have increased lipid production (i.e., de novo lipogenesis (DNL)) in their adipose tissue and this increased DNL is necessary for the improved metabolism of these mice3, 5. Elevated fatty acid levels are typically associated with poorer metabolic outcomes6 suggesting that AG4OX mice might be producing beneficial lipids in their adipose tissue. Lipidomics of these animals followed by structure elucidation of the lipids with the largest fold change between AG4OX and wild-type(WT) samples revealed the existence of a new class of lipids called Fatty Acid Esters of Hydroxy Fatty Acids (FAHFAs). FAHFAs are highly elevated in AG4OX mice8 and subsequent biological studies demonstrated that FAHFAs have potent anti-diabetic and anti-inflammatory activities8–10. Serum and adipose tissue PAHSA levels from insulin resistant people are lower, indicating that a FAHFA deficiency might underlie metabolic disease8.
FAHFA families are defined by the composition of the fatty acid and hydroxy fatty acid. Analysis of adipose tissue from AG4OX mice revealed the existence of 16 different FAHFA families8. For example, the two most highly upregulated FAHFA families in adipose tissue of AG4OX mice were palmitic acid esters of hydroxy stearic acids (PAHSAs) and oleic acid esters of hydroxy stearic acids (OAHSAs).
Within each FAHFA family discovered to date, there are multiple regioisomers, which differ by the position of the ester. The distribution and amounts of the different regioisomers can vary substantially between tissues. In mice, for example, there are eight different PAHSA regioisomers in adipose tissue three in liver, and six in serum8. The structural similarity of these regioisomers poses a challenge for their detection and quantitation. Obtaining baseline separation of different PAHSA regioisomers required extremely long isocratic gradients (90 minutes in length)8, 11.
As interest in FAHFAs grows, faster protocols for analyzing and measuring different families and regioisomers12 are needed. The current protocol for PAHSA measurements, for instance, is time-consuming because of the difficulty in separating PAHSA isomers from each other and from a contaminating ceramide8, 11. This protocol also has background PAHSA signal which is negligible for tissues with high PAHSA levels. However, this background is problematic in tissues with low PAHSA concentrations, such as serum, where this background can constitute ~10–15% of the total PAHSA signal.
Here, we optimized the protocol for endogenous FAHFA measurements to afford a method with significantly shorter run times but similar analytical performance. Our optimized protocol provides a rapid, robust method for measuring FAHFAs (PAHSAs and OAHSAs) that should increase the throughput and quality of endogenous FAHFA measurements for basic and clinical studies.
EXPERIMENTAL SECTION
Chemicals and Standards
All PAHSAs, OAHSAs, and 13C4-9-PAHSA were purchased from Cayman Chemical Company. The internal standard 13C18-12-OAHSA was synthesized as described (Supporting Information).
Samples for FAHFA Measurements
Male AG4OX mice and WT FVB littermate controls13 at 40 weeks of age were used for FAHFA measurements. Mice were fed a chow diet (Lab Diet, 5008) ad libitum. Mice were kept on a 14 hr light, 10 hr dark schedule at 22–23C. All animal care and use procedures were in accordance with and approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center, Boston, MA. Human plasma samples were obtained and prepared as previously described14. An IRB letter of exemption for the de-identified human plasma samples used in this study is on file at the Salk Institute for Biological Sciences.
Lipid Extraction
Lipid extraction was based on previous literature11, 15. Perigonadal white adipose tissue (PGWAT, 150 mg) was dounce homogenized on ice in a mixture of 1.5 mL PBS, 1.5 mL methanol, and 3 mL of chloroform. 5 pmol/sample of 13C4-9-PAHSA and 13C18-12-OAHSA and 1 pmol/sample of 13C16-5-PAHSA were added to the chloroform prior to extraction. The mixture was then centrifuged at 2,200 g, 5 min, 4°C to separate the organic (bottom) and aqueous phases. The organic phase was then transferred to a new vial and dried down under a gentle stream of nitrogen and stored at −80°C. For blood samples, 200 µL of mouse serum or human plasma was added to 1.3 mL of PBS, 1.5 mL methanol and 3 mL chloroform with internal standard (1 pmol/sample of 13C4-9-PAHSA, 13C18-12-OAHSA, and 13C16-5-PAHSA). The resulting mixture was vortexed for 30s and then centrifuged to separate the organic and aqueous phases. The organic phase was transferred, dried and stored following the same method for tissues.
Solid Phase Extraction
SPE was slightly modified from a previously reported method11. SPE was performed at room temperature using positive pressure (nitrogen) to push solvents though a Strata SI-1 Silica SPE cartridge (500 mg silica, 3 mL, Phenomenex, 8B-S012-HBJ-T). For the following steps, we added 3 × 2 mL aliquots of solvents. The SPE cartridge was first pre-washed with 6 mL of ethyl acetate and then conditioned with 6 mL hexane. Afterwards, extracted lipids, from the previous lipid extraction step, were reconstituted in 200 µL chloroform and then applied to the cartridge. Neutral lipids were eluted using 6 mL 5% ethyl acetate in hexane, followed by elution of FAHFAs using 4 mL ethyl acetate. The FAHFA fraction was dried down under a gentle stream of nitrogen and stored at −80°C.
Targeted LC/MS Analysis of PAHSAs and OAHSAs
PAHSAs and OAHSAs were measured on a TSQ Quantiva LC/MS instrument using Multiple Reaction Monitoring (MRM) in negative ionization mode. The following MS source parameters were used: spray voltage, 3.5 kV; ion transfer tube temperature, 325°C; vaporizer temperature, 275°C; sheath gas, 2.7 L/min; aux gas, 5.0 L/min; and sweep gas, 1.5 L/min. An Acquity UPLC BEH C18 column (1.7 µm, 2.1 mm × 100 mm, Waters) was used for separation of FAHFAs. PAHSAs and OAHSAs were resolved with isocratic flow at 0.2 mL/min using 93:7 methanol:water with 5 mM ammonium acetate and 0.03% ammonium hydroxide (v/v) over 30 minutes. The column was kept at a constant temperature of 25°C. Each extracted and fractionated sample was reconstituted in 40 µL methanol and 10 µL was injected for analysis. Multiple reaction monitoring (MRM) was utilized using one quantifier and one or two qualifier transitions per FAHFA (Table 1). The collision energies (CE) for the MRM transitions are as follows: FA (29 V), HFA (28 V), and HFA-H2O (27 V).
Table 1. MRM transitions for PAHSAs and OAHSAs and their respective standards.
Quantifier ion is the fatty acid (FA), and the qualifier ions are the hydroxy fatty acid (HFA) and the HFA dehydration product (HFA-H2O). The HFA-H2O ion and the FA ion for OAHSA have the same m/z. The HFA ion and the FA ion for 13C18-12-OAHSA have the same m/z. Q1 represents parent ion and Q3 represents daughter ions.
| FAHFA | Q1 [M-H]− | Q3 FA | Q3 HFA | Q3 HFA-H2O |
|---|---|---|---|---|
| PAHSA | 537.5 | 255.2 | 299.3 | 281.2 |
| 13C4-9-PAHSA | 541.5 | 259.2 | 299.3 | 281.2 |
| OAHSA | 563.5 | 281.2 | 299.3 | ----- |
| 13C18-12-OAHSA | 581.6 | 299.3 | ----- | 281.2 |
RESULTS AND DISCUSSION
Development of a faster FAHFA LC-MS gradient
In our previous studies, we established a 90-minute liquid chromatography (LC) method to resolve PAHSA isomers using a Luna C18(2) (3 µm, 250 × 2.0 mm, Phenomenex) column11. Due to the long run time of each sample, this method is not ideal for analyzing large numbers of samples. To increase throughput of FAHFA analysis, we needed to shorten the run time while maintaining chromatographic resolution of FAHFA regioisomers. Our prior method went through extensive optimization of carrier solvents, additives, temperature, and flow rates and we did not want to alter these in developing a new method11. We made a mix of different PAHSA isomers (5-, 9-, and 12-PAHSA) and OAHSA isomers (5-, 9-, and 12-OAHSA) and analyzed their retention times and resolution using different reversed phase columns. We found that the Acquity UPLC BEH C18 column (1.7 µm, 2.1 mm × 100 mm, Waters) afforded the best resolution in a 30-minute time frame (Fig. 1).
Figure 1. Development of a faster FAHFA analysis method.
(a) Structures of 5-, 9-, and 12-PAHSA and 12-OAHSA. Extracted ion chromatograms showing the retention times of 5-, 9-, and 12-PAHSA and 5-, 9-, 12-OAHSA standards on a Luna C18(2) column (3 µm, 250 × 2.0 mm, Phenomenex) (b) and an Acquity UPLC BEH C18 column (1.7 µm, 2.1 mm × 100 mm, Waters) (c).
In addition to shortening the run time, we also wanted to reduce the time and material for sample preparation. After the extraction of lipids, we enrich for FAHFAs by using solid-phase extraction (SPE). This step is critical in removing other lipids and contaminants that could impair chromatographic resolution and cause signal suppression of FAHFAs. We previously established a FAHFA enrichment setup using a silica extraction cartridge, however, this step takes nearly 4 hours. The majority of this time was waiting for flow through of solvent by gravity and drying down of the eluted FAHFAs. To reduce the sample prep time, we modified the SPE method to utilize smaller volumes of elution solvent in addition to using positive pressure (nitrogen) to push solvent through the column. Strata SI-1 Silica SPE cartridges (500 mg silica, 3 mL, Phenomenex) were conditioned with 6 mL of hexane, followed by sample addition to the cartridge. Neutral lipids were then eluted using 6 mL of 95:5 hexane:ethyl acetate, followed by the elution of FAHFAs with 4 mL of ethyl acetate. Solvents were added in 2 mL increments to avoid overfilling the SPE column. When applying positive pressure through the column, it is important not to dry out the sorbent bed mass to prevent phase collapse. This method can also be used with a vacuum manifold compatible with the SPE cartridges. Overall, this SPE method for FAHFA enrichment takes one hour.
Separation of 5-PAHSA from the ceramide contaminant
The most referenced FAHFAs with biological activity are 5- and 9-PAHSA, therefore it is essential that we can detect these two PAHSA regioisomers in biological samples with our shorter method. In addition to SPE background issues when measuring PAHSAs, another challenge to measuring PAHSAs, specifically 5-PAHSA, is the existence of a contaminating signal from a ceramide. Specifically, C16:0 ceramide shares all the major MRM transitions with PAHSAs11. This peak can be differentiated from PAHSAs by its differences in MRM transitions ratio. For example, the two most abundant product ions from the collision-induced dissociation of PAHSAs are palmitate (m/z 255.2) and dehydrated HSA (m/z 281.2), with the m/z 255.2 being greater than the m/z 281.2. For the contaminant ceramide, this ratio reversed, with the m/z 281.2 being greater than the m/z 255.2.
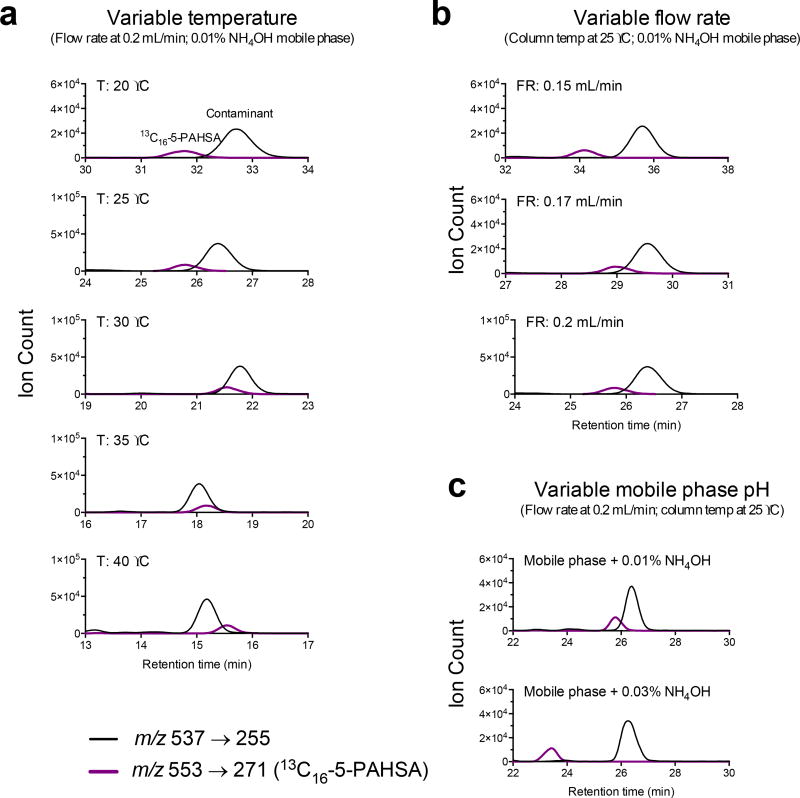
When initially measuring mouse serum PAHSAs with our faster method, we had difficulties separating the 5-PAHSA and the ceramide peak. In order to separate the 5-PAHSA from the ceramide, we tried altering the column temperature and flow rate (Fig. 2, a–b). Although lowering the flow rate from 0.2 ml/min to 0.15 ml/min allowed separation of 5-PAHSA and the ceramide peak, this came at the cost of extending the method from 30 minutes to 40 minutes. In order to keep our method under 30 minutes, we increased the pH of the mobile phase to 0.03% NH4OH from our usual 0.01% NH4OH (Fig. 3c). The increase in pH allows PAHSAs to elute faster while the neutral ceramide is unaffected, thus allowing resolution of 5-PAHSA and the ceramide.
Figure 2. Resolution of 5-PAHSA and ceramide.
(a) Traces showing the effect of altering column temperature ranging from 20 °C to 40 °C on resolution of 5-PAHSA and the ceramide. (b) Traces showing the effect of different flow rates on the resolution of 5-PAHSA and ceramide. (c) Traces showing the effect of increasing the mobile phase pH. Black lines represent the transition m/z 537 → 255 (which is shared by PAHSAs and the ceramide contaminant). Purple line represents the internal standard 13C16-5-PAHSA, which is used as a surrogate for analyzing 5-PAHSA retention time. Although 5-PAHSA is present in mouse serum, it is not shown on the chromatogram because it needs magnification to be seen.
Figure 3. Stability of PAHSAs with different NH4OH percentages in mobile phase.
Extracted ion chromatogram of 12-, 10-, and 9-PAHSA with their respective AUC with either 0.01% NH4OH (top trace) or 0.03% NH4OH (bottom trace).
With the increase in NH4OH in the mobile phase, we were concerned that this might degrade FAHFAs as esters are base-labile. To test this, we ran a PAHSA ladder consisting of 12-, 10-, and 9-PAHSA with either 0.01% NH4OH or 0.03% NH4OH and compared the area under the curves (AUC) (Fig. 3). The increased pH does not degrade FAHFAs as can be seen by comparing the integrated peak areas of the respective PAHSA standards. In addition, with the increased pH, one can also observe the earlier retention shift of PAHSAs. Another concern with the additional concentration of base is the stability of the silica within the column. We have run more than 150 samples using the higher base concentration (0.03% NH4OH) and have not observed any loss of column performance.
Solid-phase extraction contributes to the PAHSA background
We previously reported that the use of silica SPE columns and some buffers yield background when trying to detect PAHSA11. For tissues with high levels of FAHFAs such as white adipose tissue (WAT) and liver, this background is negligible. However, for other tissues or serum where FAHFA levels are low, a background signal can account for up to 15% of the total PAHSA signal11. Indeed, we find that this background is problematic when measuring serum PAHSAs using our faster protocol where we observe extremely high background that can account for up to 50% of the signal in some runs.
Previous literature has shown that SPE cartridges can be major sources of background contamination16–18. The most abundant contaminants from the SPE cartridges are phthalates and other plasticizers which are used in the manufacturing process of these cartridges. This background contamination is not constant and can vary from lot-to-lot and even column-to-column17. When we tested SPE columns from two different lots, we observed variation in PAHSA background (Fig. 4). The two lots had background PAHSAs ranging from 5- to 13/12-PAHSA, but with different PAHSA trace profiles. For example, in the cartridge from lot one, 13/12- and 9-PAHSA have similar peak heights, while in lot two, 9-PAHSA has greater peak height (Fig. 5). This high level of background PAHSAs presents serious problems when measuring PAHSAs from serum or a tissue with low PAHSA levels. The lot-to-lot variation is also problematic as the blank background levels can be significantly different.
Figure 4. SPE cartridges have variable PAHSA background from lot-to-lot.
Extracted ion chromatograms for PAHSAs (top) and internal standard 13C4-9-PAHSA (bottom). Internal standard was added during the sample loading step to account for variation in the SPE procedure.
Figure 5. Background SPE signal for OAHSAs and PAHSAs.
Extracted ion chromatograms for background PAHSAs (red) and OAHSAs (blue) in SPE cartridges from two different lots.
The general workflow for the SPE enrichment of FAHFAs begins by prepping the SPE column for sample loading by equilibration with hexanes, loading of the sample in 200 µL of chloroform, followed by stepwise elution of analytes from the SPE column. To address the background issue, we tested a SPE prewash step with different organic solvents to remove contaminants before equilibrating the column with hexanes.
We used different organic solvents in isolation or in combination to see which afforded minimal PAHSA background (Fig. 6). For example, for the “EA + MeOH Prewash” we added 6 mL of ethyl acetate (EA) followed by 6 mL methanol (MeOH) before beginning with our conditioning step of 6 mL hexanes. For these blank runs, we added 200 µL CHCL3 to the column after the conditioning step with hexanes to emulate true sample loading. We still observed some PAHSA background with these prewashes, however it was significantly reduced (Fig. 6).
Figure 6. Effects of prewashing SPE cartridge on PAHSA background.
SPE columns from the same lot number were prewashed with EA alone, MeOH alone, EA + MeOH, or EA + MeOH + DCM and their PAHSA background was compared to PAHSA background from a non-prewashed column (black line).
The triple wash with EA, MeOH, and DCM did not appear to have any added benefit compared to the MeOH prewash alone. Although the MeOH prewash appeared to lower background more than the EA prewash for this lot of SPE cartridges, when using biological samples, we noticed the EA prewash was more reliable than MeOH prewash in sample reproducibility. We believe this is due to MeOH not being completely removed before the sample is loaded, thus limiting the sample from binding to the stationary phase. We also tried an NH2 SPE cartridge to enrich for FAHFAs, but this also had high levels of background PAHSAs that could not be removed with a MeOH prewash (Fig. S1).
OAHSAs have lower background signal
Although PAHSAs have been the most studied FAHFAs to date8, 10, we reasoned that other FAHFA families might also serve as excellent markers of FAHFA levels. Indeed, during our discovery of FAHFAs, we found six FAHFA families that were highly elevated in the AG4OX mice adipose tissue8. As mentioned previously, the SPE columns can cause significant PAHSA background if the columns are not prewashed. In addition, if one alters the pH of the mobile phase for FAHFA analysis, this could lead to overlap of the 5-PAHSA and the C16:0 ceramide. Due to the potential diagnostic potential of serum FAHFA measurements, we wondered whether other FAHFAs might have a lower background to obviate this issue.
PAHSAs and OAHSAs only differ by their acyl chains. PAHSAs have a palmitoyl group (C16:0) while OAHSAs have an oleoyl group (C18:1). This physical difference can be seen chromatographically, as the OAHSA regioisomers elute later than analogous PAHSA isomers. For example, with our shorter method, the retention time of 9-OAHSA is ~19 min while the retention time of 9-PAHSA is ~18 min, showing that OAHSAs are slightly more hydrophobic than PAHSAs (Fig. 1c). This difference is more pronounced when comparing the same OAHSA and PAHSA regioisomers with the original method (Fig. 1b).
Before measuring OAHSAs in tissues, we needed to determine if SPE columns have background OAHSA contaminants. Here we analyzed background OAHSA levels from two different SPE cartridge lots (Fig. 6). As shown earlier, SPE cartridges have high and varying amounts of PAHSA background (red lines), yet the background OAHSA levels (blue lines), are not distinguishable compared to noise. The non-existent OAHSA background signal is ideal for studying the regulation of these lipids in tissues where total FAHFA levels are low, which includes blood.
FAHFA regioisomers and quantitation
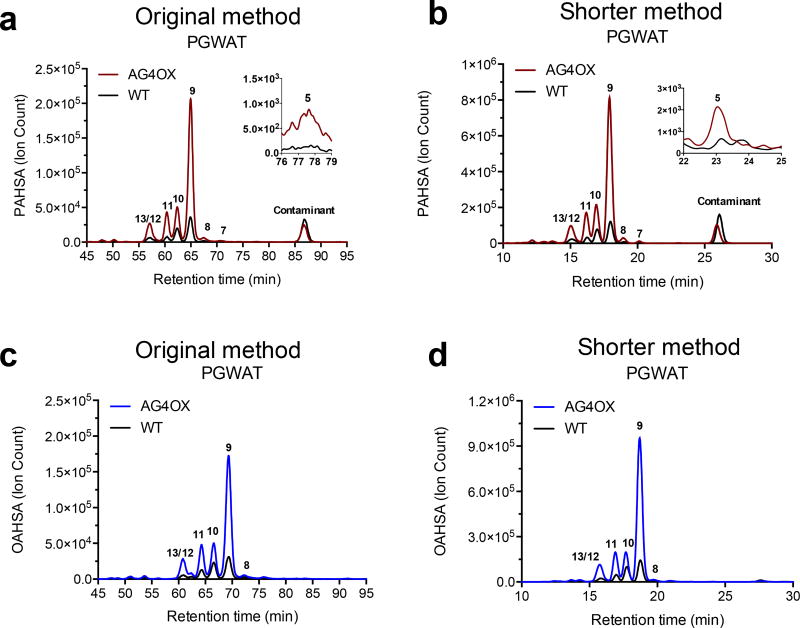
To validate that our optimized FAHFA detection method affords similar results as our previous method, we analyzed PAHSAs in PGWAT of WT and AG4OX mice using our shorter method and the original method (Fig. 7a–b). Previously, we have shown that PAHSAs are significantly upregulated in the WAT of AG4OX mice, with 9-PAHSA being the most abundant PAHSA isomer8. We observe the same profile when analyzing PAHSAs using the original and shorter method for this biological sample (Fig. 7a–b). When comparing the traces of the PGWAT from WT and AG4OX mice with the original method and the shorter method, the shape of the peaks and the separation of the isomers look identical and full resolution of PAHSA regioisomers is achieved. No prior data exists on OAHSA regioisomers and their distribution in tissues. When analyzing OAHSAs in PGWAT in WT and AG4OX mice, we observed increased levels of OAHSAs in the AG4OX mice, with 9-OAHSA having the highest degree of upregulation (Fig. 7c–d). This overall profile is similar to what is seen for PAHSAs and is also consistent when comparing OAHSAs using the original and shorter methods.
Figure 7. Analysis of OAHSAs and PAHSAs in PGWAT of WT and AG4OX mice with our original and shorter method.
Extracted ion chromatograms comparing PGWAT PAHSAs in AG4OX and WT mice using the original (a) and shorter method (b). Extracted ion chromatograms analyzing OAHSAs in PGWAT WT and AG4OX mice in the original (c) and shorter method (d).
When comparing the peak heights (the Y-axis of the chromatograms) of the same sample using the original or shorter method, one will notice they are different. The peak height is shorter for the original method due to the sample being spread out over a longer period of time compared to the shorter method. Although there is an obvious difference in peak height, the area under the curve is unaffected and is the same between both methods (Fig. S3). When the same sample, either PGWAT from WT or AG4OX mice, is analyzed using the shorter method versus the original method, the absolute quantities are the same.
We then quantified the PAHSA and OAHSA regioisomers in the PGWAT of WT and AG4OX mice using the shorter method and the longer method to determine whether there are any performance differences (Fig. 8). All measured PAHSA regioisomers measured were increased in the AG4OX mice, which is consistent with our previously reported measurements.8 We did notice that this cohort of AG4OX mice had lower levels of 9-PAHSA than what was previously reported; however, this may be due to the different age and sex of these mice. In our original report of FAHFAs, we used female mice at 8–14 weeks of age8, while in this study we used male mice at 40 weeks of age. Overall, the two methods performed equivalently with the same concentrations of PAHSAs measured regardless of the gradient utilized.
Figure 8. PAHSA and OAHSA quantification and distribution in WT and AG4OX mice PGWAT.
Quantification of PAHSA (a) and OAHSA (b) isomers in AG4OX and WT mice PGWAT. Data are expressed as means ± SEM. Differences between groups was evaluated with Mann-Whitney test. n=3–5/group, *p<0.05.
In order to accurately quantify and properly assign endogenous OAHSA isomers based on their retention times, we synthesized the internal standard 13C18-12-OAHSA (Supporting Information; Fig. S2). We detected 9-, 10-, 11-, 12, and 13-OAHSA in PGWAT, with 9-OAHSA being the mostly highly upregulated OAHSA regioisomer in AG4OX mice. As with our longer method, we could not achieve baseline resolution of the 12- and 13-OAHSA regioisomers, therefore these two regioisomers are reported as a single value (i.e. 13/12-OAHSA).
Detection of PAHSAs and OAHSAs in human plasma
Using the shorter method, we examined whether we could detect PAHSAs and OAHSAs in human plasma. To avoid the issue of dietary sources affecting endogenous plasma FAHFAs, we analyzed PAHSAs and OAHSAs in fasting humans (Fig. 9). 5–6 plasma samples were analyzed in addition to two SPE procedural blanks to show background levels from the SPE cartridge. The background for PAHSAs can be seen from the black and gray lines but are not visible when looking at plasma OAHSAs. For the PAHSA measurements, plasma was taken from 3 humans twice on different days. For example, 1A and 1B represent that same human, but with blood draws a few weeks apart. For PAHSAs, 5-, 6-, 7-, 8-, 9-, 10-, 11-, and 13/12-PAHSA regioisomers were detected in all five human plasma samples. For the OAHSAs, 8-, 9-, 10-, 11-, and 13/12-OAHSA can be identified with 8-,9-, and 10-OAHSA having the highest levels. There is also a peak at ~24 minutes which is 5-OAHSA (see alignment with 5-OAHSA standard (Fig. S4). PAHSA and OAHSA levels not only vary between different human subjects, but also from the same subject, and further human and animal studies should provide more insight into this observation.
Figure 9. PAHSA and OAHSA regioisomers in fasting human plasma.
Extracted ion chromatogram of PAHSAs (top trace) and OAHSAs (bottom trace).
CONCLUSION
We optimized the analysis of FAHFAs by chromatography to afford shorter run times without a loss in baseline resolution between FAHFA regioisomers. We discovered that solid-phase extraction (SPE) cartridges are the most significant and variable contributors to the PAHSA background signal and the background can be mitigated by additional wash steps in the SPE protocol. Other FAHFAs, in particular OAHSAs have no background signal, which might make OAHSAs the ideal markers for blood FAHFA levels. Lastly, the faster protocol was tested on fasting human plasma to determine the feasibility of this approach for clinical studies, and we detected multiple PAHSA and OAHSA regioisomers with excellent analytical performance.
The emerging importance of FAHFAs in biology and medicine will require better and faster methods for their detection. By optimizing several key steps and using a better column, we reduced the analysis time by nearly 70%. Moreover, these studies revealed that other FAHFAs, notably OAHSAs, have several qualities that improve their detection in blood, such as lower background signal and lack of any contaminating lipids. However, the physiologic and pathophysiologic regulation and the biological activities of the OAHSAs have not been reported. With this new protocol, it should now be possible to determine whether FAHFAs, PAHSAs and OAHSAs, are reliable markers of insulin resistance in people.
TEXT (Word Style "TA_Main_Text"). For full instructions, please see the journal’s Instructions for Authors. Do not modify the font in this or any other section, as doing so will not give an accurate estimate of the formatting for publication and final length of the paper.
FIGURES (Word Style "VA_Figure_Caption"). Each figure must have a caption that includes the figure number and a brief description, preferably one or two sentences. The caption should follow the format "Figure 1. Figure caption." All figures must be mentioned in the text consecutively and numbered with Arabic numerals. The caption should be understandable without reference to the text. Whenever possible, place the key to symbols in the artwork, not in the caption. To insert the figure into the template, be sure it is already sized appropriately and paste before the figure caption. For formatting double-column figures, see the instructions at the end of the template. Do NOT modify the amount of space before and after the caption as this allows for the rules, space above and below the rules, and space above and below the figure to be inserted upon editing.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grants R01 DK043051 and P30DK57521 (to B. B. K.), R01 DK106210 (to B. B. K. and A. S.), R56 DK110150-01A1 (D.S.),F31 DK112604 (M.J.K.), F31 DK112622 (A.T.N.), and F32 DK111159 (M.E.E.), a grant from the JPB Foundation (to B. B. K.).,NCI Cancer Center Support Grant P30 (CA014195 MASS core, A.S.), and Dr. Frederick Paulsen Chair/Ferring Pharmaceuticals (A.S.).
Footnotes
ASSOCIATED CONTENT
Detailed description of synthetic procedures for 13C18-12-OAHSA is available.
Author Contributions
M.J.K and A.S. conceived the project. A.T.N. and M.P.C. synthesized the 12-OAHSA standard. M.J.K., T.C. and M.E.E. performed lipid extraction and SPE. M.J.K and A.S. analyzed and interpreted data. L.O. and M.H. provided human plasma samples. M.J.K. and A.S. wrote the paper. A.T.N., T.C., M.E.E., M.P.C., L.O., M.H., B.B.K., and D.S. edited the paper.
References
- 1.Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. Journal of Biological Chemistry. 1993;268(30):22243–22246. [PubMed] [Google Scholar]
- 2.Gnudi L, Tozzo E, Shepherd PR, Bliss JL, Kahn BB. High level overexpression of glucose transporter-4 driven by an adipose-specific promoter is maintained in transgenic mice on a high fat diet, but does not prevent impaired glucose tolerance. Endocrinology. 1995;136(3):995–1002. doi: 10.1210/endo.136.3.7867610. [DOI] [PubMed] [Google Scholar]
- 3.Tozzo E, Shepherd PR, Gnudi L, Kahn BB. Transgenic GLUT-4 overexpression in fat enhances glucose metabolism: preferential effect on fatty acid synthesis. Am J Physiol. 1995;268(5 Pt 1):E956–64. doi: 10.1152/ajpendo.1995.268.5.E956. [DOI] [PubMed] [Google Scholar]
- 4.Tozzo E, Gnudi L, Kahn BB. Amelioration of insulin resistance in streptozotocin diabetic mice by transgenic overexpression of GLUT4 driven by an adipose-specific promoter. Endocrinology. 1997;138(4):1604–11. doi: 10.1210/endo.138.4.5043. [DOI] [PubMed] [Google Scholar]
- 5.Herman MA, Peroni OD, Villoria J, Schon MR, Abumrad NA, Bluher M, Klein S, Kahn BB. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484(7394):333–8. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winzell MS, Ahrén B. The High-Fat Diet–Fed Mouse. Diabetes. 2004;53(suppl 3):S215–S219. doi: 10.2337/diabetes.53.suppl_3.s215. [DOI] [PubMed] [Google Scholar]
- 7.US Department of Health and Human Services. Centers for Disease Control and Prevention. National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States. 2014 [Google Scholar]
- 8.Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, Peroni OD, Dhaneshwar AS, Hammarstedt A, Smith U, McGraw TE, Saghatelian A, Kahn BB. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159(2):318–32. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuda O, Brezinova M, Rombaldova M, Slavikova B, Posta M, Beier P, Janovska P, Veleba J, Kopecky J, Kudova E. Docosahexaenoic Acid–Derived Fatty Acid Esters of Hydroxy Fatty Acids (FAHFAs) With Anti-inflammatory Properties. Diabetes. 2016;65(9):2580–2590. doi: 10.2337/db16-0385. [DOI] [PubMed] [Google Scholar]
- 10.Lee J, Moraes-Vieira PM, Castoldi A, Aryal P, Yee EU, Vickers C, Parnas O, Donaldson CJ, Saghatelian A, Kahn BB. Branched Fatty Acid Esters of Hydroxy Fatty Acids (FAHFAs) Protect against Colitis by Regulating Gut Innate and Adaptive Immune Responses. J Biol Chem. 2016;291(42):22207–22217. doi: 10.1074/jbc.M115.703835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang T, Chen S, Syed I, Stahlman M, Kolar MJ, Homan EA, Chu Q, Smith U, Boren J, Kahn BB, Saghatelian A. A LC-MS-based workflow for measurement of branched fatty acid esters of hydroxy fatty acids. Nat Protoc. 2016;11(4):747–63. doi: 10.1038/nprot.2016.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson AT, Kolar MJ, Chu Q, Syed I, Kahn BB, Saghatelian A, Siegel D. Stereochemistry of Endogenous Palmitic Acid Ester of 9-Hydroxystearic Acid and Relevance of Absolute Configuration to Regulation. Journal of the American Chemical Society. 2017;139(13):4943–4947. doi: 10.1021/jacs.7b01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem. 1993;268(30):22243–6. [PubMed] [Google Scholar]
- 14.Ohlsson L, Rosenquist A, Rehfeld JF, Harrod M. Postprandial effects on plasma lipids and satiety hormones from intake of liposomes made from fractionated oat oil: two randomized crossover studies. Food Nutr Res. 2014;58 doi: 10.3402/fnr.v58.24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–7. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 16.Eichelberger J, Behymer T, Budde W, Munch J. Method 525.2, Determination Of Organic Compounds In Drinking Water By Liquid-Solid Extraction And Capillary Column Gas Chromatography. Mass Spectrometry. National Eeposure Research Laboratory Office of Research and Development USEPA CINCINNATI, OHIO. 1995;45268 [Google Scholar]
- 17.Junk GA, Avery MJ, Richard JJ. Interferences in solid-phase extraction using C-18 bonded porous silica cartridges. Analytical Chemistry. 1988;60(13):1347–1350. [Google Scholar]
- 18.Stiles R, Yang I, Lippincott RL, Murphy E, Buckley B. Potential sources of background contaminants in solid phase extraction and microextraction. J Sep Sci. 2007;30(7):1029–36. doi: 10.1002/jssc.200600358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.