Prion-like proteins overlap with intrinsically disordered and low-complexity sequence families. These proteins are widespread, especially among mRNA-binding proteins. A salient feature of these proteins is the ability to form protein assemblies with distinct biophysical and functional properties. While prion-like proteins are involved in myriad of cellular processes, we propose potential roles for protein assemblies in regulated protein synthesis. Since proteins are the ultimate functional output of gene expression, when, where, and how much of a particular protein is made dictates the functional state of a cell. Recent finding suggest that prion-like proteins offer unique advantages in translation regulation raising questions regarding the role of prion-like protein assembly in translational regulation.
Defining prion-like proteins
Prion-like proteins are defined by their ability to assume distinct conformational states, with one of these states leading to a self-assembling higher-order structure. Once formed, the self-assembled state can be sustained not only within a cell but can stably propagate across cell divisions by its ability to seed and recruit its monomer counterparts. Although initially discovered in the context of disease, prion-like proteins have been found to serve normal physiological functions [1-3].
Prion-like proteins often contain a modular domain, referred to as the prion-like domain. Certain prion-like domains (PrDs) have a signature Q/N-rich stretch like well-known yeast prions [4], and ~1.2% of human protein-coding genes harbor PrDs by such definition [5, 6]. However, many proteins that possess Q/N-rich domains do not behave like prions, and certain proteins lacking any obvious prion-like domain still display prion-like behavior [7]. Nonetheless, ~30% of both the Q/N-rich PrDs and non-Q/N-rich PrDs reside in RNA-binding proteins [5, 7], suggesting prevalent prion-like mechanisms in mRNA translation, transport, and metabolism.
Low-complexity sequences (LCSs) are defined by sequence composition: long stretches of low overall amino acid diversity. Although seemingly simple, in silico analysis suggests that LCSs are evolutionary features of eukaryotic proteins, which harbor three-fold more internal repeats of amino acids and amino acid motifs than prokaryotic proteins [8-10]. This suggests that they may function in cellular processes that are unique to or more advanced in eukaryotes, such as subcellular compartmentalization or complex protein-protein and protein-nucleic acid interactions. Proteins with LCSs or repetitive modular motifs (an expansion of LCSs) tend to participate in multivalent interactions, which can lead to phase separations, a proposed biophysical mechanism for the formation of membrane-less organelles including stress granules, P-bodies, neuronal granules, germ granules and nucleoli, all of which belong to RNA-protein particles (RNPs) [11]. Some of these RNP-granules are tightly packed, while others are loosely associated and more dynamic [12-16].
Although defined by unrelated criteria, PrDs and LCSs surprisingly converge in two ways: (i) ~30% of each are RNA-binding proteins that can regulate translation in various ways and thereby the protein composition of the cell [5], and (ii), they are structurally predicted to be intrinsically disordered, excluded from globular regions and lacking known structural motifs [17-19]. One important thing to keep in mind is that in silico prediction of disordered domains or the behavior of proteins in isolation is not necessarily predictive of their in vivo state. Recent comparative sequence analysis suggests that at least some ‘disordered domains’ could have structured states or may assume unknown structural states [20]. Moreover, proteomics-based protein conformation analyses indicate that in the cellular environment, the number of proteins in a disordered state may be significantly less than what in silico analysis predicts [21, 22]. Nonetheless, given that RNP structures formed by RNA-binding proteins manifest distinct dynamics, and that prion-like domains allow proteins to adopt various physical states ranging from liquid droplet to hydrogel to amyloid [23], it is important to explore what circumstances—and how and why—proteins adopt a particular type of protein assembly. In addressing these two questions—how proteins assemblies are formed, and, once formed, what specific purpose they serve—we first focus on the functional consequences.
We focus here on prion-like proteins because of their unique features: (i) a conformational change of a small population of proteins can template non-converted monomers, allowing a persistent alteration in function; (ii) the same protein can give rise to a spectrum of outputs based on its conformational differences and seeding capacities [24-27]; and (iii) the structure can be homotypic and ordered (but not always), allowing stereotypic organization of the various domains [28]. We have used specific examples of prion-like RNA-binding proteins to illustrate how these features of RNP assemblies contribute to translational regulation. This review does not intend to cover the general properties of “intrinsically disordered” proteins [29] or the physical principles that guide assembly [30], since a number of excellent reviews have already been written on these subjects.
Role of protein assemblies in translation regulation
RNA-binding proteins influence all aspects of RNA fate: synthesis, transport, stability, and translation. Here, we focus on certain RNA-binding proteins involved in the control of mRNA translation. Translation regulation implies an increase or decrease in the rate of either global translation or translation of specific mRNAs, translation in a specific place, or holding certain mRNAs in translation-competent or -incompetent states for future use. The change in translation can be transient or persistent. How do conformational plasticity and various types of protein assemblies formed by prion-like proteins contribute to translation regulation? Recent reports suggest that internal or external environmental changes can induce global or selective translation changes by rapidly changing protein conformation and assembly. Additionally, the duration of translational control can be encoded in the nature of the assembly. These findings lend support for prion-like proteins in translation regulation.
Translation regulation by amyloid-like assembly
One of the earliest examples of a physiological prion-like protein is the yeast Sup35 protein, a translation terminator[31]. The Sup35 protein in its monomeric form helps translation to cease at stop codons and confers translational fidelity. When Sup35 forms a self-sustaining amyloidogenic assembly, it loses the translation termination activity, allowing translational read-through. Translation read-through is mostly disadvantageous; however, under certain environmental conditions protein diversity generated by translational read-through can be advantageous[32]. In multicellular eukaryotes an early example of a prion-like RNA-binding protein whose higher-order assemblies serve a physiological function is the cytoplasmic polyadenylation element binding protein (CPEB) family. Some members of this family, such as CPEB3 in vertebrates, Orb2 in Drosophila, and ApCPEB in Aplysia, harbor prion-like domains and form stable protein assemblies, with some features of an amyloid-like state [33-35]. Blocking assembly interferes with synaptic facilitation and long-term memory [35, 36] and facilitating assembly enhances target mRNA translation as well as memory consolidation [37]. Interestingly, all CPEBs studied thus far seem to have dual functions whether they possess prion-like domains (Orb2, CPEB3) or not (CPEB1), switching between translational repression and activation in response to external signals [38]. In neurons, this functional switch likely generates asymmetry in translation in different synapses within the same neuron; synapses with monomeric CPEB would repress the translation of specific mRNAs, while the same mRNAs would be translated in synapses with the conformationally distinct higher-order assembly of CPEB. A stable and ordered assembly of CPEB provides a self-sustaining local change in protein state and translation, with the ability to recreate the same state from a few molecules that can render memory immune to the turnover of individual protein molecules over long periods (Figure 1B) [39, 40]. However, the exact biophysical properties of synaptic CPEB and its function remain contentious. For instance, it has been proposed that instead of directly activating translation, aggregated CPEB sequesters heterologous translational repressors and thus disengages the target mRNA from a repressive state [41]. Whether these alternative models are mutually exclusive or reflect the spectrum of functional assemblies formed by the same protein remains to be seen.
Figure 1. Translation regulation by stable amyloidogenic assembly: prion-like CPEB.
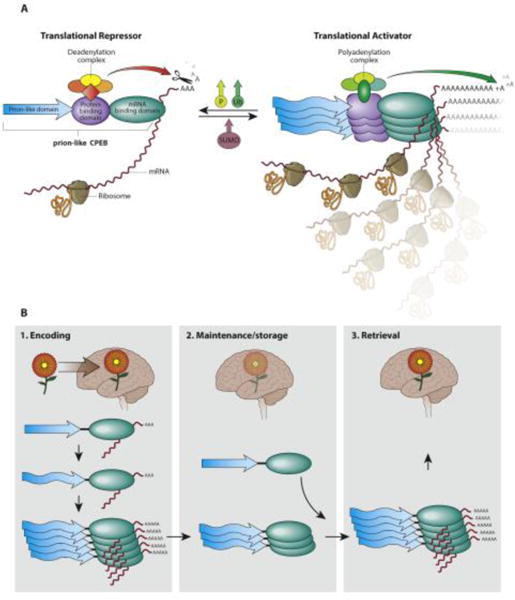
(A) Schematic organization of various domains of the mRNA binding protein CPEB and the consequence of conversion from monomer to polymer. The prion-like CPEB regulates target mRNA translation bidirectionally: when it’s in monomeric form, the protein recruits mRNA deadenylation complex and represses translation; when the protein adopt amyloid-like assembly, it recruits polyadenylation complex and enhance translation. The conversion between two states can be regulated by protein phosphorylation (P), ubiquitination (Ub) and SUMOylation (SUMO). (B) The connection between CPEB amyloid-like assembly and various stages of memory. During memory encoding, CPEB is being converted from a monomer to an amyloidogenic form. Overtime the memory is maintained by the self-sustaining nature of the assembled state. During recall it likely activates translation of specific mRNA to aid expression of memory.
Although amyloid formation was thought to be a ‘one way’ process, recent discoveries suggest that these assemblies can also be dynamically regulated. An example of this is the translation regulator Rim4 in yeast (Figure 2A). Precise and acute control of mRNA stability and translation is required in stage-specific production of new proteins during yeast meiosis [42]. The mRNA for one of the critical meiosis determinants, the B-type cyclin Clb3, is produced at the onset of meiosis I but its protein expression is restricted to meiosis II [43]. The timely burst of Clb3 translation is regulated by the prion-like mRNA-binding protein Rim4, which forms amyloidogenic aggregates that bind to and inhibit translation of Clb3 mRNA. At the onset of meiosis II, Rim4 is dissolved upon phosphorylation, releasing Clb3 mRNA, prompting rapid translation [44]. The Rim4 amyloidogenic aggregates’ formation and more importantly disappearance is quite rapid (within minutes). In this case, phosphorylation of the PrD is the mechanism that leads to clearance of aggregates at the onset of meiosis II [44]. What remains unclear is how the Rim4 aggregates repress translation and how phosphorylation dissolves Rim4 aggregates.
Figure 2. Translation regulation by dynamic amyloidogenic protein assembly: Rim4 and Xvelo.
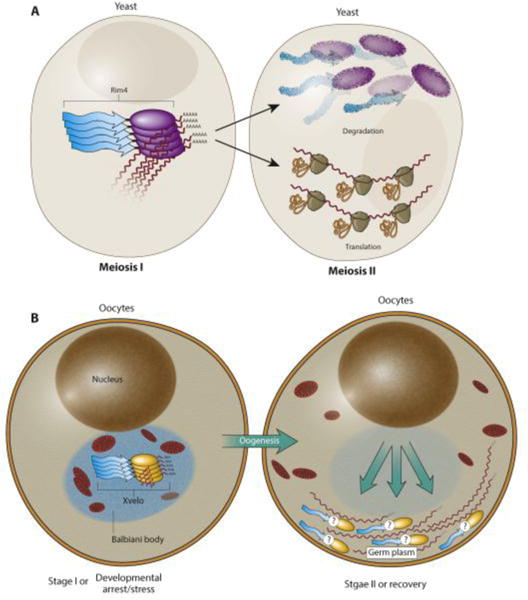
(A) Rim4 is a RNA-binding protein, which regulates meiosis progression in yeast. In addition to RNA-binding domain, it also contains a prion-like domain. Rim4 proteins adopt an amyloidogenic assembly during meiosis I and bind and repress translation of the B-type cyclin CLB3 mRNA. Upon onset of meiosis II, phosphorylation of Rim4 results in disassembly and degradation of the Rim4 amyloids, allowing translation of CLB3 and progression of meiosis II. (B) The RNA-binding protein Xvelo is an abundant constituent of the Balbiani Body of the vertebrate oocytes. It harbors a prion-like domain that can adopt an amyloidogenic assembly when the oocyte is in stage I. Germline specific mRNAs are trapped and translationally-repressed in Balbiani Body, partially by Xvelo. During oogenesis, Balbiani body disappears and germline specific mRNAs are dispersed towards the vegetal pole for germline establishment.
The use of an amyloid-like state to hold mRNA dormant for future activation could be a common mechanism of translation regulation. In Xenopus, for example, the gene products that establish the germline of the next generation are maternally inherited, and these germ-line specific maternal mRNAs are deposited into a non-membranous compartment termed the Balbiani body (Figure 2B) [45-47]. One function of the Balbiani body is to protect maternal RNAs from degradation and pass them on to the next generation [48]. The core protein that organizes the Balbiani body, Xvelo in Xenopus and Bucky Ball in zebrafish, forms amyloid-like assemblies [46, 49]. Somewhat similar to Rim4, formation and dispersal of the Balbiani body is developmental stage-specific [47]. The mechanism of dispersal is yet unclear. An important unanswered question is what specific advantage amyloid-like states offer for Rim4 or Balbiani body over other forms of protein assemblies for this purpose. One possibility is that given that developmental arrest is very common during oogenesis, amyloidogenic assemblies could persist across long-term dormancy and protect critical mRNA from environmental fluctuation. If this is true, then one would suspect such amyloidogenic mRNPs may exist for other physiological processes that require holding mRNAs in dormant state for long time.
Translation regulation by non-amyloidogenic superassembly
mRNA-binding proteins with prion-like domains can also form protein assemblies that are distinct from amyloid-like states. An example of such a translation regulator is the mRNA binding protein Whi3 [50]. In budding yeast S. cerevisiae, the cell cycle is arrested when haploid cells encounter pheromones from a mating partner (Figure 3). Whi3 inhibits G1 cyclin cln3 translation, therefore arresting cell cycle progression [51, 52]. However, if the mating is deceptive, cell cycle progression resumes and this process is regulated by super-assembly of Whi3 into a single structure, disinhibiting translation of cln3 [50]. A transient exposure to pheromone can induce the refractory state, and assembly of Whi3 creates a “molecular memory” for the stable maintenance of the refractory state in the absence of pheromone. The Whi3 super-assembly differs from other prion-like assemblies in several ways: one, while the Whi3 super-assembly is protease- and detergent-resistant (properties associated with amyloidogenic aggregates), it does not bind to thioflavinT, a common feature of amyloids. Second, the prion-like state of Whi3 is asymmetrically retained in mother cells and not inherited by the daughter cells. Finally, while the poly-Q stretch of Whi3 is necessary for its super-assembly in S. cerevisiae, in vitro experiments with Ashbya gossypii suggest that Whi3 alone cannot form homomeric protein assemblies under physiological salt concentrations [53]. It therefore appears that Whi3 assembly requires additional factors and that these additional factors could attach it to some structure, confining its function within the mother cells. The structural bases of these assemblies and, given their function, the advantage of this structure over amyloids remains to be addressed.
Figure 3. Translation regulation by non-amyloidogenic stable protein assembly.
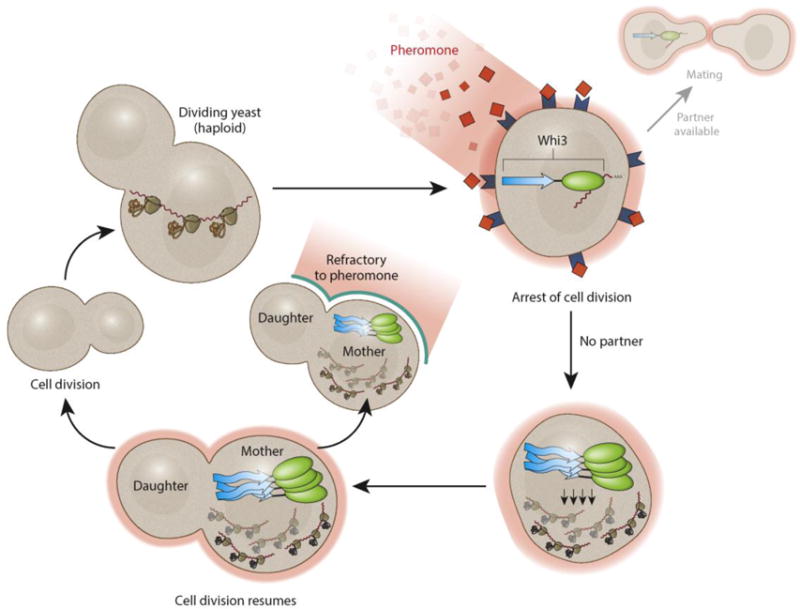
Whi3 regulates cell cycle progression through inhibiting translation of a G1 cyclin cln3. During mating, haploid yeast cells encounter pheromones. Whi3 binds and inhibits cln3 translation and arrests cell division and prepares the cell for mating. However, prolonged pheromone exposure without a mating partner induces conformational change of Whi3 prion-like domain. Whi3 proteins therefore form a stable super-assembly and release cln3 mRNA for translation. Interestingly, Whi3 super-assembly is non-amyloidogenic and retains in the mother cell possibly through attaching to some cellular organelles.
Translation regulation by heterogeneous assembly
Q/N-rich amyloidogenic PrDs occupy a small part of the range of sequences found in various PrDs [6]. They adopt structures of different dynamics, likely tuned to the specific physiological functions they serve. TIA-1 is one of the core nucleating RNA-binding proteins whose PrD is essential for stress granule formation [54]. Stress granules (SG) are microscopically visible deposits of stalled translation machinery, formed when cells encounter unfavorable conditions; they disassemble when the cell returns to its normal state [55, 56]. Several functions of stress granules have been proposed, including facilitation of translation initiation complex assembly for future use, translation of stress-responsive mRNA, and stabilization or sequestration of mRNA, which would also indirectly influence translation [55]. Some studies suggest that in yeast the stress granules behave like non-amyloid solid aggregates via promiscuous interactions between low-complexity domains, while in mammalian cells stress granules form liquid-like structures mediated by weak interaction among proteins [57]. Another model proposed that the stress granule adopts a stable core and dynamic shell structure [58]. The core proteins, together with stalled pre-initiation complexes, nucleate first into RNAase resistant stable assemblies [59], then the shell, with more dynamic features, starts to wrap around the core [58]. Overexpression of full length TIA-1 induces, while overexpression of just the TIA-1 PrD inhibits, stress granule formation [54], suggesting a role of RNA binding domain in proper formation of stress granule. The advantage of dynamic assemblies during stress is perhaps in allowing cells to rapidly sequester or utilize proteins and mRNAs. When the stress is relieved, the assembly rapidly disperses, allowing cells to revert to pre-stressed conditions. Interestingly, when cells are missing key protein components of stress granules, they no longer form canonical stress granule in response to moderate stress[60]. Instead, smaller cellular foci form when cells are challenged with extreme stress. The components and function of the small aggregates are unknown; however, it is consistent with the notion that that RNP aggregates are dynamically regulated based on cellular demand.
Cellular mechanisms that control formation of physically and functionally distinct RNP assemblies
In the cellular milieu, an mRNA-binding protein and its target mRNA exist in non-equilibrium states amid various degrees of interaction with millions of other molecules. It is remarkable that in this crowded cellular milieu, the prion-like proteins and LCSs, though occupying a large percentage of the proteome and being multivalent, do not coalesce into a giant organelle; instead they give rise to distinct protein assemblies with specific compositions in specific compartments [61]. Some studies indicate, at least for RNP particles, that RNA can act as a possible guide. However, most RNA-binding proteins have multiple RNA targets, and different RNA-binding proteins can share the same RNA-binding domains. How then do RNP particles of distinct compositions exist in the same compartment?
PrDs and LCS proteins can also generate different assemblies depending on the cellular conditions. In the case of Whi3 neither its poly-Q stretch nor its RNA-binding motif is required for its recruitment to stress granules. This suggests that under stress conditions, Whi3 may be passively sequestered into stress granules by proteins or mRNA components in the stress granules to suppress target mRNA translation [51, 52]. However, the formation of Whi3 super-assemblies in response to pheromones does require its poly-Q domain [50]. Therefore, the super-assembly is distinct from stress granules: the super-assembly requires Whi3′s poly-Q domain to initiate an ordered structure, while the poly-Q domain is dispensable for loosely wrapping around the shell of stress granules. Likewise, the prion-like NM domain of Sup35 can substitute the prion-like domain of TIA-1 and the chimeric TIA-1 function in forming stress granules. However, subsequent overexpression of NM does not interfere with endogenous stress granule formation, suggesting the absence of hetero-oligomerization between TIA-1 and Sup35′s prion-like domain. In yeast, although Sup35 can be recruited to the stress granule, its prion domain is dispensable for SG formation [62], suggesting that the function of PrDs can also be context dependent. How is then this context-specific assemblies of RNP particles achieved?
Assembly of ribosomes—RNP particles critical for protein synthesis— provides some general guidance towards assembly of RNP particles. Many ribosomal proteins (26% of the S. cerevisiae 60S ribosomal proteins for instance) have a disordered region rich in arginine and lysine that contributes to their high positive charge [63] and like most LCS proteins they are aggregate-prone by themselves [64]. Since they are abundant in every cell, and since ribosomal proteins are made in the cytosol but ribosome assembly takes place in the nucleus, cells have evolved stringent regulation on transport, assembly, and degradation of ribosomal proteins to avoid forming spurious assemblies and to control the amount of mature ribosomes: first, specific interacting partners such as importins in the cytoplasm and escortin Tsr2 in the nucleoplasm safeguards the solubility of ribosomal proteins, possibly by shielding the basic region [65, 66]; second, during assembly both rRNA and ribosomal proteins are concentrated at nucleoli, a structure reportedly utilizes liquid-liquid phase separation into distinct compartments[67] or subcompartments [16], which facilitates the interaction between rRNA and ribosomal proteins; and third, excess unassembled ribosomal proteins are degraded by the ubiquitin-proteasome system mediated by a specific E3 ubiquitin ligase Tom1, which recognizes exposed LCSs [68, 69]. Therefore, specificity in RNP assembly can be achieved by facilitating or favoring correct interaction; dis-favoring or preventing non-specific interaction; and eliminating excess or unwanted proteins. Below we briefly discuss factors that can contribute to these general features.
Composition of PrD
Although the Prds and LCSs do not assume a specific three-dimensional structure, they nonetheless vary in their amino acid composition and often have short repeat sequences. The amino acid composition of PrDs and LCSs influences intra- or inter-molecular long-range electrostatic interactions between charged residues, short-range cation−π interactions between aromatic residues and charged residues, dipole−dipole interactions between polar residues, and π-π interaction between aromatic residues [70]. For example, ribosomal proteins are enriched in highly charged residues favoring electrostatic interactions with rRNAs or repetitive motifs such as FG, RG, SY, and Q/N in LCS proteins, which can mediate weak, short-range local interaction. In other words, PrDs or other disordered domains could be a new type of protein-protein recognition domain as has been suggested [29] and like other protein domains amino acid composition and sequence features would favor certain interaction over others.
Interacting partners, protein modifications and expression level
The protein assemblies discussed so far likely undergo an initial nucleating process: ribosomal proteins and rRNA form the core of ribosomes, 43S pre-initiation complexes together with several core proteins nucleate stress granule formation, and in the case of prion-like assemblies it’s the switch of one or a few monomeric proteins to an alternative conformational state. The initiation stage can also determine what kind of protein assembly will be formed, because the PrDs/LCS domain-containing proteins, before committing to a certain assembly, are considered ‘naked’, and the exposed structure-less region seeks partners either to counterbalance or to adopt specific conformations. Thus, the same proteins could potentially form different protein assemblies based on their associating partners during the initiating process.
In addition to protein, the role of mRNA itself in protein assembly is becoming more evident [14, 53]. One possibility is that mRNAs with distinct cis-elements dictate the nature as well as location of assembly. For example, the Whi3 protein super-assembly discussed before indeed could adopt different assemblies with varying stabilities. This is best illustrated in the highly polarized filamentous and multinucleate fungi Ashbya gossypii, where Whi3 controls the localized translation of the protein formin Bni1 at branching sites [71], as well as the perinuclear translation of G1 cyclin Cln3 [72]. How is such compartmentalization achieved? In vitro, adding the mRNA targets facilitates liquid-like droplet formation consisting of both Whi3 protein and target mRNA [53]. Interestingly, the biophysical properties of Whi3-CLN3 mRNA droplet and Whi3-BNI1 mRNA droplet appear different in protein/RNA ratio, as well as in surface tension and viscosity, likely explaining how the two different RNP complexes do not intermingle [53]. However, it is still unclear whether they form liquid-droplet phases in vivo and, if so, how they avoid droplet demixing.
What else may prevent non-specific assembly? Controlling the expression of the protein at the right time and in the correct amount would also reduce the probability of non-specific assembly. Indeed, there appears to be a tight control over the expression of prion-like protein and other LCS proteins [73, 74]. Small proteins, such as ribosomal components, don’t have a large surface area to interact with other proteins, which can reduce non-specific interactions; however, such a logic is not applicable for most cellular proteins. Recently, a “nanny model” for protein assembly has been proposed [75], in which intrinsically disordered proteins are prevented from degradation to allow them to form protein complex. In fact such “nanny” proteins can also prevent non-specific interactions, and binding to mRNA/protein or post-translational modification of the target protein releases the nanny proteins, unmasking the domain to form either homotypic or heterotypic assembly. While any protein can serve as a nanny protein, protein chaperones are ideal for such “nanny” protein function. In addition to helping proteins attain correct conformation, chaperones are already known to aid self-templating behavior of prion-like proteins [76]. A role for chaperones would also provide a possible link between changes in cells’ proteostatic capacity and problems with unregulated protein assembly, particularly in the nervous system [77].
Cellular milieu
For all protein assemblies mentioned above, we have tried to focus on their functional behavior in vivo since that takes into consideration the surrounding cellular environment that different protein assemblies inhabit. The interior cellular space is estimated to be packed with 200-300mg/ml of macromolecules [78]; every biochemical activity inside cells is embedded in this molecularly crowded environment, which could impede, facilitate or alter protein-protein interaction. It is also important to consider that majority of proteins are made in a relatively crowded place on the surface of the endoplasmic reticulum and yet these proteins wait until they reach their particular subcellular location to form specific assemblies. For technical reasons, most biophysical analysis of proteins is performed in isolation using proteins purified from heterologous systems. However, over the last few decades, using in vitro systems, efforts have been made to understand how molecular crowding affects protein folding, the shape of molecules, enzymatic activities, molecular interactions, protein assembly dynamics, and dynamics of unstructured protein regions [79]. These in vitro studies give a quantitative view of how molecular crowding affects specific protein assemblies; nevertheless, the crowding agents used to mimic a dense cellular milieu are often inert, in contrast to real biological systems. Recent evidence shows that, upon energy depletion, the bacterial cytoplasm becomes glass-like [80], and similar phenomena are also observed in starved or dormant yeast, in which cytoplasm become less fluid and the movement of mRNPs is restricted [81, 82]. And even within a single cell, molecular crowding is heterogeneous [80]. The nucleus and cytoplasm vary in their macromolecule concentrations and composition—as do polarized neurons, where molecular crowding effects could differ between the soma and synapses. Future work emphasizing proteins’ behavior in vivo will allow a better understanding of the unique biophysical properties of different prion-like proteins and their role in translational regulation and cellular biology more broadly.
Concluding Remarks
The protein assemblies discussed above share some common features: spatial confinement, high local concentration, a structured platform, and a configuration that is stable but can nonetheless be altered non-autonomously. These features are particularly relevant for controlling translation in space and time. Moreover, prion-like translation regulators can function without the prion-like domain. For example, Sup35 can terminate translation and Orb2 can suppress translation in the absence of their prion-like domains[83, 84]. However, without a prion domain, these proteins cannot modify their translation regulatory activity to accommodate altered cellular states or to serve a specific purpose. This suggests that the addition of prion-like domains expands the existing functional repertoire of a given protein. While these features of prion-like proteins are generally accepted, there are areas of contention. One such issue concerns the material properties of the various prion-like RNA-protein assemblies: whether they are liquid, gel, glass, fiber, or amyloid fiber[41, 85]. Advances in cryo-EM and in-cell NMR technologies, which allows structural analysis of proteins in more natural conditions may help in resolving this issue. Another area of contention is the transmissibility of prion-like assemblies. It will be important to define clearly what transmissibility means in various contexts. The transmissibility of conformational information encompasses three distinct processes: between proteins within a cell, between cells, or between organisms. The transmission of pathological prions involves all three processes, while for functional prion-like proteins the transmissibility often refers to transfer of information from protein to protein within a cell or transfer between cells that were at some point cytoplasmically connected. Advances in live imaging technologies and the increasing ease of modifying proteins in natural contexts via genome editing should provide insight into how protein conformational information transmits (see Outstanding questions).
Outstanding questions box.
What are the biophysical properties of the proteins assemblies formed by prion-like proteins in vivo?
How prion-like proteins undergo conformational switch? Do preexisting proteins are reconfigured or newly synthesized proteins are guided to assume the altered protein conformation? If so, what cellular factors are involved in this process?
How the cellular context influence the properties of the protein assemblies? Does same prion-like protein forms different types of proteins assemblies in different cell types? If so, does it lead to alteration of the pre-existing function or gain of a new function?
How in the same cellular compartment different prion-like proteins can co-exist and form different type of protein assemblies with unique composition?
How cellular machinery discriminate between functional protein assemblies from non-functional or abnormal assemblies?
Trends Box.
Prions, originally discovered in several transmissible diseases, are proteins with multiple conformations, at least one of which is infectious to other protein species. Recently it has been discovered proteins with a prion-like domain can serve physiological functions.
Of all the characterized physiological processes that prion-like proteins are involved, many of them are related to RNA biology, regulating RNA synthesis, stability, translation and degradation.
Based on the cell types, the temporal and spatial scale of a physiological process, prion-like proteins adopt various structures raging from stable amyloid-like assembly, non-amyloid superassembly, and heterogeneous labile protein assembly.
The amino acid composition of each prion-like domain, the abundance of the protein in a defined compartment, post-translational modification, together with factors in the cellular environment determines the biophysical properties of the protein assembly.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nature Reviews Molecular Cell Biology. 2015;16(1):18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halfmann R, et al. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Si K. Prions: what are they good for? Annu Rev Cell Dev Biol. 2015;31:149–69. doi: 10.1146/annurev-cellbio-100913-013409. [DOI] [PubMed] [Google Scholar]
- 4.Alberti S, et al. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–58. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.March ZM, et al. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 2016;1647:9–18. doi: 10.1016/j.brainres.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King OD, et al. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakrabortee S, et al. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell. 2016;167(2):369–381 e12. doi: 10.1016/j.cell.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcotte EM, et al. A census of protein repeats1. Journal of Molecular Biology. 1999;293(1):151–160. doi: 10.1006/jmbi.1999.3136. [DOI] [PubMed] [Google Scholar]
- 9.Alba MM, et al. Amino acid repeats and the structure and evolution of proteins. Genome Dyn. 2007;3:119–30. doi: 10.1159/000107607. [DOI] [PubMed] [Google Scholar]
- 10.Toll-Riera M, et al. Role of Low-Complexity Sequences in the Formation of Novel Protein Coding Sequences. Molecular Biology and Evolution. 2011 doi: 10.1093/molbev/msr263. [DOI] [PubMed] [Google Scholar]
- 11.Pak CW, et al. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol Cell. 2016;63(1):72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324(5935):1729–32. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 13.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753–67. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163(1):123–33. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Protter DSW, Parker R. Principles and Properties of Stress Granules. Trends in Cell Biology. 2016;26(9):668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feric M, et al. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 2016;165(7):1686–97. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weathers EA, et al. Insights into protein structure and function from disorder-complexity space. Proteins. 2007;66(1):16–28. doi: 10.1002/prot.21055. [DOI] [PubMed] [Google Scholar]
- 18.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293(2):321–31. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 19.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–84. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 20.Toth-Petroczy A, et al. Structured States of Disordered Proteins from Genomic Sequences. Cell. 167(1):158–170.e12. doi: 10.1016/j.cell.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ovchinnikov S, et al. Protein structure determination using metagenome sequence data. Science. 2017;355(6322):294–298. doi: 10.1126/science.aah4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leuenberger P, et al. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science. 2017;355(6327) doi: 10.1126/science.aai7825. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Fuxreiter M. The Structure and Dynamics of Higher-Order Assemblies: Amyloids, Signalosomes, and Granules. Cell. 2016;165(5):1055–1066. doi: 10.1016/j.cell.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chien P, Weissman JS. Conformational diversity in a yeast prion dictates its seeding specificity. Nature. 2001;410(6825):223–7. doi: 10.1038/35065632. [DOI] [PubMed] [Google Scholar]
- 25.Uptain SM, et al. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 2001;20(22):6236–45. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derkatch IL, et al. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144(4):1375–86. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka M, et al. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428(6980):323–8. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 28.Saibil HR, et al. Heritable yeast prions have a highly organized three-dimensional architecture with interfiber structures. Proc Natl Acad Sci U S A. 2012;109(37):14906–11. doi: 10.1073/pnas.1211976109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tompa P, et al. Close encounters of the third kind: disordered domains and the interactions of proteins. Bioessays. 2009;31(3):328–35. doi: 10.1002/bies.200800151. [DOI] [PubMed] [Google Scholar]
- 30.Banani SF, et al. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18(5):285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickner RB, et al. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–85. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- 32.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–83. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 33.Fioriti L, et al. The Persistence of Hippocampal-Based Memory Requires Protein Synthesis Mediated by the Prion-like Protein CPEB3. Neuron. 2015;86(6):1433–1448. doi: 10.1016/j.neuron.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Si K, et al. Aplysia CPEB Can Form Prion-like Multimers in Sensory Neurons that Contribute to Long-Term Facilitation. Cell. 2010;140(3):421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 35.Majumdar A, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148(3):515–29. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Hervas R, et al. Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation. PLoS Biol. 2016;14(1):e1002361. doi: 10.1371/journal.pbio.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, et al. A Putative Biochemical Engram of Long-Term Memory. Curr Biol. 2016;26(23):3143–3156. doi: 10.1016/j.cub.2016.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivshina M, et al. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol. 2014;30:393–415. doi: 10.1146/annurev-cellbio-101011-155831. [DOI] [PubMed] [Google Scholar]
- 39.Krüttner S, et al. Drosophila CPEB Orb2A Mediates Memory Independent of Its RNA-Binding Domain. Neuron. 2012;76(2):383–395. doi: 10.1016/j.neuron.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruttner S, et al. Synaptic Orb2A Bridges Memory Acquisition and Late Memory Consolidation in Drosophila. Cell Rep. 2015;11(12):1953–65. doi: 10.1016/j.celrep.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudhakaran IP, Ramaswami M. Long-term memory consolidation: The role of RNA-binding proteins with prion-like domains. RNA Biol. 2017;14(5):568–586. doi: 10.1080/15476286.2016.1244588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brar GA, et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335(6068):552–7. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carlile TM, Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133(2):280–91. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berchowitz LE, et al. Regulated Formation of an Amyloid-like Translational Repressor Governs Gametogenesis. Cell. 2015;163(2):406–18. doi: 10.1016/j.cell.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pepling ME, et al. Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proceedings of the National Academy of Sciences. 2007;104(1):187–192. doi: 10.1073/pnas.0609923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bontems F, et al. Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol. 2009;19(5):414–22. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 47.Kloc M, et al. The Balbiani body and germ cell determinants: 150 years later. Curr Top Dev Biol. 2004;59:1–36. doi: 10.1016/S0070-2153(04)59001-4. [DOI] [PubMed] [Google Scholar]
- 48.Houston DW, King ML. Germ plasm and molecular determinants of germ cell fate. Curr Top Dev Biol. 2000;50:155–81. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- 49.Boke E, et al. Amyloid-like Self-Assembly of a Cellular Compartment. Cell. 2016;166(3):637–650. doi: 10.1016/j.cell.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caudron F, Barral Y. A Super-Assembly of Whi3 Encodes Memory of Deceptive Encounters by Single Cells during Yeast Courtship. Cell. 2013;155(6):1244–1257. doi: 10.1016/j.cell.2013.10.046. [DOI] [PubMed] [Google Scholar]
- 51.Cai Y, Futcher B. Effects of the yeast RNA-binding protein Whi3 on the half-life and abundance of CLN3 mRNA and other targets. PLoS One. 2013;8(12):e84630. doi: 10.1371/journal.pone.0084630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gari E, et al. Whi3 binds the mRNA of the G1 cyclin CLN3 to modulate cell fate in budding yeast. Genes Dev. 2001;15(21):2803–8. doi: 10.1101/gad.203501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, et al. RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60(2):220–30. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gilks N, et al. Stress Granule Assembly Is Mediated by Prion-like Aggregation of TIA-1. Molecular Biology of the Cell. 2004;15(12):5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36(6):932–41. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kedersha N, Anderson P. Regulation of translation by stress granules and processing bodies. Prog Mol Biol Transl Sci. 2009;90:155–85. doi: 10.1016/S1877-1173(09)90004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kroschwald S, et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wheeler JR, et al. Distinct stages in stress granule assembly and disassembly. eLife. 2016;5:e18413. doi: 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain S, et al. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164(3):487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kedersha N, et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212(7):845–60. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Souquere S, et al. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci. 2009;122(Pt 20):3619–26. doi: 10.1242/jcs.054437. [DOI] [PubMed] [Google Scholar]
- 62.Grousl T, et al. Heat Shock-Induced Accumulation of Translation Elongation and Termination Factors Precedes Assembly of Stress Granules in S. cerevisiae. PLoS ONE. 2013;8(2):e57083. doi: 10.1371/journal.pone.0057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fedyukina DV, et al. Charge Segregation and Low Hydrophobicity Are Key Features of Ribosomal Proteins from Different Organisms. The Journal of Biological Chemistry. 2014;289(10):6740–6750. doi: 10.1074/jbc.M113.507707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.David DC, et al. Widespread Protein Aggregation as an Inherent Part of Aging in C. elegans. PLOS Biology. 2010;8(8):e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jakel S, et al. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21(3):377–86. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schütz S, et al. A RanGTP-independent mechanism allows ribosomal protein nuclear import for ribosome assembly. eLife. 2014;3:e03473. doi: 10.7554/eLife.03473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brangwynne CP, et al. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proceedings of the National Academy of Sciences. 2011;108(11):4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sung MK, et al. Ribosomal proteins produced in excess are degraded by the ubiquitin-proteasome system. Mol Biol Cell. 2016;27(17):2642–52. doi: 10.1091/mbc.E16-05-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sung MK, et al. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. eLife. 2016;5:e19105. doi: 10.7554/eLife.19105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brangwynne CP, et al. Polymer physics of intracellular phase transitions. Nat Phys. 2015;11(11):899–904. [Google Scholar]
- 71.Lee C, et al. PolyQ-dependent RNA–protein assemblies control symmetry breaking. The Journal of Cell Biology. 2015;208(5):533–544. doi: 10.1083/jcb.201407105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee C, et al. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev Cell. 2013;25(6):572–84. doi: 10.1016/j.devcel.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gsponer J, et al. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322(5906):1365–8. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gill J, et al. Regulated Intron Removal Integrates Motivational State and Experience. Cell. 2017;169(5):836–848 e15. doi: 10.1016/j.cell.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsvetkov P, et al. The nanny model for IDPs. Nat Chem Biol. 2009;5(11):778–81. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- 76.Landreh M, et al. Specific chaperones and regulatory domains in control of amyloid formation. J Biol Chem. 2015;290(44):26430–6. doi: 10.1074/jbc.R115.653097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brandvold KR, Morimoto RI. The Chemical Biology of Molecular Chaperones–Implications for Modulation of Proteostasis. J Mol Biol. 2015;427(18):2931–47. doi: 10.1016/j.jmb.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends in Biochemical Sciences. 2001;26(10):597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 79.Kuznetsova IM, et al. What Macromolecular Crowding Can Do to a Protein. International Journal of Molecular Sciences. 2014;15(12):23090–23140. doi: 10.3390/ijms151223090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parry Bradley R, et al. The Bacterial Cytoplasm Has Glass-like Properties and Is Fluidized by Metabolic Activity. Cell. 2014;156(1–2):183–194. doi: 10.1016/j.cell.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Munder MC, et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife. 2016;5:e09347. doi: 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Joyner RP, et al. A glucose-starvation response regulates the diffusion of macromolecules. eLife. 2016;5:e09376. doi: 10.7554/eLife.09376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ter-Avanesyan MD, et al. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–92. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 84.Keleman K, et al. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10(12):1587–93. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- 85.Wu H, Fuxreiter M. The Structure and Dynamics of Higher-Order Assemblies: Amyloids, Signalosomes, and Granules. Cell. 2016;165(5):1055–66. doi: 10.1016/j.cell.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


