Abstract
Background
Poly purine.pyrimidine sequences have the potential to adopt intramolecular triplex structures and are overrepresented upstream of genes in eukaryotes. These sequences may regulate gene expression by modulating the interaction of transcription factors with DNA sequences upstream of genes.
Results
A poly purine.pyrimidine sequence with the potential to adopt an intramolecular triplex DNA structure was designed. The sequence was inserted within a nucleosome positioned upstream of the β-galactosidase gene in yeast, Saccharomyces cerevisiae, between the cycl promoter and gal 10Upstream Activating Sequences (UASg). Upon derepression with galactose, β-galactosidase gene expression is reduced 12-fold in cells carrying single copy poly purine.pyrimidine sequences. This reduction in expression is correlated with reduced transcription. Furthermore, we show that plasmids carrying a poly purine.pyrimidine sequence are not specifically lost from yeast cells.
Conclusion
We propose that a poly purine.pyrimidine sequence upstream of a gene affects transcription. Plasmids carrying this sequence are not specifically lost from cells and thus no additional effort is needed for the replication of these sequences in eukaryotic cells.
Background
Stretches of poly purine.pyrimidine (poly pur.pyr) sequences are overrepresented in the eukaryotic genome [1,2] and often positioned upstream of genes [3,4]. Many eukaryotic genes contain a poly pur.pyr box upstream of the coding region [5]. Transcription factors may interact specifically with those regions as occurs upstream of the Drosophila hsp26 gene [6,7] where transcription is enhanced during a heat shock.
Under torsional stress poly pur.pyr sequences have the potential to adopt an intramolecular triplex formation as HY-3 and H-Y5 [8-10]. A proposed triplex model fits with the regular poly pur.pyr tract with mirror symmetry [11] around a loop sequence. Shimizu et al.[10] have shown that base composition in loop sequences affects intramolecular triplex formation while orientation of the insert in supercoiled plasmid has no effect on isomerisation of the two types of triplex DNA. Nonmirror symmetric poly pur.pyr sequences can also adopt an intramolecular triplex structure [12].
The in vivo existence of poly pur.pyr sequences as an intramolecular triplex has been detected in Escherichia coli[13,14]. Modulation of gene expression by insertion of designed poly pur.pyr sequences within the E. coli genes [15,16] and upstream of a gene in mouse cells has been demonstrated [17]. Poly pur.pyr sequences in active regions of transcription form a DNA-RNA hybrid triplex and make the template transcriptionally inert [18]. Poly homo purine.pyrimidine sequences stop in vitro replication and pause in vivo replication when cloned into a plasmid under the control of the SV40 promoter [19]. Peleg et al.[20] showed that the unwinding activity of the SV40 large T-antigen helicase is decreased in the presence of a triple helix in vitro. When the triplex structure is stabilized with BeP1 [benzo(c) pyridoindole], replication elongation is inhibited in plasmids by prokaryotic DNA polymerase [16]. Friedreich Ataxia-associated poly pur.pyr expansion (GAA.TTC) when cloned in plasmids, shows inhibition of transcription and replication in cos-7 cells [21]. However, structural aspects of the poly pur.pyr sequences as intramolecular triplex DNA and their role in different biological processes in transcription regulation, replication, recombination and interaction with the nucleosome in vivo have not been fully determined [3].
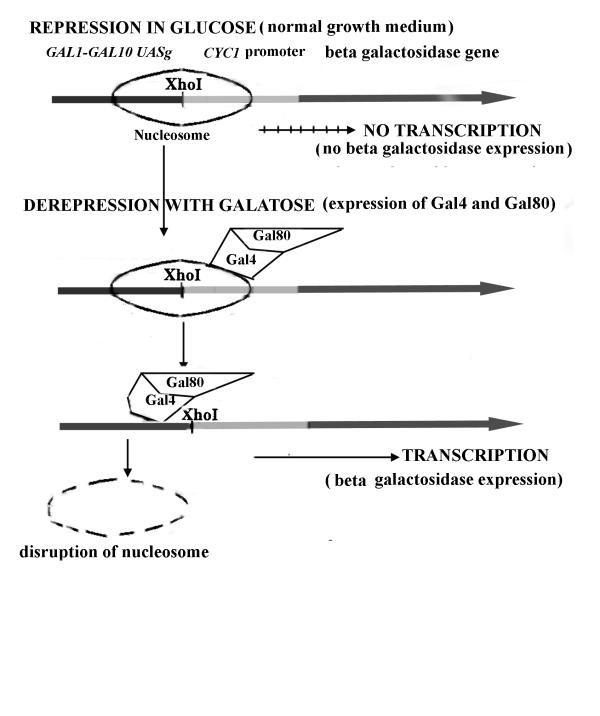
A systematic search for poly pur.pyr sequences within yeast chromosome III indicates that they frequently occur upstream of yeast genes [4]. The presence of these sequences in the promoter regions suggests they are involved in gene regulation. Here, we examine the effect of poly pur.pyr sequences on gene regulation in a position (UASg), upstream of the β-galactosidase gene where enhancer proteins are known to disrupt nucleosomes to activate transcription initiation. The E. coli (β-galactosidase gene is under the control of upstream activating sequences of gall-gal 10 and the cycl promoter (Fig. 1). Yeast cells carrying this plasmid [22] grow normally within medium supplemented with glucose as the carbon source and do not express (β-galactosidase (repressed state). When cells are transferred to a medium containing galactose (derepression), two proteins, Gal4 and Gal80, are expressed from chromosomal genes, disrupt nucleosomes, bind at the UASg and activate transcription [23,24]. We inserted poly pur.pyr sequences and a duplex control sequence within a positional nucleosome at the XhoI site between the UASg and the cycl promoter.
Figure 1.
A β-galactosidase expression system in yeast. The E. coli β-galactosidase gene is under the control of gall-gal 10 UASg and the cycl promoter. In glucose medium, this gene is not expressed and remains repressed, while activated in galactose medium by the expression of chromosomal proteins Gal4 and Gal80. The Gal4-Gal80 complex disrupts nucleosomes and binds DNA at UASg to activate transcription.
Results
Poly pur.pyr sequences affect gene expression in yeast cells
A 61-mer Xho1-Xho1 fragment containing a 58-mer poly pur.pyr sequence with a loop sequence at the center capable of adopting a triplex DNA structure was designed (Fig. 2). The fragment was cloned (single copy, pAMTS61; multiple copies, pAMTM61a, pAMTS61b, pAMTMR61; Fig. 3a) into the Xho1 site of pLGSDS36. The latter differs from the original pLGSD5 [22,25] in having a polylinker. A random sequence of the same length that will take up a normal duplex structure was designed and included in the study (pAMDC61).
Figure 2.
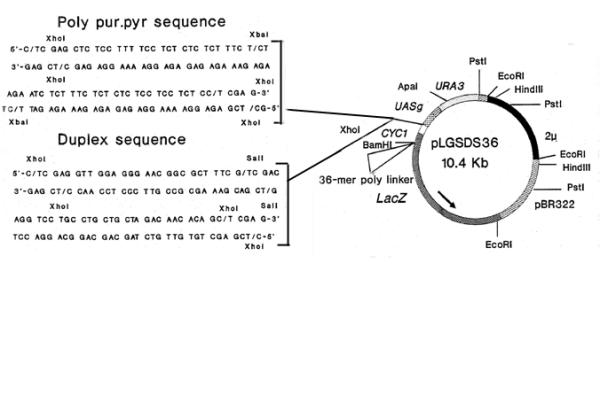
Poly purine, duplex sequences and construction of yeast expression vectors with these sequences. (a) Poly pur.pyr and duplex control sequences flanked by a Xho1 site were cloned at the junction of the cycl promoter and UASg.
Figure 3.
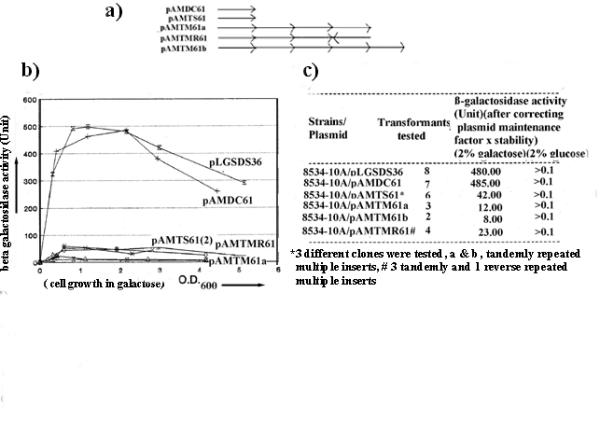
Measurement of β-galactosidase expression in the yeast cells. (a) Clones generated are: single copy, pAMTS61; multiple tandem copies, pAMTM61a (4 copies); pAMTM61b (5 copies), multiple, 3 tandem and 1 reverse copy, pAMTMR61; single copy 61-mer duplex sequences, pAMDC61. (b) Graph showing β-galactosidase expression in yeast cells carrying different plasmids in the growth phase. After transferring cells from glucose to galactose medium, aliquots of culture were removed and (β-galactosidase expression measured intermittently from starting point of culture to the exponential phase of growth. Numbers in parentheses represent transformants from two separate clones. The experiments were repeated at least 5 times for all transformants and best curves are shown for each. The maximum error observed was ± 5 units, (c) Table shows the number of independent transformants tested for β-galactosidase expression at OD600 = 1. Activity unit is corrected in all instances by the plasmid maintenance factor (stability).
We have taken advantage of the induction system of the β-galactosidase gene under the control of gall-gal 10 UASg. Upon derepression with galactose, enzyme expression is reduced about 12–50 fold (Fig. 3b and table 3c) in cells carrying single to multiple copies of poly pur.pyr inserts containing plasmids compared with duplex-carrying plasmids. No marked differences were observed in the enzyme activities in cells containing pLGSDS36 (480 units) and pAMDC61 (485 units) (i.e. insertion of extra DNA sequences of the same length as the poly pur.pyr sequence or separation of adjacent DNA sequences around the Xho1 site has no effect on β-galactosidase expression). Therefore, the presence of the poly pur.pyr sequence in the plasmid causes a reduction in β-galactosidase expression during derepression.
Reduced gene expression is correlated with reduced transcription
After derepression with galactose, transcripts of the β-galactosidase gene were analyzed and quantitated from yeast cells carrying pAMTS61 and pAMDC61. Cells were aliquoted during the expression measurement (Table 3c) at OD600 = 1. For northern blot analysis, total cellular RNA was hybridized with the 4.8 kb EcoR1 fragment carrying the ura3 and lacZ genes. There was reduced β-galactosidase (lacZ) transcript (Fig. 4b lane 2) in cells carrying pAMTS61 compared with cells carrying pAMDC61 (lane 1). There was 10-fold less transcript in cells harboring pAMTS61 (single copy poly pur.pyr sequence lacZ band/ura3 band = 0.05) than those harboring pAMDC61 (normal duplex lacZ band/ura3 band = 0.5). There was about a 12-fold reduction in β-galactosidase expression in cells carrying the poly pur.pyr sequence (42 units) compared with cells carrying the duplex (485 units). Measurements of protein and transcript indicate that reduced expression in cells carrying the poly pur.pyr sequence is almost correlated with reduced transcription.
Figure 4.
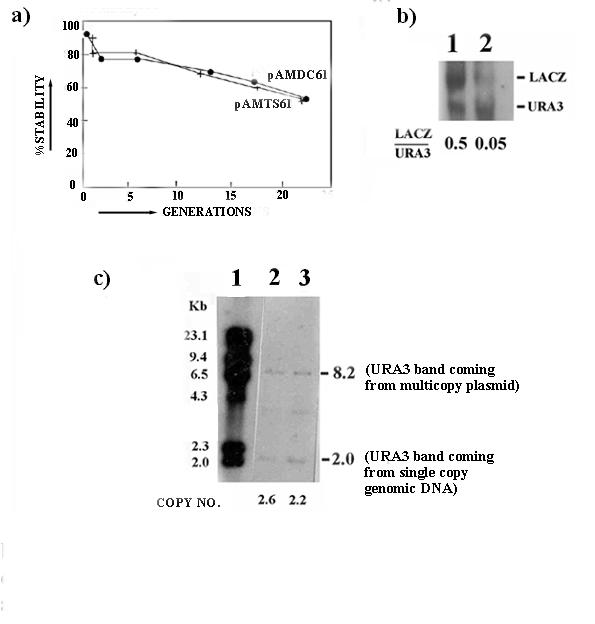
Stability, copy number and northern blots showing β-galactosidase expression in duplex and triplex carrying cells. (a) Stabilities of pAMDC61- and pAMTS 61-carrying cells are shown upto 24 generations. A graph depicting the percentage of stabilities for both plasmids is presented. After subsequent generations of growth, both type of plasmids show almost equal stabilities and stability of triplex carrying plasmids do not fall drastically (b) Copy number of cells carrying plasmids containing duplex and poly pur.pyr sequences. Lane 1, lambda/HindIII; lane 2, pAMDC61; lane 3, pAMTS61. (c) Northern blot of total RNA from duplex- and triplex-carrying cells. Duplex-carrying cells have 10-fold higher level of β-galactosidase expression than triplex carrying cells.
Poly pur.pyr insert carrying plasmids are not specifically lost from the cells
To determine whether plasmids carrying the poly pur.pyr sequence are specifically lost from cells after subsequent generations of growth in nonselective medium, plasmid stabilities and copy numbers were measured. Yeast cells do not maintain [26] these minichromosome vectors in 100% efficiency and typically 80–90% of cells carry these plasmids in high copy number (20–30 copies/cell) after transformation. Cells loose plasmids slowly during subsequent generations of growth in nonselective medium. However, it has been shown that when replication is impaired at the plasmid origin, copy number/cell rapidly decreases during subsequent culture (within 10–25 generations) and cells completely loose the plasmid [26]. The percentage of cells maintaining plasmids is denoted as % stability. The % stability of each plasmid could be quantitated by the loss of a marker (ura3) from the cells, which will not grow in minimal medium devoid of uracil (CM-URA).
If replication of poly pur.pyr sequences in the plasmid were affected, a reduction in stability as well as in copy number would be expected [26] typically within 15–20 generations of growth. Figure 4a shows little difference in stabilities. Almost equal stability has been observed in both the cases after 20 generations of growth (i.e. cells are not preferentially loosing plasmids carrying a poly pur.pyr sequence). The copy number (Fig. 4c) of plasmids carrying a poly pur.pyr sequence (copy number, 2.6) compared with those carrying a duplex (copy number, 2.2) after 20 generations of growth does not differ significantly. If replication of the plasmid is impaired, a marked decrease in copy number of plasmids carrying a poly pur.pyr sequence would have been observed. These results suggest that plasmid replication may not be significantly reduced or affected by the presence of a poly pur.pyr sequence.
Discussion
Poly pur.pyr sequences affect β-galactosidase expression
The yeast expression system has been used to illustrate the role of poly pur.pyr sequences in gene expression where the β-galactosidase gene is under the control of the cycl promoter and gall-gal 10 UASg.
The poly pur.pyr sequence designed has the potential to form an intramolecular triplex DNA structure and mimic sequences present naturally in the upstream region of various genes in yeast genome. Reduced β-galactosidase expression is due to reduced transcription initiation and poly pur.pyr sequences are involved in transcription initiation in vivo. The 61-mer sequence containing the poly pur.pyr sequence is S1 nuclease sensitive in supercoiled plasmid in vitro[27]. In addition to their presence in the S. cerevisiae genome, loop sequences containing poly pur.pyr stretches are also known to be present upstream of various genes in eukaryotes [4,28,29] and are implicated to be involved in gene regulation.
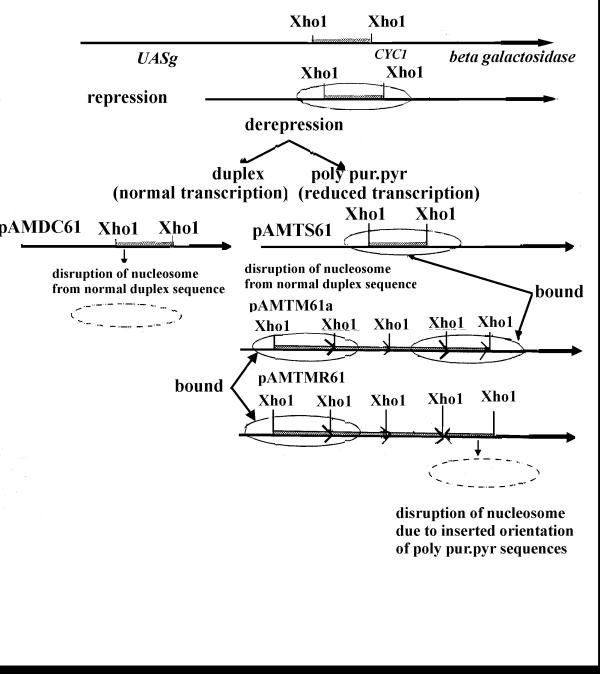
It has been reported that poly pur.pyr sequences within genes down-regulate gene expression in E. coli[15]. A homogenous d (CT) n. d (AG) n stretch (which does not form an intramolecular triplex in vivo) together with Heat Shock Elements (HSE) upstream of the Drosophila hsp26 gene plays an integral role in resetting chromatin structure [6,7]. It is possible that heat shock facilitates nucleosome disruption from tightly bound dCT.dAG stretch by GAGA transcription factor [30] and transcription initiation occurs upon binding with Heat Shock Factors (HSF). The fine-tuning of eukaryotic gene regulation also depend upon the DNA sequences located upstream of genes. The decreased transcription, observed in yeast cells, in this study, may be due to the presence of poly pur.pyr sequences that might bind more tightly to a positional nucleosome(s) present at the junction of UASg and the cycl promoter [23] than the control DNA sequences. This binding of nucleosome with a poly pur.pyr sequence may inhibit the binding of protein factors [24,31] on the DNA. Here, poly pur.pyr and duplex sequences are inserted in DNA where a positional nucleosome is reorganizable and is normally disrupted before transcription initiation by the Gal4-Gal80 complex. We analyzed the fate of this nucleosome in vivo during repression with glucose and derepression with galactose in both duplex- and poly pur.pyr-carrying cells (unpublished observation). Preliminary results suggest that the single copy poly pur.pyr sequence binds with the positional nucleosome with higher affinity than the normal duplex sequence and inhibits disruption by the Gal4-Gal80 complex before transcription initiation. Also, in multiple tandem copies the poly pur.pyr sequence (pAMTAM61a) keeps the nucleosome binding and interestingly disruption of nucleosome occurs from the inverted orientation of the poly pur.pyr sequence (pAMTMR61) at the other end. From these data, the reduced gene expression may be explained in the context of nucleosome binding with the poly pur.pyr sequence and a model is presented (Fig. 5). However, this observation is contrary to the evidence that an intermolecular triplex repels the nucleosome in vitro[32]. We believe that attraction or repulsion of the nucleosome in vivo depends on the different poly pur.pyr sequences and their conformation at the nucleosomal surface (i.e. as an intramolecular triplex). This sequence forms an intramolecular triplex in vitro[27], however its conformation in vivo requires further investigation.
Figure 5.
A model is suggested for the reduction of β-galactosidase gene expression in the presence of a poly pur.pyr sequence. Before transcription initiation, nucleosome disruption is necessary from DNA sequences by the Gal4-Gal80 complex. This protein complex may not disrupt the nucleosome from the single copy and tandemly repeated multiple copy poly pur.pyr sequence due to the higher affinity of binding with the nucleosome while these may disrupt nucleosomes from normal duplex carrying sequences and from the inverted orientation of the poly pur.pyr sequence. It is possible that the single and tandem multiple copy of the poly pur.pyr sequence may form an intramolecular triplex while inverted orientation disrupts the structure.
No large decrease in stability and copy number of plasmids carrying the poly pur.pyr sequence was observed during subsequent generations of growth suggesting that the presence of this sequence does not affect replication in eukaryotic cells unlike the observation of Duval-Valentine et al.[16] where prokaryotic DNA polymerases pause during replication in vitro. This supports the suggestion that no extra effort is needed by DNA polymerases to replicate such sequences that are abundant in the genome of eukaryotes. A plausible explanation is that the intramolecular triplex structure does not exist on naked DNA in eukaryotes rather such looped structure exists on the nucleosomal surface [33] and as replication proceeds, the nucleosome makes way [34] for the DNA polymerase, the intramolecular triplex structure resolves into duplex DNA and facilitates replication. It is possible that extra replication machinery in eukaryotic cells could resolve the intramolecular structure during replication. It has been observed that the SV40 large T-antigen has the unwinding capacity of intramolecular triplex during replication [35]in vitro and replication termination in poly dT tract is relieved by mitochondrial single-strand binding proteins [36]. It would be interesting to investigate whether these sequences form an intramolecular triplex structure at the nucleosomal surface in vivo. Further characterization of superhelical stress and other conditions of adopting an intramolecular triplex structure in the chromosome of eukaryotes are under investigation [27].
Materials and Methods
Strains
Saccaromyces cerevisiae strains [26] 8534-10A (Cir+ mat a ura3-52, leu2-3 his 4Õ34) and (cir ° Mat a ura3-52, leu2-3 his 4534) were used in the study. Escherichia coli strain HB101 was used in all cloning procedures.
Design and cloning of poly pur.pyr stretch with a loop sequence
pLGSD5 [22,25] is a yeast expression vector containing the β-galactosidase gene under the control of the upstream activating sequence of gall-gal 10 (UASg) and the cycl promoter. A multiple cloning site containing a 36 bp poly linker is cloned at the BamH1 site in the 5' of β-galactosidase gene to create pLGSDS36.
The 61-mer sequence containing a 58-mer poly pur.pyr sequence flanked by a XhoI site was synthesized and cloned at the XhoI site (-154 bp from translation start site; -130 bp from transcription initiation site) of pLGSDS36 at the junction of UASg and the CYC1 promoter to create pAMTS61 (single copy), pAMTM61 (a and b; multiple tandemly repeated; 4 and 5 copies) and pAMTMR61 (3 copies tandemly and 1 copy reversibly repeated) (Fig. 3a). The status and copy numbers were confirmed by restriction mapping and sequencing. A 61-mer duplex sequence flanked by a XhoI site was also synthesized and cloned at the XhoI site of pLGSDS36 to create pAMDC61.
β-galactosidase assay
β-galactosidase expression was measured in transformants grown in glucose medium (repressed state) and in galactose medium (derepressed state). Yeast cells transformants carrying the plasmids were grown in 10 mL of complete medium (YEPD) in the presence of glucose (2%) as the carbon source. After exponential growth, cells were centrifuged and resuspended in medium (500 mL) containing galactose (2%) as a carbon source to activate transcription. At specific time intervals during exponential growth beginning when the cells were transferred to galactose medium (OD600 about 0–0.04), (β-galactosidase expression was measured in aliquots of culture. Fewer cells (10 mL, OD600 = 2) were transferred to a large volume (500 mL) to allow the cells more divisions in galactose medium to attain subsequent growth to deplete the internal cellular glucose, which may partially repress transcription in their initial phase of growth in galactose. (β-galactosidase expression was measured as described by Reynolds and Lundablad [37] and expressed in Miller's unit [38]. An aliquot of culture was plated on complete medium (YEPD) and replica plated onto minimal medium (CM-URA) to determine the percentage of cells carrying plasmids (% stability). Actual (β-galactosidase activities were obtained by multiplying average (β-galactosidase activity with the stability factor of each plasmid.
Stability and copy number determination
Plasmid stabilities and copy numbers were determined essentially as described earlier [26]. Transformants carrying plasmids containing the poly pur.pyr sequence and the duplex sequence were grown in YEPD (rich medium) from a single colony to saturation and plated on YEPD plates after dilution. After sufficient growth, plates were replica plated onto CM-URA synthetic medium.
% Stability = (Number of colonies grown in CM-URA / number of colonies grown in YEPD) × 100.
Plasmid copy number was determined by quantifying ura3, which is present in single copy in yeast genome and presence of it in the multicopy plasmids. Total celullar DNA from yeast transformants was digested with Hindlll. A Southern blot was performed on the digested DNA using a 1.1 kb ura3 fragment as a probe. Autoradiograms were scanned using an Ultroscan Broma Laser Densitometer and average plasmid copy number was determined as the ratio of the area of ura3 on the plasmid band (8.2 kb coming from the multicopy plasmid) to the chromosomal band (2.0 kb; single copy). Actual copy number was determined by multiplying with the stability factor of each plasmid maintenance (actual number of cells carrying the plasmid).
Isolation of total RNA and northern hybridisation
Total celullar RNA was isolated by the hot phenol method [39] from each culture (grown in selective media with 2% galactose) of cells carrying pAMTS61 and pAMDC61. An equal amount of RNA was run in a MOPS-formaldehyde agarose gel and transferred to a nylon membrane with 20 × SSC. A 4.8 kb EcoR1 fragment carrying the entire lacZ and ura3 genes of pLGSDS36 was labeled with alpha-32P dATP by random priming and used as a probe for hybridization at 42°C in 5 × SSC, 50% formamide. Filters were washed twice with 2 × SSC (37°C) for 15 min and twice with 0.5 × SSC (42°C) and autoradiographed. Autoradiograms were scanned and the ratio of the intensities of lacZ transcripts to ura3 transcripts was calculated.
Acknowledgments
Acknowledgement
We thank Dr. P. S. Sarkar for designing poly pur.pyr sequences. Dr. L. Guarante for generous gift of plasmid, pLGSD5 and K. Usha for synthesizing oligonucleotides. We are thankful to "Sciencemanager" for careful editing of this manuscript. AKM is supported by DBT postdoctoral fellowship (R (1A) RA (NBTB)/ MBUAKM/92–777). SKB is supported by a grant from DBT, Govt of India. We are grateful to anonymous reviewers for their helpful comments to improve this manuscript.
Contributor Information
Amit K Maiti, Email: amit@dplanet.ch.
Samir K Brahmachari, Email: skb@cbt.res.in.
References
- Tripathy J, Brahmachari SK. Distribution of simple repetative (TG/CA) n and (CT/AG) n sequences in human and Rodent genomes. J Biomol Struc & Dyn. 1991;9:387–397. doi: 10.1080/07391102.1991.10507919. [DOI] [PubMed] [Google Scholar]
- Behe MJ. An overabundance of long oligopurine tracts occurs in the genome of simple and complex eukaryotes. Nucl Acids Res. 1995;23:689–95. doi: 10.1093/nar/23.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RD, Collier DA, Hanvey JC, Shimizu M, Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine. oligopyrimidine sequences. FASEB J. 1988;2:2939–49. [PubMed] [Google Scholar]
- Raghavan S, Burma PK, Brahmachari SK. Positional preferences of polypurine/polypyrimidine tracts in Saccharomyces cerevisiae genome: implications for cis regulation of gene expression. J Mol Evol. 1997;45:485–98. doi: 10.1007/pl00006253. [DOI] [PubMed] [Google Scholar]
- De Martinoff G, Pohl V, Mercken L, van Ommen GJ, Vassert G. Structural organization of the bovine thyroglobulin gene and its 5'-flanking region. Eur J Biochem. 1987;164:591–599. doi: 10.1111/j.1432-1033.1987.tb11168.x. [DOI] [PubMed] [Google Scholar]
- Glaser RL, Thomas GH, Siegfried E, Elgin SC, Lis JT. Optimal heat-induced expression of the Drosophila hsp26 gene requires a promoter sequence containing (CT) n. (GA) n repeats. J Mol Biol. 1990;211:751–61. doi: 10.1016/0022-2836(90)90075-W. [DOI] [PubMed] [Google Scholar]
- Lu Q, Wallrath LL, Granok H, Elgin SC. (CT) n (GA) n repeats and heat shock elements have distinct roles in chromatin structure and transcriptional activation of the Drosophila hsp26 gene. Mol Cell Biol. 1993;5:2802–2814. doi: 10.1128/mcb.13.5.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H, Dahlberg JE. Topology and formation of triple-stranded H-DNA. Science. 1989;243:1571–6. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- Johnston BH. The S1-sensitive form of d(C-T)n.d(A-G)n: chemical evidence for a three-stranded structure in plasmids. Science. 1988;241:1800–4. doi: 10.1126/science.2845572. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Kubo K, Matsumoto U, Shindo H. The loop sequence plays crucial roles for isomerization of intramolecular DNA triplexes in supercoiled plasmids. J Mol Biol. 1994;235:185–97. doi: 10.1006/jmbi.1994.1036. [DOI] [PubMed] [Google Scholar]
- Mirkin SM, Lyamichev VI, Drushlyak KN, Dobrynin VN, Filippov SA, Frank-Kamenetskii MD. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature. 1987;330:495–7. doi: 10.1038/330495a0. [DOI] [PubMed] [Google Scholar]
- Klysic J. An intramolecular triplex structure from non mirror repeated sequence containing both py:pu.py and pu:pu.py triads. J Mol Biol. 1995;245:499–507. doi: 10.1006/jmbi.1994.0041. [DOI] [PubMed] [Google Scholar]
- Karlovsky P, Pecinka P, Vojtiskova M, Makaturova E, Palecek E. Protonated triplex DNA in E. coli cells as detected by chemical probing. FEES Lett. 1990;274:39–42. doi: 10.1016/0014-5793(90)81324-H. [DOI] [PubMed] [Google Scholar]
- Kohwi Y, Malkhosyan SR, Kohwi-Shigematsu T. Intramolecular dG.dG.dC triplex detected in Escherichia coli cells. J Mol Biol. 1992;223:817–822. doi: 10.1016/0022-2836(92)90242-c. [DOI] [PubMed] [Google Scholar]
- Sarkar PS, Brahmachari SK. Intramolecular triplex potential sequence within a gene downregulates its expression in vivo. Nucl Acids Res. 1992;20:5713–5718. doi: 10.1093/nar/20.21.5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval-Valentin G, de Bizemont T, Takasugi M, Mergny JL, Bisagni E, Helene C. Triple-helix specific ligands stabilize H-DNA conformation. J Mol Biol. 1995;247:847–58. doi: 10.1006/jmbi.1995.0185. [DOI] [PubMed] [Google Scholar]
- Kohwi Y, Kohwi-Shigematsu T. Altered gene expression correlates with DNA structure. Genes Dev. 1991;5:2547–54. doi: 10.1101/gad.5.12b.2547. [DOI] [PubMed] [Google Scholar]
- Reaban ME, Griffin JA. Induction of RNA stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature. 1990;348:342–344. doi: 10.1038/348342a0. [DOI] [PubMed] [Google Scholar]
- Rao BS. Regulation of DNA replication by homopurine/homopyrimidine sequences. Mol Cell Biochem. 1996;156:163–8. doi: 10.1007/BF00426339. [DOI] [PubMed] [Google Scholar]
- Peleg M, Kopel V, Borowiec JA, Manor H. Formation of DNA triple helices inhibits DNA unwinding by the SV40 large T-antigen helicase. Nucleic Acids Res. 1995;23:1292–9. doi: 10.1093/nar/23.8.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J Biol Chem. 1998;273:14588–95. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- Guarente L, Yocum RR, Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci USA. 1982;79:7410–4. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor MJ, Lue NF, Kornberg RD. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988;204:109–27. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Morse RH. Nucleosome disruption by transcription factor binding in yeast. Science. 1993;262:1563–6. doi: 10.1126/science.8248805. [DOI] [PubMed] [Google Scholar]
- Guarente L, Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:2199–203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti AK, Sinha P. The mcm2 mutation of yeast affects replication, rather than segregation or amplification of the two micron plasmid. J Mol Biol. 1992;224:545–558. doi: 10.1016/0022-2836(92)90543-s. [DOI] [PubMed] [Google Scholar]
- Brahmachari SK, Meera G, Sarkar PS, Balagurumoorthy P, Tripathi J, Raghavan S, Shaligram U, Pataskar S. Simple repetitive sequences in the genome: structure and functional significance. Electrophoresis. 1995;16:1705–14. doi: 10.1002/elps.11501601283. [DOI] [PubMed] [Google Scholar]
- Yagil G. Paranemic structures of DNA and their role in DNA unwinding. Crit Rev Biochem & Mol Biol. 1991;26:475–559. doi: 10.3109/10409239109086791. [DOI] [PubMed] [Google Scholar]
- Brahmachari SK, Sarkar PS, Raghavan S, Narayan M, Maiti AK. Polypurine/ polypyrimidine sequences as cis-acting transcriptional regulators. Gene. 1997;190:17–26. doi: 10.1016/S0378-1119(97)00034-6. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–32. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- Chasman DI, Lue NF, Buchman AR, LaPointe JW, Lorch Y, Kornberg RD. A yeast protein that influences the chromatin structure of UASg and functions as a powerful auxiliary gene activator. Genes Dev. 1990;4:503–14. doi: 10.1101/gad.4.4.503. [DOI] [PubMed] [Google Scholar]
- Westin L, Blomquist P, Milligan JF, Wrange O. Triple helix DNA alters nucleosomal histone-DNA interactions and acts as a nucleosome barrier. Nucleic Acids Res. 1995;23:2184–91. doi: 10.1093/nar/23.12.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A, Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982;2:609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Bonne-Andrea C, Wong ML, Alberts BM. In vitro replication through nucleosomes without histone displacement. Nature. 1990;343:719–26. doi: 10.1038/343719a0. [DOI] [PubMed] [Google Scholar]
- Kopel V, Pozner A, Baran N, Manor H. Unwinding of the third strand of a DNA triple helix, a novel activity of the SV40 large T-antigen helicase. Nucleic Acids Res. 1996;24:330–5. doi: 10.1093/nar/24.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov VS, Bogenhagen DF. Termination within oligo(dT) tracts in template DNA by DNA polymerase gamma occurs with formation of a DNA triplex structure and is relieved by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 1996;271:30774–80. doi: 10.1074/jbc.271.48.30774. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Lundablad V. In current protocols in Molecular Biology (ed Ausubel et al; Green publishing associates, inc and John willey and Sons. 1993.
- Miller JH. experiments in molecular Genetics. (Cold spring harbour laboratory Press, Cold spring harbour, NY), 1972.
- Collart M, Oliviero S. Preparation of yeast RNA. In current protocols in Molecular Biology (ed Ausubel et al; Green publishing associates, inc and John willey and Sons) 1993. [DOI] [PubMed]