Abstract
DNA oligonucleotides containing site-specific N7-guanine monoadducts of cisplatin, diepoxybutane, and epichlorohydrin were used as templates for DNA synthesis by two bacterial DNA polymerases and human polymerase β. These polymerases were able to bypass the lesions effectively, although the efficiency was decreased, with inhibition increasing with the size of the lesion. Fidelity of incorporation was essentially unaltered, suggesting that N7-guanine monoadducts do not significantly contribute to the mutational spectra of these agents.
Keywords: DNA alkylation, diepoxybutane, cisplatin, epichlorohydrin, mutagenesis, primer extension assay
1. Introduction1
Many first-line anticancer drugs, including alkylating agents and platinum compounds, act by covalently modifying DNA, leading to genotoxic and cytotoxic effects. Some bifunctional agents are particularly potent drugs, first forming monoadducts that can react further with solvent, protein, or another nucleophilic site on the DNA [1]. If the second reaction occurs with the same DNA strand, the result is an intrastrand cross-link, whereas if it occurs with the opposite strand, an interstrand cross-link is formed [2]. The latter are believed to be the most cytotoxic lesions, with as few as 20 interstrand cross-links comprising the mean lethal dose in some mammalian cell lines [3].
Cross-linking agents have been used therapeutically since about 1950, contributing to increased survival rates for many types of cancer [4]. For example, the development of platinum drugs has made testicular cancer largely curable, with a 5-year relative survival rate of more than 90% [5]. However, DNA-modifying drugs also increase the long-term risk of secondary malignancies [6,7,8], with one study of testicular cancer survivors showing about a 5-fold increase in the risk of acute myelogenous leukemia over 20 years [5]. Even decades after chemotherapy for Hodgkin’s lymphoma, survivors remain at increased risk for subsequent malignant neoplasms [9]. Mutations that arise in surviving cancer cells following the formation of DNA lesions may also contribute to drug resistance [10]. Furthermore, germ line mutations following chemotherapy potentially pose a heritable genetic risk [11].
Mutations can arise during replication of covalently modified DNA when DNA polymerase encounters the damaged template. Alkylating agents and platinum compounds preferentially target the N7 position of guanine, often leading to transitions and transversions at guanine [7]. However, minor products are also formed, and different lesions vary in their biological effects. For example, N3 methylation of adenine, but not N7 methylation of guanine, inhibits DNA polymerase and terminates replication [12].
The mutagenic effects of a specific lesion type can be determined by introducing an oligonucleotide containing that lesion into cells and monitoring for base substitution events. Such studies have revealed that minor adducts can be significantly more mutagenic than the major adduct. For example, the most abundant cisplatin intrastrand cross-link occurs at 5′-GG sites, forming a highly lethal adduct [13]. However, the intrastrand cross-link at 5′-AG is 5–10-fold more mutagenic than that at 5′-GG [13,14].
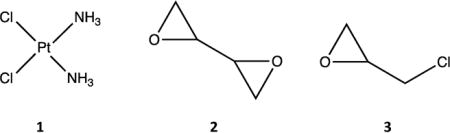
Another method for characterizing the mutagenicity of specific DNA adducts is to monitor directly the ability of DNA polymerase to replicate a damaged template accurately. Our goal was to use such an assay to assess the potential mutagenicity of guanine-N7 monoadducts for three bifunctional agents: cisplatin (1), diepoxybutane (DEB; 2), and epichlorohydrin (ECH; 3). These compounds vary widely in their carcinogenic potentials, with TD50 values in rodents of 0.0667 mmol/kg body weight, 18.5 mmol/kg body weight, and 148 mmol/kg body weight, respectively [15]. Cisplatin is widely used in cancer chemotherapy, with particular effectiveness against testicular and ovarian cancer [13,14]. DEB is the active form of the anti-cancer prodrug treosulfan [16], used to treat ovarian cancer, as well as a major metabolite of the widely used industrial compound 1,3-butadiene [17]. Epichlorohydrin is not a cancer drug, but it is widely used in the synthetic polymer industry and is classified as “probably carcinogenic to humans” [18]. All three agents preferentially target the N7 position of guanine, with a variety of secondary reaction sites, primarily at adenine [19,20,21,22]. Because alkylated N7-guanine adducts are quite labile, the mutagenicities of the primary DEB and ECH adducts have not previously been reported [23].
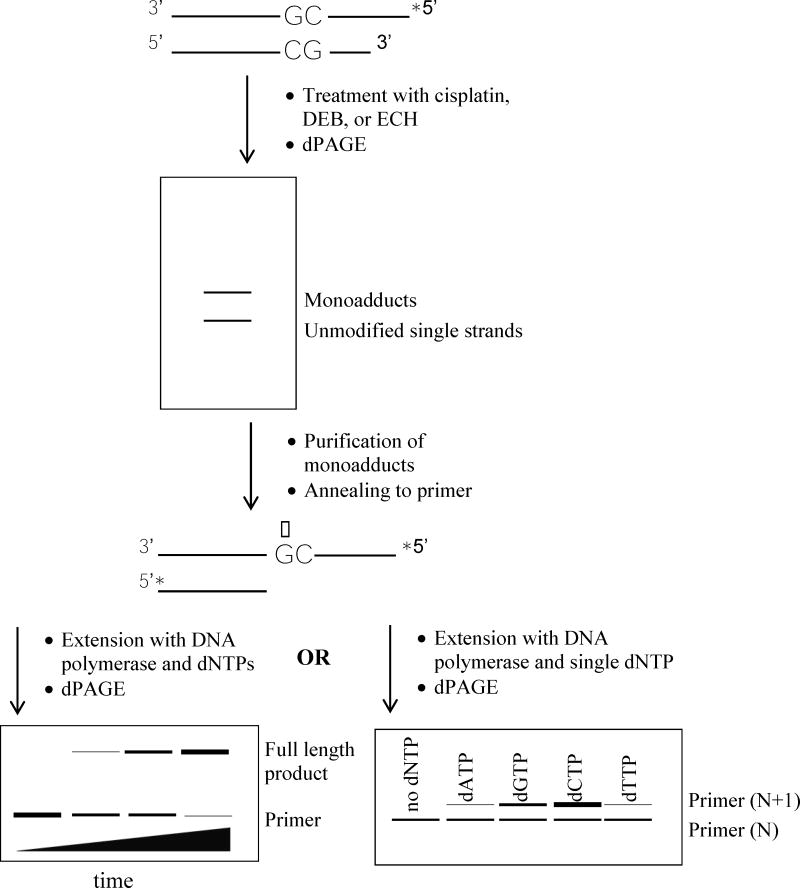
Our methods are summarized in Scheme 1. Briefly, monoadducted template DNA was purified through denaturing polyacrylamide gel electrophoresis (DPAGE) after treatment of a synthetic DNA duplex containing a single guanine target site with cisplatin, DEB, or ECH. The single-stranded template was then annealed to a primer, and DNA synthesis studies were performed with three different polymerases: Klenow fragment from Escherichia coli (KF), the large fragment of DNA Polymerase I from Bacillus stearothermophilus (BF), and recombinant human polymerase β (hPol β). The former are two related A-family bacterial polymerases commonly used as model enzymes in lesion bypass studies [24, 25], and the latter is a replicative X-family polymerase. Primers terminated two nucleotides before the lesion, one nucleotide before the lesion, or at the lesion in order to assess whether any effect of the lesion is highly local or also affects synthesis at flanking bases. The results of these studies suggest that the major N7-guanine monoadducts of these agents are not significantly mutagenic although they are somewhat blocking to the DNA polymerases used here.
Scheme 1.
Steps involved in assessing DNA polymerase activity when replicating a template containing a monoadduct (represented by the starred guanine residue). Duplex A (Chart 1) was used to generate the monoadducted template. Asterisks denote 32P radiolabels.
2. Materials and methods
2.1 Materials
Recombinant hPol β was expressed and purified according to published procedures [26]. BF, KF, and other enzymes were purchased from New England Biolabs (Ipswich, MA). Adenosine 5′-triphosphate [γ-32P] was from PerkinElmer Health Sciences (Shelton, CT). Synthetic oligonucleotides (Integrated DNA Technologies in Coralville, IA) were purified prior to use by DPAGE (19:1 acrylamide: bisacrylamide; 40% urea) followed by UV shadowing and the crush-and-soak procedure [27]. Cisplatin and racemic DEB and ECH were from Sigma-Aldrich Chemical Company (St. Louis, MO). (Caution: cisplatin, DEB, and ECH are suspect carcinogens and must be handled appropriately.) All other chemicals were of reagent grade.
2.2 Preparation of monoadducts
Radiolabeling at the 5′-terminus of Strand 1 of Duplex A (see Chart 1) was achieved by incubating 1 OD260 of the single strand with [γ-32P]-ATP (10 μCi) and T4 polynucleotide kinase under standard conditions. Following ethanol precipitation [27], the dry pellet was mixed with 1 OD260 of Strand 2 in reaction buffer as follows: for cisplatin, 40 mM sodium perchlorate, 2 mM monobasic potassium phosphate, pH 7.6; for DEB, TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH 7.4); and for ECH, 0.3 M sodium acetate, pH 5.2. The two strands were annealed by incubating at 60°C for 15 min and then at ambient temperature for 15 min. Reactions were initiated by addition of cisplatin (12.5 μL of a 1 mM solution prepared with stirring in water 24 h prior; final concentration 0.25 mM), DEB (2 μL; final concentration 250 mM), or ECH (2 μL; final concentration 250 mM). Following initial time course studies to determine optimal reaction times, incubation was at 37oC for 3 h (cisplatin and ECH) or 1 h (DEB), followed by ethanol precipitation and lyophilization.
Chart 1.
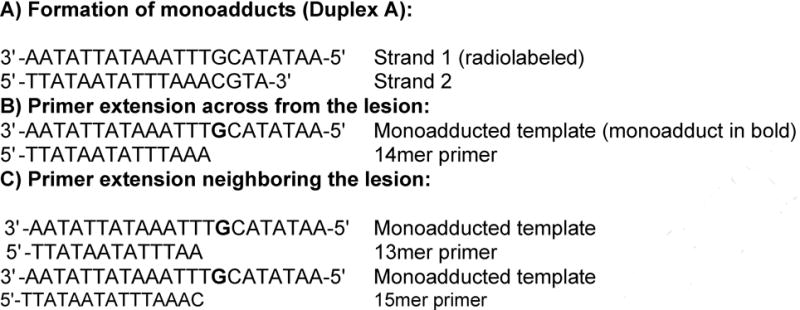
Oligonucleotides used in this study.
2.3 Purification of monoadducts
Drug-treated samples were dissolved in 10 μL of 5 M aqueous urea/0.1% xylene cyanol and loaded onto a 20% denaturing polyacrylamide gel (19:1 acrylamide/bisacrylamide, 50% urea) run at 65 W. Monoadducts were located with x-ray film as bands just above Strand 1, excised from wet gels, and purified by the crush-and-soak procedure followed by ethanol precipitation. Alkylation at N7 of guanine was confirmed by heating at 90°C in 100 μL of 10% aqueous piperidine for 2 h, lyophilization, and analysis on 20% denaturing polyacrylamide gels (19:1 acrylamide/bisacrylamide, 50% urea). Maxam-Gilbert G reactions were used as references for cleavage at N7-alkylated guanine residues [28].
2.4 Primer extension reactions
Purified monoadducted Strand 1 was mixed with 0.3 OD of radiolabeled primer (see Chart 1) in 100 μL of the appropriate polymerase buffer (for KF: 50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.9; for BF: 20 mM Tris-HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton® X-100, pH 8.8; for hPol β: 50 mM Tris-HCl, 5 mM MgCl2, 10 mM NaCl, 4 mM DTT, 0.5 mg/mL BSA, pH 8.0). Annealing was achieved by incubating at 50°C for 15 min and then at ambient temperature for 15 min. Reaction conditions (concentration of DNA polymerase and concentrations of dNTPs) were optimized as follows. The annealed product was either incubated with DNA polymerase (KF, 1.0 unit; BF, 8.0 units; hPol β, 3.5 μg) in the presence of all four dNTPs (500 μM), or the annealed product was split into five equal aliquots, four of which were used for primer extension reactions and one of which was the control. In the latter case, extension was initiated by the addition of a single dNTP (20 μM) and DNA polymerase (KF, 0.25 units; BF, 1.2 units; hPol β, 0.2 μg). Reactions were stopped by the addition of an equal volume of loading dye (95% formamide, 10 mM EDTA, 0.03% xylene cyanol). Samples were run on 20% polyacrylamide gels (as above) and visualized via phosphorimagery (Amersham Biosciences STORM 860). Primer and product bands were quantified using ImageQuant version 5.2. Quantitative experiments were run in triplicate.
2.5 Steady state kinetics
Steady state kinetics experiments were performed with hPol β and all four dNTPs under the same conditions as for the full-length extension reactions except that eight different concentrations of dNTPs were used (100 – 800 μM; times ranged from 0 – 15 min). Initial rates were obtained from the disappearance of primer bands, quantified as above, and used to generate Michaelis-Menten curves, from which Km and kcat values were obtained in replicate.
3. Results
3.1 Preparation of monoadducts
A radiolabeled single-stranded oligonucleotide containing a single guanine residue was annealed to a shorter complementary strand and then reacted with cisplatin, DEB, or ECH under previously reported conditions [29,30,31]. This duplex lacked suitable sites for formation of either intrastrand [32] or interstrand cross-links [2,30,31], suggesting that the monoadduct would be a terminal product. A double-stranded target was used because formation of monoadducts by other alkylating agents has been shown previously to be highly dependent on a duplex structure [33].
DPAGE of reaction products showed bands of slightly lower mobility than unmodified DNA that increased in intensity with reaction time (Figure 1). The position of these bands was consistent with previous reports of monoalkylated DNA [33,34]. At short reaction times, there was a single well-defined low mobility band, but smeared bands at higher molecular weight appeared at longer times. The homogeneity of the initial band suggests one major monoalkylated product, with the later “ladder” of bands resulting from the formation of multiple adducts on a single strand [33].
Figure 1.
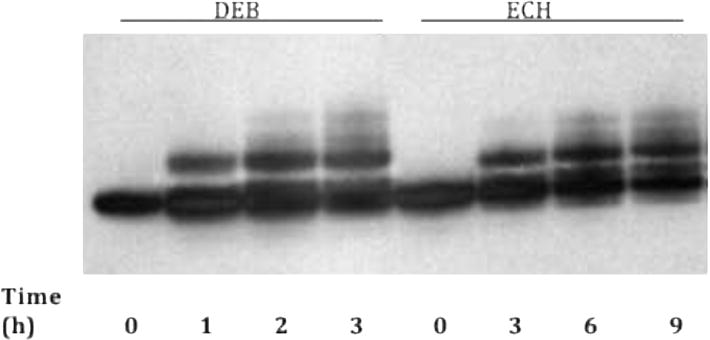
Representative time course of monoalkylation reactions with Duplex A. Shown here are the results for DEB (250 mM) and ECH (250 mM). Presumed monoadducts appear as bands with lower mobility than the zero time point band.
Reaction times were optimized for each agent to yield the maximum amount of the major low-mobility product, which was produced with average yields of ~30%, while minimizing higher molecular weight material. The band presumed to correspond to the major monoadduct showed an increase in electrophoretic retardation with the molar mass of the agent, further supporting the identification of this band as monoadduct. Gel purification of this product was achieved with minimal degradation, and we verified that the presumed monoadducts are stable for at least 30 min at 50°C (Supplementary Material, Figure S1) and for several days at −20°C (Figure 2A; lanes 1–3).
Figure 2.
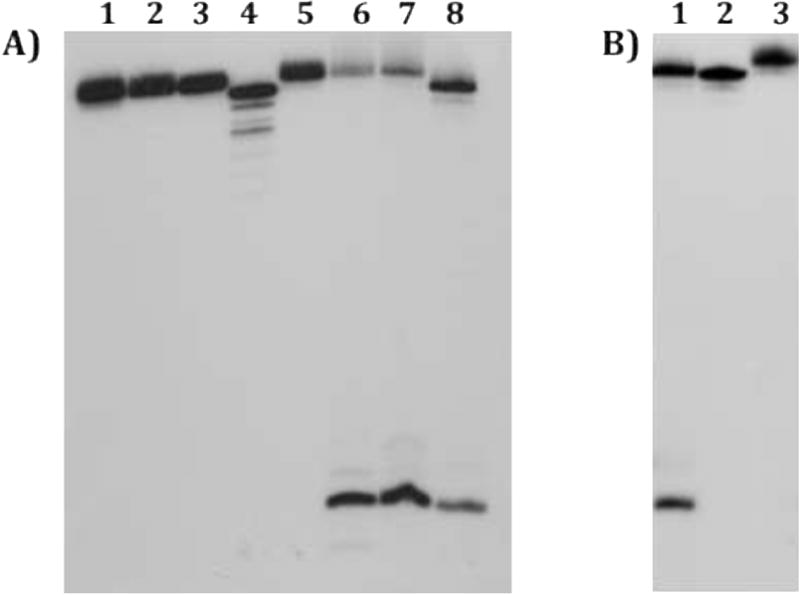
Characterization of purified monoadducts. Panel A) Lane 1: purified cisplatin monoadducts; Lane 2: purified DEB monoadducts; Lane 3: purified ECH monoadducts; Lane 4: unmodified single-stranded DNA; Lane 5: piperidine-treated cisplatin monoadducts; Lane 6: piperidine-treated DEB monoadducts; Lane 7: piperidine-treated ECH monoadducts: Lane 8: G-reaction. Monoadducts were several days old. Alkylation, but not platination, at the N7-position results in cleavage at the single G residue of Strand 1. Panel B) Lane 1: G-reaction; Lane 2: unmodified single-stranded DNA; Lane 3: DMS- and piperidine-treated cisplatin monoadduct. Cisplatin protects from DMS attack, verifying linkage at the N7 of guanine.
Because ECH and DEB have secondary targets in addition to guanine, the site of alkylation for these agents was confirmed through piperidine cleavage, which results in depurination and strand cleavage at N7-alkylated guanines [28]. Presumed DEB and ECH monoadducts were gel purified, heated in piperidine, and then analyzed on a sequencing gel. For both agents, a single major cleavage band that comigrated with a Maxam-Gilbert G reaction was visible, corresponding to cleavage at the sole dG residue of Strand 1 (Figure 2A, lanes 6 and 7). These results are consistent with the major low mobility band corresponding to a single product linked at the N7 position of guanine. The site of cisplatin modification was verified by treating purified monoadducts with DMS and then piperidine (Figure 2B). The single guanine was resistant to DMS attack (lane 3), showing that the monofunctional cisplatin adduct is also bound to the N7 position of guanine.
3.2 Primer extension efficiency with all dNTPs
Gel-purified monoadducts were annealed to a complementary 14mer primer (Chart 1) with a 3′-end terminating one nucleotide upstream from the lesion. This duplex was used as the substrate for bacterial DNA Polymerase I (either KF or BF) with all four dNTPs present. Both enzymes generated full-length products (22mers), with some 23mer products also visible (presumably from blunt-end addition [35]). However, the rate of synthesis was lower for monoadducted templates than for control unmodified template. For BF, virtually complete conversion of primer to full-length products was observed at longer time points (Figure 3A), with a relative rate of primer extension for monoadducts about 30% of that of the control. Klenow showed a greater reduction in its extension efficiency, as evidenced by incomplete primer extension and the presence of shorter products (Figure 3B). This reduction varied with the type of monoadduct, with the average rate of primer extension by KF relative to control following the order ECH (91%) > DEB (20%) > cisplatin (10%).
Figure 3.
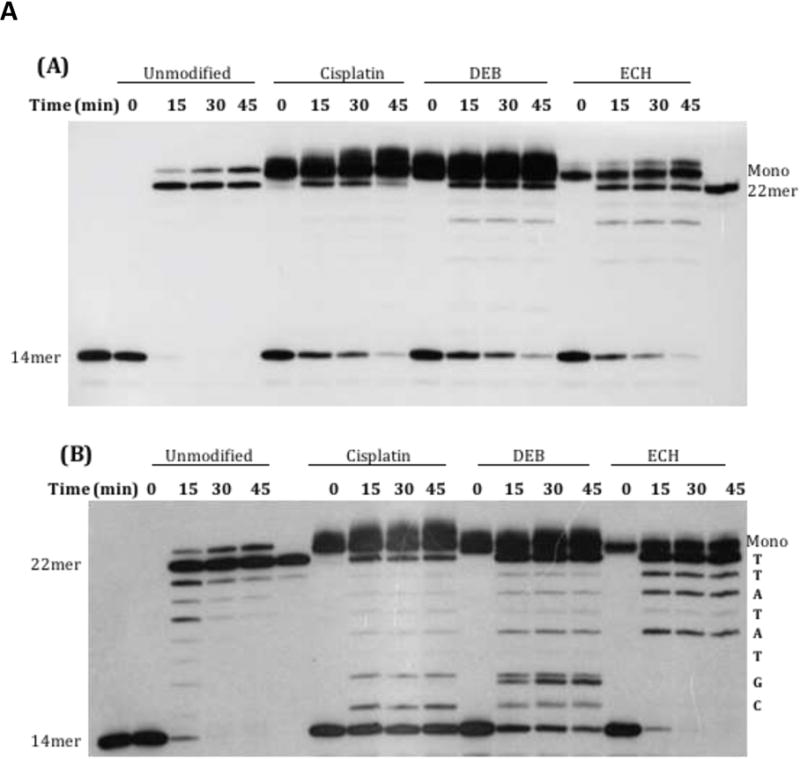
Extension of a 14mer primer annealed to Strand 1 (22 nucleotides long) by bacterial polymerases in the presence of all four dNTPs. Strand 1 was unmodified or modified with cisplatin, DEB, or ECH. Incubation was at 37°C for the indicated time points. The band in the t= 0 drug-treated lanes with lower mobility than the 22mer corresponds to monoadducted Strand 1 (“Mono”). Note that at long time points, the enzyme appears to add an extra base to the new strand. (A) Extension with BF. Control 14mer and full-length 22mer are in the first and last lane, respectively. (B) Extension with KF. Control 14mer and full-length 22mer are in lanes 1 and 6, respectively. The sequence of the newly synthesized strand is shown along the right margin.
The same experiment was repeated using recombinant hPol β (Figure 4). The general trend was similar to KF, with the average rate of primer extension relative to control following the order ECH (60%) > DEB (44%) > cisplatin (30%).
Figure 4.
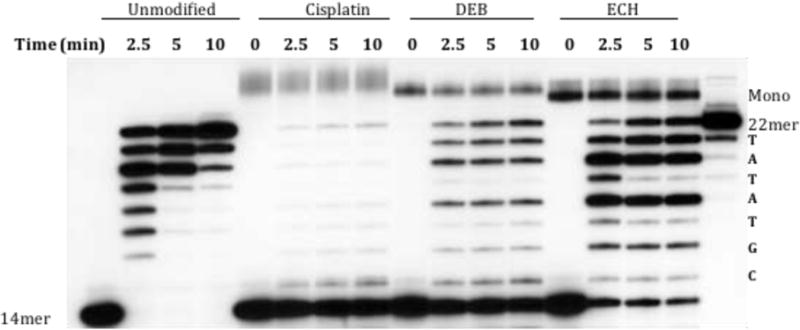
Primer (14mer) extension by hPol β. Strand 1 was unmodified or modified with cisplatin, DEB, or ECH. Incubation was at 37°C for the indicated time points. The band in the t= 0 drug-treated lanes with lower mobility than the 22mer corresponds to monoadducted Strand 1 (“Mono”). Control 14mer and full-length 22mer are in the first and last lane, respectively. The sequence of the newly synthesized strand is shown along the right margin.
3.3 Steady state kinetics with human polymerase β
Steady-state kinetics experiments with hPol β were performed for each monoadduct annealed to the 14mer primer. The concentrations of the dNTPs were varied and the rate of extension was calculated by monitoring the disappearance of primer over time. The kinetic results are summarized in Table 1, where the relative efficiency reflects the ratio of the specificity constant (kcat/Km) of the monoadduct-containing template versus the unmodified template.
Table I.
Steady-state kinetic parameters for primer extension by hPol β.
| Template | kcat (min−1) | Km (dNTP)(μM) | kcat/Km (1/min-μM) | Relative Efficiencya |
|---|---|---|---|---|
| Unmodified | 6.3 ± 1.1 | 111 ± 77 | 0.057 | 1.0 |
| Cisplatin monoadduct | 0.23 ± 0.22 | 48 ± 25 | 0.0048 | 0.084 |
| DEB monoadduct | 0.31 ± 0.16 | 42 ± 32 | 0.0074 | 0.13 |
| ECH monoadduct | 0.38 ± 0.24 | 51 ± 55 | 0.0075 | 0.13 |
The ratio of kcat/Km of the monoadduct-containing template versus the unmodified template.
3.3 Primer extension with individual dNTPs
Extension assays were also performed with all three polymerases in the presence of each dNTP individually on duplexes consisting of monoadducted strand 1 annealed to either the 14mer primer or another primer that terminated in the vicinity of the lesion (a 13mer or 15mer, see Chart 1). These studies allowed us to assess the preference for incorporating each of the four nucleotides across from the monoadduct or immediately upstream or downstream. Control reactions with unmodified template showed incorporation of the expected dNTP, with only traces of incorporation of the other dNTPs visible with long exposures under our reactions conditions, which had relatively low concentrations of dNTP and enzyme. For all three agents, each polymerase showed a strong preference (>20-fold) for inserting the correct nucleotide in the vicinity of the monoadduct (Figure 5 and Supplementary Material, Figures S2 and S3).
Figure 5.
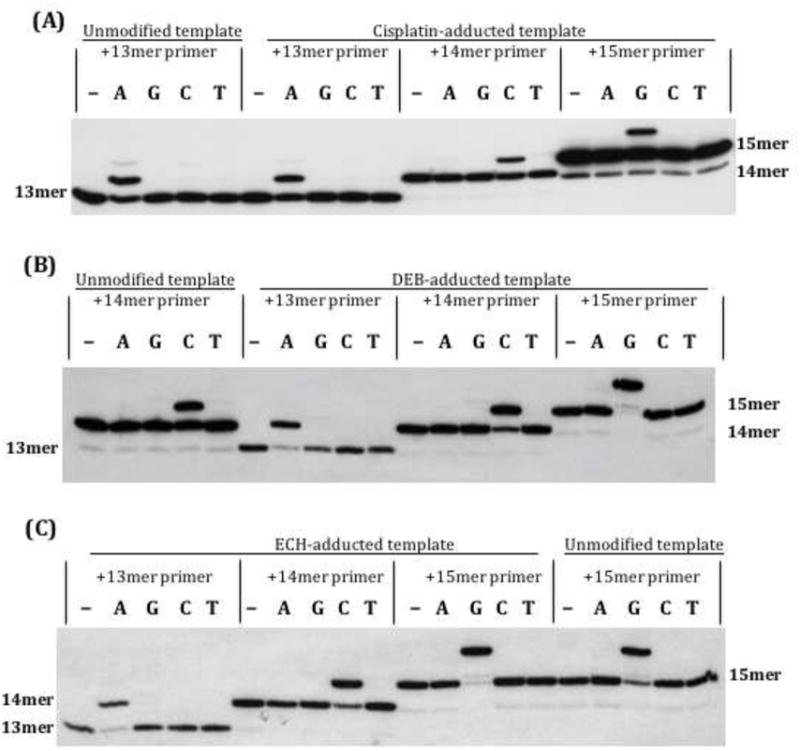
Single nucleotide addition with KF. Strand 1, either unmodified or monoadducted, was first annealed with primer. The first lane in each set is the no-dNTP control. The 13mer primer reveals nucleotide addition one nucleotide before the adduct (across from T); the 14mer reflects addition across from the adduct (at G), and the 15mer reflects addition one nucleotide after the adduct (across from C). (A) Cisplatin-adducted template; (B) DEB-adducted template; (C) ECH-adducted template.
4. Discussion
We used a polymerase bypass assay to assess potential blocking and/or mutagenic effects by the N7-guanine monoadducts formed by three DNA cross-linkers: cisplatin, diepoxybutane, and epichlorohydrin. Many DNA adducts induce structural changes that inhibit replication or make it more error-prone, contributing to such outcomes as carcinogenesis. Studies to date are consistent with increased blocking and/or mutagenicity of adducts that cause large structural changes or interfere with normal Watson-Crick base pairing. N7-guanine monoadducts are not generally considered to be mutagenic because the N7-position does not participate in Watson-Crick hydrogen bonding [36]. However, the bulky N7-guanine adduct formed by the potent liver carcinogen aflatoxin B1 (8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1) does cause mutations at the lesion site and its 5′ side [37].
Although alkylated N7-guanine adducts can be experimentally challenging to study because of their instability and tendency to undergo depurination during sample preparation, we were able to gel purify monoadducts prepared from a duplex containing a single guanine residue. Because of the power of DPAGE to resolve isomers that differ only in the position of alkylation or radiolabel [38, 39], the sharp band migrating just above the native single strands is likely to comprise a discrete monoalkylated product. Furthermore, these agents have a strong preference for reacting at N7 of guanine [40]. For example, DEB alkylates at N7 of guanine with a five-fold preference over alkylation at N3 of adenine in calf thymus DNA [21]. Therefore, it is likely that that under our single-hit reaction conditions, the major product corresponds to N7-guanine monoadducts.
We experimentally verified sites of alkylation for the DEB and ECH monoadducts with piperidine treatment, which results in strand cleavage at the 5′ side of an N7-alkylated guanine [28]. Indeed, a single major cleavage band of similar mobility to a Maxam-Gilbert G reaction resulted from piperidine cleavage of purified DEB and ECH monoadducts. As expected, the cisplatin product was not piperidine cleaved [41], but its linkage was verified through DMS protection. Small amounts of full-length DEB and ECH product remained after piperidine cleavage, which may have resulted from conversion of alkylated guanine residues to ring-opened formamidopyrimidine (FAPY) lesions. FAPY lesions do not spontaneously depurinate and are therefore resistant to piperidine-induced strand cleavage [36]. Because adducts at N3 of adenine (the major secondary reaction site) do undergo spontaneous depurination [21], it is unlikely that these monoadducts resulted from alkylation at adenine residues.
When the purified monoadducted strands were annealed to a primer that terminated one base before the lesion, the rate of DNA synthesis was inhibited relative to a control unmodified duplex. This inhibition was lowest for the epichlorohydrin monoadduct, supporting the idea that small N7-adducts are not highly blocking, such as previously reported for dimethylsulfate [12]. However, BF was able to bypass all monoadducts, leading to full-length product, at longer times (45 minutes, as compared to 15 minutes for the control). Klenow and hPol β were also able to bypass the lesions, although there was evidence of some pause sites. For example, DNA synthesis was inhibited opposite the cisplatin and DEB monoadducts (at dC) and at the next nucleotide (at dG). ECH monoadducts resulted in a strong KF pause site two bases past the lesion, as has been previously observed for this enzyme with other damaged templates (42). The observation that a significant amount of primer remained unextended for the cisplatin and DEB lesions, even at the longest times, is consistent with bulkier lesions impeding bypass by KF and hPol β. Steady state kinetics analyses for extension by hPol β suggest that the reduced efficiency of adding a base opposite the monoadducts is primarily a kcat effect.
The fidelity of DNA synthesis in the vicinity of the lesion was monitored by providing only a single dNTP in the presence of primers that monitored addition of a single nucleotide opposite the lesion or directly before or after it. All three polymerases added the correct nucleotide virtually exclusively in all cases. In some trials, trace amounts (< 5%) of product from addition of the incorrect nucleotide were visible. Because FAPY adducts are significantly more mutagenic than the N7 lesions from which they are derived [43], these trace amounts of misincorporation could have resulted from low levels of FAPY conversion for the DEB and ECH monoadducts.
Our finding that cisplatin monoadducts were somewhat blocking to the bacterial polymerases used in these studies is consistent with previous findings [44]. Other studies report that platinated monoadducts are not significantly mutagenic [14], which is also consistent with our results. Human polymerase β has also been shown to accurately bypass the bulky cisplatin 5′-GG intrastrand cross-link, although with reduced rates, suggesting a role for this enzyme in cisplatin resistance [45].
Because of the lability of N7 adducts, previous work on DEB has focused on other, minor adducts. N2-guanine [46,47] and several different N6-adenine monoadducts of butadiene metabolites [48,49] are only weakly mutagenic, although somewhat blocking to bacterial polymerases in vitro, as we found for the N7-guanine adducts. However, adducts that interfere with Watson-Crick hydrogen bonding, such as doubly alkylated N6-adenine adducts, are highly blocking and mutagenic to several DNA polymerases in vitro [49,50]. These adducts are likely to contribute to the A→T and A→C transversions induced by DEB and butadiene. Furthermore, experimentally observed GC→AT transitions may be due to cross-links, rather than monoadducts [51,52]. Our findings that epichlorohydrin monoadducts are weakly blocking and minimally, if at all, mutagenic suggest that the major N7 guanine adducts are also not significant contributors to this agent’s genotoxic effects.
In conclusion, we have found that N7-guanine monoadducts of cisplatin, diepoxybutane, and epichlorohydrin are bypassed effectively by the bacterial polymerases BF and KF and human Pol β with minimal errors, although these lesions inhibit the rate of DNA synthesis. To our knowledge, this is the first report on the bypass of purified DEB and ECH N7-monoadducts. Further work is necessary to determine the impact of these lesions on error-prone human polymerases involved in translesion DNA synthesis.
Supplementary Material
Highlights.
DNA adducts of cisplatin, diepoxybutane, and epichlorohydrin have relevance in cancer therapy and carcinogenesis.
We investigated the integrity of DNA synthesis from templates containing N7-guanine monoadducts of these agents.
Two bacterial polymerases and recombinant human polymerase β successfully bypassed the lesions, although with reduced rate of DNA synthesis.
The polymerases did not show significant misincorporation near the lesion.
The N7-guanine monoadducts, although the principal reaction products of these agents, do not appear to contribute significantly to the mutational spectra of cisplatin, DEB, and ECH.
Acknowledgments
We thank Emma Bartlett, Wasif Hussain, Phuong Le, Chuck Jones, and Darcy Ahern for technical assistance, Professor Kevin Rice (Colby College) for helpful comments, and Professor Joann Sweasy (Yale University) for the hPol β vector. Research reported in this publication was supported by the Colby College Research Grant Program and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM0103423.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: DEB, 1,2,3,4-diepoxybutane; ECH, epichlorohydrin; DPAGE, denaturing polyacrylamide gel electrophoresis; KF, DNA Polymerase I, Klenow fragment from E. coli; BF, DNA Polymerase I, large fragment from Bacillus stearothermophilus; hPol β, human polymerase beta; TE, 10 mM Tris buffer, 1 mM EDTA, pH 7.4; FAPY, formamidopyrimidine.
References
- 1.Rajski SR, Williams RM. DNA cross-linking agents as antitumor drugs. Chem Rev. 1998;98:2723–2795. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 2.Hopkins PB, Millard JT, Woo J, Weidner MF, Kirchner JJ, Sigurdsson STh, Raucher S. Sequence preferences of DNA interstrand cross-linking agents: Importance of minimal DNA structural reorganization in the cross-linking reactions of mechlorethamine, cisplatin, and mitomycin C. Tetrahedron. 1991;47:2475–2489. [Google Scholar]
- 3.Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutat Res. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- 4.Wingo PA, Cardinez CJ, Landis SH, Greenlee RT, Ries LA, Anderson RN, Thun MJ. Long-term trends in cancer mortality in the United States, 1930-1998. Cancer. 2003;97(Suppl 12):3133–3275. doi: 10.1002/cncr.11380. [DOI] [PubMed] [Google Scholar]
- 5.Travis LB, Curtis RE, Storm H, Hall P, Holowaty E, Van Leeuwen FE, Kohler BA, Pukkala E, Lynch CF, Andersson M, Bergfeldt K, Clarke EA, Wiklund T, Stoter G, Gospodarowicz M, Sturgeon J, Fraumeni JF, Jr, Boice JD., Jr Risk of second malignant neoplasms among long-term survivors of testicular cancer. J Natl Cancer Inst. 1997;89:1429–1439. doi: 10.1093/jnci/89.19.1429. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson LR, Pearson AE. The clinical use of mutagenic anticancer drugs. Mutat Res. 1996;355:1–12. doi: 10.1016/0027-5107(96)00019-x. [DOI] [PubMed] [Google Scholar]
- 7.Sanderson BJS, Shield AJ. Mutagenic damage to mammalian cells by therapeutic alkylating agents. Mutat Res. 1996;355:41–57. doi: 10.1016/0027-5107(96)00021-8. [DOI] [PubMed] [Google Scholar]
- 8.Choi DK, Helenowski I, Hijiya N. Secondary malignancies in pediatric cancer survivors: Perspectives and review of the literature. Int J Cancer. 2014;135:1764–1773. doi: 10.1002/ijc.28991. [DOI] [PubMed] [Google Scholar]
- 9.Schaapveld M, Aleman BMP, van Eggermond AM, Janus CPM, Krol ADG, van der Maazen RWM, Roesink J, Raemaekers JMM, de Boer JP, Zijlstra JM, van Imhoff GW, Petersen EJ, Poortmans PMP, Beijert M, Lybeert ML, Mulder I, Visser O, Louwman MWJ, Krul IM, Lugtenburg PJ, van Leeuwen FE. Second cancer risk up to 40 years after treatment for Hodgkin’s lymphoma. N Engl J Med. 2015;373:2499–2511. doi: 10.1056/NEJMoa1505949. [DOI] [PubMed] [Google Scholar]
- 10.Szikriszt B, Póti Á, Pipek O, Krzystanek M, Kanu N, Molnár J, Ribli D, Szeltner Z, Tusnády GE, Csabai I, Szallasi Z, Swanton C, Szüts D. A comprehensive survey of the mutagenic impact of common cancer cytotoxics. Genome Biol. 2016;17:99–115. doi: 10.1186/s13059-016-0963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ståhl O, Boyd HA, Giwercman A, Lindholm M, Jensen A, Kjær SK, Anderson H, Cavallin-Ståhl E, Rylander L. Risk of birth abnormalities in the offspring of men with a history of cancer: A cohort study using Danish and Swedish national registries. J Natl Cancer Inst. 2011;103:398–406. doi: 10.1093/jnci/djq550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 13.Yarema KJ, Lippard SJ, Essigmann JM. Mutagenic and genotoxic effects of DNA adducts formed by the anticancer drug cis-diamminedichloroplatinum(II) Nucleic Acids Res. 1995;23:4066–4072. doi: 10.1093/nar/23.20.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley LJN, Yarema KJ, Lippard SJ, Essigmann JM. Mutagenicity and genotoxicity of the major DNA adduct of the antitumor drug cis-diamminedichloroplatinum(II) Biochemistry. 1993;32:982–988. doi: 10.1021/bi00054a031. [DOI] [PubMed] [Google Scholar]
- 15.Vogel EW, Nivard MJM, Ballering LAB, Bartsch H, Barbin A, Nair J, Comendador MA, Sierra LM, Aguirrezabalaga I, Tosal L, Ehrenberg L, Fuchs RPP, Janel-Bintz R, Maenhaut-Michel G, Montesano R, Hall J, Kang H, Miele M, Thomale J, Bender K, Engelbergs J, Rajewsky MF. DNA damage and repair in mutagenesis and carcinogenesis: implications of structure-activity relationships for cross-species extrapolation. Mutat Res. 1996;113:177–218. doi: 10.1016/0027-5107(96)00032-2. [DOI] [PubMed] [Google Scholar]
- 16.Hartley JA, O’Hare CC, Baumgart J. DNA alkylation and interstrand cross-linking by treosulfan. Br J Cancer. 1999;79:264–266. doi: 10.1038/sj.bjc.6690043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bond JA, Medinsky MA. Insights into the toxicokinetics and toxicodynamics of 1,3-butadiene. Chem Biol Interact. 2001;135–136:599–614. doi: 10.1016/s0009-2797(01)00199-5. [DOI] [PubMed] [Google Scholar]
- 18.IARC. Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 71. IARC; Lyon: 1999. pp. 603–628. [PMC free article] [PubMed] [Google Scholar]
- 19.Baik M-H, Friesner RA, Lippard SJ. Theoretical study of cisplatin binding to purine bases: Why does cisplatin prefer guanine over adenine? J Am Chem Soc. 2003;125:14082–14092. doi: 10.1021/ja036960d. [DOI] [PubMed] [Google Scholar]
- 20.Tretyakova NY, Sangaiah R, Yen T-Y, Swenberg JA. Synthesis, characterization, and in vitro quantitation of N-7-guanine adducts of diepoxybutane. Chem Res Toxicol. 1997;10:779–785. doi: 10.1021/tx970004q. [DOI] [PubMed] [Google Scholar]
- 21.Tretyakova NY, Sangaiah R, Yen T-Y, Gold A, Swenberg JA. Adenine adducts with diepoxybutane: Isolation and analysis in exposed calf thymus DNA. Chem Res Toxicol. 1997;10:1171–1179. doi: 10.1021/tx9700681. [DOI] [PubMed] [Google Scholar]
- 22.Kolman A, Chovanec M, Osterman-Golkar S. Genotoxic effects of ethylene oxide, propylene oxide and epichlorohydrin in humans: update review (1990–2001) Mutat Res. 2002;512:173–194. doi: 10.1016/s1383-5742(02)00067-4. [DOI] [PubMed] [Google Scholar]
- 23.Carmical JR, Zhang M, Nechev L, Harris CM, Harris TM, Lloyd RS. Mutagenic potential of guanine N2 adducts of butadiene mono- and diolepoxide. Chem Res Toxicology. 2000;13:18–25. doi: 10.1021/tx9901332. [DOI] [PubMed] [Google Scholar]
- 24.Hamm ML, Crowley KA, Ghio M, Del Giorno L, Gustafson MA, Kindler KE, Ligon CW, Lindell MAM, McFadden EJ, Siekavizza-Robles C, Summers MR. Importance of the C2, N7, and C8 positions to the mutagenic potential of 8-oxo-2′-deoxyguanosine with two A family polymerases. Biochemistry. 2011;50:10713–10723. doi: 10.1021/bi201383c. [DOI] [PubMed] [Google Scholar]
- 25.Miller H, Grollman AP. Kinetics of DNA polymerase I (Klenow fragment exo-) activity on damaged DNA templates: effect of proximal and distance template damage on DNA synthesis. Biochemistry. 1997;36:15336–15342. doi: 10.1021/bi971927n. [DOI] [PubMed] [Google Scholar]
- 26.Frederick AM, Davis ML, Rice KP. Inhibition of human DNA polymerase β activity by the anticancer prodrug Cloretazine. Biochem Biophys Res Commun. 2009;378:419–423. doi: 10.1016/j.bbrc.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Second. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 28.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millard JT, Wilkes EE. cis- and trans-Diamminedichloroplatinum(II) interstrand cross-linking of a defined sequence nucleosomal core particle. Biochemistry. 2000;39:16046–16055. doi: 10.1021/bi0022285. [DOI] [PubMed] [Google Scholar]
- 30.Millard JT, White MM. Diepoxybutane cross-links DNA at 5′-GNC sequences. Biochemistry. 1993;32:2120–2124. doi: 10.1021/bi00059a034. [DOI] [PubMed] [Google Scholar]
- 31.Romano KP, Newman AG, Zahran RW, Millard JT. DNA interstrand cross-linking by epichlorohydrin. Chem Res Toxicol. 2007;20:832–838. doi: 10.1021/tx700066h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eastman A. Reevaluation of interaction of cis-dichloro(ethylenediamine)platinum(II) with DNA. Biochemistry. 1986;25:3912–3915. doi: 10.1021/bi00361a026. [DOI] [PubMed] [Google Scholar]
- 33.Paz MM, Sigurdsson STh, Hopkins PB. Monoalkylation of DNA by reductively activated FR66979. Bioorg Med Chem. 2000;8:173–179. doi: 10.1016/s0968-0896(99)00270-9. [DOI] [PubMed] [Google Scholar]
- 34.Williams RM, Rajski SR, Rollins SB. FR900482, a close cousin of mitomycin C that exploits mitosene-based DNA cross-linking. Chem Biol. 1997;4:127–137. doi: 10.1016/s1074-5521(97)90256-8. [DOI] [PubMed] [Google Scholar]
- 35.Fiala KAl, Brown JA, Ling H, Kshetry AK, Zhang J, Taylor J-S, Yang W, Suo Z. Mechanism of template-independent nucleotide incorporation catalyzed by a template-dependent DNA polymerase. J Mol Biol. 2007;365:590–602. doi: 10.1016/j.jmb.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boysen G, Pachkowski BF, Nakamura J, Swenberg JA. The formation and biological significance of N7-guanine adducts. Mutat Res. 2009;678:76–94. doi: 10.1016/j.mrgentox.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey EA, Iyer RS, Stone MP, Harris TM, Essigmann JM. Mutational properties of the primary aflatoxin B1-DNA adduct. Proc Natl Acad Sci USA. 1996;93:1535–1539. doi: 10.1073/pnas.93.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millard JT, Weidner MF, Kirchner JJ, Ribeiro S, Hopkins PB. Sequence preferences of DNA interstrand crosslinking agents: quantitation of interstrand crosslink locations in DNA duplex fragments containing multiple crosslinkable sites. Nucleic Acids Res. 1991;19:1885–1891. doi: 10.1093/nar/19.8.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romero RM, Rojsittisak P, Haworth IS. Electrophoretic mobility of duplex DNA cross-linked by mechlorethamine at a cytosine-cytosine mismatch pair. Electrophoresis. 2013;34:917–924. doi: 10.1002/elps.201200543. [DOI] [PubMed] [Google Scholar]
- 40.Lawley PD, Brookes P. Interstrand cross-linking of DNA by difunctional alkylating agents. J Mol Biol. 1967;25:143–160. doi: 10.1016/0022-2836(67)90285-9. [DOI] [PubMed] [Google Scholar]
- 41.Johnson NP, Macquet JP, Wiebers JL, Monsarrat B. Structures of the adducts formed between [Pt(dien)Cl]Cl and DNA in vitro. Nucleic Acids Res. 1982;10:5255–5271. doi: 10.1093/nar/10.17.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller J, Grollman AP. Kinetics of DNA polymerase I (Klenow fragment exo-) activity on damaged DNA templates: effect of proximal and distal template damage on DNA synthesis. Biochemistry. 1997;36:15336–15342. doi: 10.1021/bi971927n. [DOI] [PubMed] [Google Scholar]
- 43.Smela ME, Hamm ML, Henderson PT, Harris CM, Harris TM, Essigmann JM. The aflatoxin B1 formamidopyrimidine adduct plays a major role in causing the types of mutations observed in human hepatocellular carcinoma. Proc Natl Acad Sci USA. 2002;99:6655–6660. doi: 10.1073/pnas.102167699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaller W, Reisner H, Holler E. Kinetic investigation of the DNA platination reaction: Evidence for a transient adduct between deoxyribonucleic acid and cis-platinum(II) Biochemistry. 1987;26:943–950. doi: 10.1021/bi00377a039. [DOI] [PubMed] [Google Scholar]
- 45.Koag M-C, Lai L, Lee S. Structural basis for the inefficient nucleotide incorporation opposite cisplatin-DNA lesion by human DNA polymerase β. J Biol Chem. 2014;289:31341–31348. doi: 10.1074/jbc.M114.605451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minko IG, Washington MT, Prakash L, Prakash S, Lloyd RS. Translesion DNA synthesis by yeast DNA polymerase η on templates containing N2-guanine adducts of 1,3-butadiene metabolites. J Biol Chem. 2001;276:2517–2522. doi: 10.1074/jbc.M007867200. [DOI] [PubMed] [Google Scholar]
- 47.Carmical JR, Zhang M, Nechev L, Harris CM, Harris TM, Lloyd RS. Mutagenic potential of guanine N2 adducts of butadiene mono- and diolepoxide. Chem Res Toxicology. 2000;13:18–25. doi: 10.1021/tx9901332. [DOI] [PubMed] [Google Scholar]
- 48.Carmical JR, Nechev LV, Harris CM, Harris TM, Lloyd RS. Mutagenic potential of adenine N6 adducts of monoepoxide and diolepoxide derivatives of butadiene. Environ Mol Mutagen. 2000;35:48–56. [PubMed] [Google Scholar]
- 49.Kotapati S, Wickramaratne S, Esades A, Boldry EJ, Dorr DQ, Pence MG, Guengerich FP, Tretyakova N. Polymerase bypass of N6-deoxyadenosine adducts derived from epoxide metabolites of 1,3-butadiene. Chem Res Toxicol. 2015;28:1496–1507. doi: 10.1021/acs.chemrestox.5b00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kotapati S, Maddukuri L, Wickramaratne S, Seneviratne U, Goggin M, Pence MG, Villalta P, Guengerich FP, Marnett L, Tretyakova N. Translesion synthesis across 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2′-deoxyadenosine (1,N6-γ-HMHP-dA) adducts by human and archebacterial DNA polymerases. J Biol Chem. 2012;287:38800–38811. doi: 10.1074/jbc.M112.396788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carmical JR, Kowalczyk A, Zou Y, Van Houten B, Nechev LV, Harris CM, Harris TM, Lloyd RS. Butadiene-induced intrastrand DNA cross-links: A possible role in deletion mutagenesis. J Biol Chem. 2000;275:19482–19489. doi: 10.1074/jbc.M002037200. [DOI] [PubMed] [Google Scholar]
- 52.Klug AR, Harbut MB, Lloyd RS, Minko IG. Replication bypass of N2-deoxyguanosine interstrand cross-links by human DNA polymerases η and ι. Chem Res Toxicol. 2012;25:755–762. doi: 10.1021/tx300011w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.