Abstract
Saccharomyces cerevisiae mitoribosomes are specialized in the translation of a few number of highly hydrophobic membrane proteins, components of the oxidative phosphorylation system. Mitochondrial characteristics, such as the membrane system and its redox state driven mitoribosomes evolution through great diversion from their bacterial and cytosolic counterparts. Therefore, mitoribosome presents a considerable number of mitochondrial-specific proteins, as well as new protein extensions. In this work we characterize temperature sensitive mutants of the subunit bL34 present in the 54S large subunit. Although bL34 has bacterial homologs, in yeast it has a long 65 amino acids mitochondrial N-terminal addressing sequence, here we demonstrate that it can be replaced by the mitochondrial addressing sequence of Neurospora crassa ATP9 gene. The bL34 temperature sensitive mutants present lowered translation of mitochondrial COX1 and COX3, which resulted in reduced cytochrome c oxidase activity and respiratory growth deficiency. The sedimentation properties of bL34 in sucrose gradients suggest that similarly to its bacterial homolog, bL34 is also a later participant in the process of mitoribosome biogenesis.
Keywords: cytochrome c oxidase, mitoribosome, mtDNA, respiratory chain, translation, yeast
Introduction
Saccharomyces cerevisiae mitoribosome is a 74S ribonucleoprotein complex essential for organellar protein synthesis. It is composed of a 54S large subunit (mtLSU) and a 37S small subunit (mtSSU), a total of 73 mitoribosome proteins (MRPs) and two ribosomal RNAs (rRNAs (Desai et al., 2017). The biogenesis of mitochondrial ribosomes depends on two genomes, with the rRNAs and the 37S protein Var1 encoded by the mitochondrial DNA and all other MRPs encoded in the nuclear genome (Terpstra et al., 1979; Tzagoloff and Myers, 1986). Despite their endosymbiotic origin, yeast mitoribosomes have diverged greatly from their bacterial and cytosolic counterparts by acquiring new protein and RNA segments (Mears et al., 2006; Amunts et al., 2014; Desai et al., 2017). At any rate, biogenesis of mitoribosome remains poorly understood. Ribosome assembly involves the transcription, processing, and modification of rRNA; the translation and modification of ribosomal proteins; the proper folding of rRNA and ribosomal proteins; the binding of ribosomal proteins; and the binding and release of assembly factors (Shajani et al., 2011). To date, in addition to rRNA modification enzymes, only a few factors have been identified in yeast for mitoribosomes assembly. They include three GTPases and two DEAD-box helicases. MTG1 codes for a member of the YIqF GTPase family (Barrientos et al., 2003); MTG2 is a suppressor of rRNA methyltransferase mutant, the product of this gene is a member of the Obg GTPase family that binds to the large ribosomal subunit. (Datta et al., 2005); and MTG3 is a conserved member of the YqeH family of GTPases that jointly with uL29, a component of the 54S subunit, functions in regulating assembly of the 37S subunit by modulating the processing of its 15S rRNA precursor (Paul et al., 2012). MRH4 codes for a DEAD Box helicase with an essential role during the late stages of mitoribosome assembly by probably promoting remodeling of the 21S rRNA-protein interactions (De Silva et al., 2013). Finally, Mss116 is another DEAD-box helicase required to COX1 mRNA splicing and translation, that is also required for an undefined early step during 54S biogenesis (De Silva et al., 2017).
The large 54S subunit catalyzes peptide bond formation during protein synthesis and it has a tunnel exit for the growing nascent polypeptide chain. This exit has a strong functional specialization related to the synthesis of the highly hydrophobic mitochondrial inner membrane proteins (Greber et al., 2014). As part of an effort to better understand mitochondrial translation and the mitoribosome biogenesis, here we characterize bL34 temperature sensitive mutants encoded in yeast by MRPL34 (ORF YDR115w). Mitoribosome bL34 associates with a specific mitoribosome protein mL41, involved in the stabilization of rRNA 16S helix 8-ES1, and uL29 present in the mitoribosome exit tunnel site (Brown et al., 2014; Desai et al., 2017). bL34 is also present in bacteria (L34) and it is considered a late participant in the well-studied process of bacterial 50S biogenesis (Shajani et al., 2011). Bacterial mutants with impaired biogenesis of subunit 50S did not assemble L34 protein into intermediates, suggesting a loose association of L34 as well as its late association in 50S biogenesis (Charollais et al., 2003, 2004; Akanuma et al., 2014). In contrast to the bacterial counterpart, we do not have much information about the order of events occurring during the biogenesis of the mitoribosome. The study of mrpl34 temperature sensitive mutants unveiled some important aspects of the mitoribosome biogenesis. First, the mrpl34-ts mutants present reduced translation of Cox1p and Cox3p two hydrophobic protein encoded by the mitochondrial genome, and important constituents of cytochrome c oxidase. This result suggests that the mitoribosome exit tunnel, which is adapted for the exit of highly hydrophobic protein, is compromised in the mrpl34-ts mutants. Second, we have observed that mrpl34-ts mutants are able to accumulate fully assembled 54S subunit, but putative assembly intermediates are also detected in our sedimentation assays suggesting that likewise its bacterial counterpart bL34, it is a late peripheral participant in the biogenesis of mitochondrial ribosome. Therefore, in this work we characterize bL34-ts mutants showing deficient translation of cytochrome c oxidase subunits and accumulation of mitoribosome assembly intermediates.
Material and methods
Yeast strains and growth media
The genotypes and sources of yeast strains used in this study are listed in Table 1. Yeast strains were maintained in YPD (1% yeast extract, 2% peptone, 2% glucose), YPGal (1% yeast extract, 2% peptone, 2% galactose), YPEG (1% yeast extract, 2% peptone, 2% glycerol, 2% ethanol), minimal glucose (2% glucose, 0.67% yeast nitrogen base without amino acids, supplemented with auxotrophic requirements), and sporulation media (0.5% yeast extract, 1% potassium acetate, 0.05% glucose).
Table 1.
Genotypes and sources of yeast strains
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | a |
| W303-1B | α ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | a |
| W303/I0 | a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 mitI0 | Barros et al. (2006) |
| W303 ΔMRPL34 | a/α ade2-1ade2-1 his3-1,5 his2-1,5 leu2-3,112 leu2-3,112 trp1-1 trp1-1 ura3-1 ura3-1 MRPL34 mrpl34::HIS3 | This study |
| W303ΔMRPL34/TS1/I0 | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 mrpl34-ts1 mitI0 | This study |
| W303 ΔMRPL34/TS1 | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 mrpl34-ts1 | This study |
| W303 ΔMRPL34/TS2 | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 mrpl34-ts2 | This study |
| W303 ΔMRPL34/TS3 | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 mrpl34-ts3 | This study |
| W303 ΔMRPL34/F79A | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 MRPL34-F79A | This study |
| W303 ΔMRPL34/W101R | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 MRPL34/W101R | This study |
| W303 ΔMRPL344/65 | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 mrpl34-T65 | This study |
| W303 ΔMRPL34/65HA | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 mrpl34-T65HA | This study |
| W303 ΔMRPL34/TS65HA | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 mrpl34-tsT65HA | This study |
| W303 ΔMRPL34/55 | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 mrpl34-G55 | This study |
| W303 ΔMRPL34/61 | α ade2-1 his3-1,5 leu2-3,112 trp1-1 ura3-1 mrpl34::HIS3 mrpl34-S61 | This study |
Dr. Rodney Rothstein, Department of Genetics and Development, Columbia University.
Cloning and disruption of MRPL34
The pair of primers 496 5′-cacccgggtacacgagttttttgacg-3′ and 497 5′-gggagctcgtcggcaaatgctacttc-3′ were used to amplify MRPL34 from total yeast nuclear DNA. The 1,100 bp fragment containing the gene flanked by 400 and 390 nucleotides of 5′ and 3′ untranslated sequences, respectively, was double digested with SmaI and SacI transferred to pUC19 and YEp351 (Hill et al., 1986) resulting, respectively, in the recombinant plasmids pDR115/1 and pDR115/2. The resultant plasmid pDR115/1 was used as a template to delete the MRPL34 reading frame with the bidirectional primers 5′-ggcagatc-tagagcagagaaagaagat-3′ and 5′-ggcagatctggtggtttttgtctcatt-3′. The gapped product was digested with BglII and ligated to a 1 kb BamHI fragment containing the yeast HIS3 gene. The MRPL34::HIS3 allele was isolated from this plasmid (pDR115/3) as a linear 1.8 kb BamHI fragment and used to substitute by homologous recombination (Rothstein, 1983) the wild-type gene in the respiratory competent diploid strains a/aW303. After sporulation and tetrad dissection respiratory deficient and histidine-independent transformants were confirmed to have the null allele by genetic crosses to a mutant with a deletion in YDR115w, obtained from the Genome Deletion strain collection and by specific diagnostic by PCR amplification.
Construction of a temperature-sensitive allele of mrpl34 and site-directed mutagenesis
MRPL34 was mutagenized by PCR amplification of the gene under conditions causing misincorporation of deoxynucleotides (Paul et al., 2012). The gene was amplified from pDR115/1 with primers 496 and 497 described above in four separate reactions containing 0.03 mM MnCl2, 1.25 mM MgCl2, and 0.2 mM dNTP but having a three-fold lower concentration of one of the four deoxynucleotides. The products of the four reactions were pooled and double digested with SmaI and SacI and cloned into YCplac22, a centromeric plasmid containing the TRP1 marker (Gietz and Sugino, 1988). The resultant library was used to transform the heterozygous diploid a/αW303ΔMRPL34. Tryptophan prototrophic transformants were sporulated in potassium acetate media and after 2 or 3 days the spores obtained were spread on minimal selective media without histidine and tryptophan supplementation and replicated on rich ethanol/glycerol media (YPEG), and incubated either at 30° C or 37° C. The ts mutants mrpl34/ts1 and mrpl34/ts2 were selected based on their clear ts growth phenotype on YEPG.
MRPL34W101R, MRPL34F79A, and MRPL34R95W alleles were generated by PCR amplification of the gene in pDR115/1 in two separated reactions. For MRPL34W101 the first fragment of 714 bp was amplified with oligonucleotides 5′-ggcttaagccattgaaatttcacatac-3′ and primer 497, which was cut with AflII and SacI. In the second reaction, oligonucleotides 5′-gggcttaagaaccgcctacctttgagcttcc-3′ and 5′-ggcctgcagggtcatttgaagctcggatc-3′ amplified a product of 504 bp, which was cut with AflII and PstI. The fragment used for the synthesis of MRPL34F79A was amplified with 5′-ggcgctagctaggccaagagcaaacagggc-3′ and primer 497 generating a product of 714 bp that was cut with AflII and SacI. The second 354 bp fragment was generated with oligonucleotides 5′-ggcgctagcgcaccaaaagttctcttccgtttc-3′ and 5′-ggcctgcagggtcatttgaagctcggatc-3′, and subsequently cut with NheI and PstI. Finally, for the MRPL34R95W mutant primers pairs 5′-ggccttaagcgatggaagctcaaaggtaggtgg-3′ and primer 497, and 5′-ggccttaagtatcttagagccctgtttgctcttgg-3′ and 5′-ggcctgcagggtcatttgaagctcggatc-3′, respectively, yielded a fragment of 415 bp, which was cut with SacI and AflII and a second 477 bp product cut with AflII and PstI. The two digested fragments for each cloning were combined and ligated to YIp351 (Hill et al., 1986) previously digested with SacI and PstI. Each construction was sequenced and the corresponding recombinant plasmids linearized with BstXI site inside the LEU2 gene of YIp351 and recombined at the LEU2 chromosomal locus of the heterozygous diploid a/a W303ΔMRPL34. The diploids transformants were sporulated and single haploid cells obtained by tetrads dissection.
Construction of truncated variants of Mrpl34p as well as tagged versions
N-terminal truncated variants of Mrpl34p were constructed through the fusion of MRPL34 with the mitochondrial signal sequence of Neurospora crassa ATP9 obtained in the plasmid pATP8/ST4 (Barros et al., 2011), which was previously cloned underthe control of GPD promoter in YCp22 (Zampol et al., 2010). The MRPL34-G55 truncated allele was obtained through PCR of pDR115/1 with oligonucleotides 5′-gggggatccggtcaaagaagatggaatc-3′ and 5′-ccgctgcaggcgaatcagttttgtcttg-3′, and yielded a fragment of 373 bp. MRPL34-S61 truncated allele was amplified with oligonucleotides 5′-gcggatcctcaaggggtaacacctatc-3′ and 5′-ccgctgcaggcgaatcagttttgtcttg-3′ resulting in a 353 bp product. Finally, MRPL34-T65 was amplified with oligonucleotides 5′-ccgggatccacctatcaacctagtacattg-3′ and 5′-ccgctgcaggcgaatcagttttgtcttg-5′ resulting in a 342 bp product. All PCR products were digested with BamHI and PstI and inserted into the YCp22/GPD/NcATP9 (pNATP9-22), recombinant plasmid previously digested with the same enzymes.
An N-terminal hemagglutinin A (HA) tagged version of MRPL34 was obtained through PCR of the wild-type MRPL34, mrpl34-ts1, and mrpl34/ts2 alleles with oligonucleotides 5′-ggcggatcctacccatacgacgtcccagactacgctacctatcaacctagtacattg-3′ and 5′-ccgctgcaggcgaatcagttttgtcttg-3′ resulting in a 369 bp product. The products from each template were digested with BamHI and PstI and inserted into YCp22/GPD/NcATP9.
Construction of a plasmid expressing a trpE/Mrpl34 fusion protein for antibody production
The MRPL34 coding sequence was PCR amplified with primers 5′-ccgggatccatgccactatttgcaagg-3′ and 5′-ggcctgcagatgagacaaaaccacctacc-5′ from pDR115/1, digested with a combination of BamHI and PstI and cloned into pATH11 (Koerner et al., 1991). E. coli RR1 transformed with this construct expressed a 44 kDa fusion protein as an inclusion body. The insoluble protein fraction, consisting mainly of the fusion protein, was solubilized with in 1% sodium dodecyl sulfate, size-fractionated on SDS–PAGE and used to immunize rabbits (VBP Biotecnologia LTDA, Campinas/Brazil).
Mitochondrial protein synthesis
Mitochondrial gene products were labeled in whole cells for 10 min with [35S]-methionine-cysteine mixture (7 mCi/mmol) in the presence of cycloheximide. The conditions for extraction and separation the radiolabeled proteins on a 17.5% polyacrylamide-SDS and on a 12% polyacrylamide-6M urea gel was described previously (Barros et al., 2011; Moda et al., 2016). Radiolabeled proteins were transferred to a nitrocellulose membrane and visualized by exposure to an X-ray film. Bands intensities in each film were quantified using the software BioRaD Image Lab 6.0.
Mitochondria isolation, ribosome extraction and fractionation
Yeast mitochondria were prepared by the method of Faye et al. (1974) (25) except that Zymolyase 20T, instead of Glusulase, was used to obtain spheroplasts. Mitoribosomes were extracted by addition to the mitochondrial suspension at a protein concentration of 20 mg/mL of an equal volume of 2× AMT (1M ammonium acetate, 20 mM Tris-Cl, pH 7.5, 20 mM MgCl2 and 12 mM 2-mercaptoethanol) and 0.1 volume of 10% potassium deoxycholate pH 8. The partially solubilized mixture was centrifuged at 16,000g for 10 min and the supernatant was layered over 4.5 mL of 50% sucrose in 1xAMT. Ribosomal subunits were pelleted by centrifugation at 229,000g for 16 h. They were suspended in 0.2 mL of AMT and layered on top of 4 mL of 10–30% linear sucrose gradient in AMT buffer. The gradients were centrifuged at 370,000g for 1.5 h at 4° C in a Beckmann SW55Ti rotor and were fractionated into 14 equal fractions (Paul et al., 2012). Four milligrams of mitochondrial preparation were also solubilized in 400 mL of an extraction buffer (20 mM Hepes pH 7.4, 25 mM KCl, 0.5 mM PMSF, 0.8% Triton X100, 5 mM EDTA or 0.5 mM MgCl2) which was centrifuged at 27,000g for 15 min and applied onto a linear sucrose gradient 0.3M–1.0M, with the same constitution of the correspondent extraction buffer (De Silva et al., 2013).
General procedures
Standard methods were used for plasmid manipulations and transformation of yeast (Schiestl and Gietz, 1989). Protein concentrations were determined by the method of Lowry et al. (1951). The buffer system of Laemmli (1970) was used for SDS-PAGE electrophoresis with the modifications indicated in figure legends. Spectral analyzes of mitochondrial cytochromes and measurements of respiratory enzymes were performed as described previously (Tzagoloff et al., 1975).
Results
mrpl34 mutants present growth impairment on selective media for respiratory activity
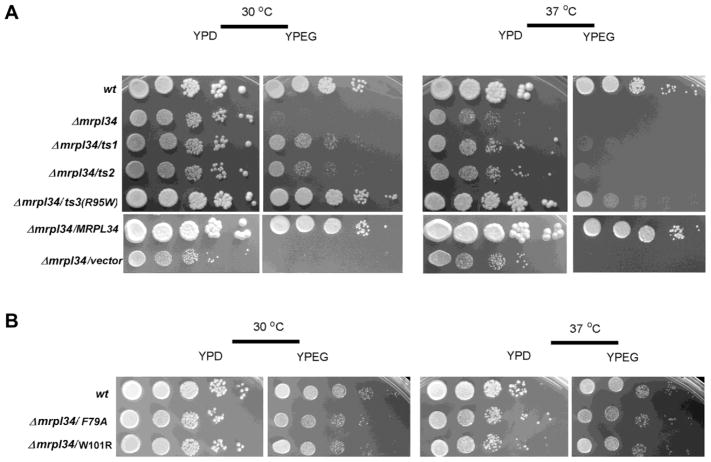
As expected for mitoribosome components, the deletion of mrpl34 resulted in respiratory deficiency as observed by the absence of growth in media containing non-fermentable carbon sources (ethanol/glycerol—Figure 1A). As well documented, the impairment of mitochondrial translation machinery leads to partial or complete deletions in mitochondrial DNA (Myers et al., 1985). This property makes it difficult to gather better details of the direct defect resulted by the null mutant. This problem has been circumvented by the generation of temperature sensitive (ts) alleles, which present a more stable mitochondrial genome (Barros et al., 2011; Paul et al., 2012; Barros and Tzagoloff, 2017). A plasmid library with mutations in mrpl34 was obtained by PCR mutagenesis of MRPL34 under low-stringency conditions. This library was used to transform a heterozygous mrpl34 null mutant and the resultant diploid transformants were sporulated and haploid progeny tested for temperature sensitive growth in non-fermentable carbon sources. Two independent mutants were obtained from this screening: mrpl34/ts1 has two amino acid changes at positions K90R and R95W and mrpl34/ts2 with three changes R58G, K90R, and R95S. The non-conservative changes at position R95 in the two mutants prompt us to investigate whether this was a critical residue for thermal stability in bL34. Through site direct mutagenesis, a R95W single mutant was generated and the resultant allele named mrpl34/ts3. Both ts1, ts2 mutants shown slow growth on non-fermentable carbon sources (YPEG) at the permissive temperature (30° C) and no growth at non-permissive temperature (37° C) after 48 h of incubation while ts3 showed a growth similar to the wild-type at 30° C and slow growth at 37° C (not shown). Nevertheless, after another 24 h-period, totalizing 3 days of growth the ts1 and ts2 growth on YPEG at 30° C were clearly detected while at 37° C their growth was still hampered. The longer incubation of ts3, on the other hand, resulted in medium-sized yeast colonies compared to the wild-type individual colonies observed in the more diluted spots (Figure 1A). Altogether the growth properties of these ts mutants indicate that R95 is important for thermal stability of bL34, but the mutation R95W slow growth in respiratory selective media rather than impeding it. The combination of R95W mutation together with K90R present in the ts1 and ts2 alleles impacts negatively on the respiratory capacity of these mutants, but it did not prevent growth in YPEG at the permissive temperature as observed by comparing with the null mutant spots (Figure 1A).
Figure 1. Growth properties of mrpl34/ts mutants.
(A) Comparative growth of wild-type cells (wt) and mrpl34 null mutant alone (Δmrpl34), or harboring plasmids expressing different mrpl34 mutants: ts alleles ts1, ts2 and ts3 (Δmrpl34/ts1, Δmrpl34/ts2, Δmrpl34/ts3(R95W) and the point mutants F79A (Δmrpl34/F79A) W101R (Δmrpl34/W101R) as well as an empty plasmid (Δmrpl34/vector) and a plasmid with the wild-type MRPL34 gene (Δmrpl34/MRPL34). Cultured cells were serially diluted and spotted in rich glucose media (YPD) and rich ethanol-glycerol media (YPEG) plates. Plates were incubated at 30° C and 37° C as indicated and photographed after three days of incubation. (B) Comparative growth of the point mutants F79A (Δmrpl34/F79A) and W101R (Δmrpl34/W101R) was also performed as in “A”.
Analyses of bL34 family sequences indicate that R95 is highly conserved (present in 99% of 367 analyzed sequences. Another very conserved residue is F79 (96.2%) that was also subjected to site-directed mutagenesis as well as W101, which has 57% of conservation and has probable interactions with uL23 and uL24 (Amunts et al., 2014). Likewise R95, F79, and W101 were mutated to W101R and F79A; however, differently from R95W, none of these mutants resulted in any detectable growth impairment in selective media for respiratory activity (Figure 1B).
Translation of newly synthesized mitochondrial products and enzymatic assays of mrpl34-ts mutants
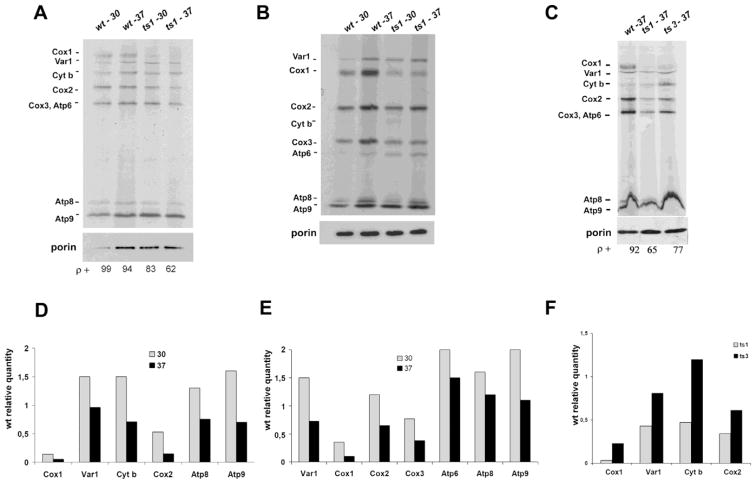
Curiously, depletion of bL34 product in mrpl34-ts1 mutants at the non-permissive temperature has not fully impaired the translation of newly synthesized mitochondrial products. This result suggests that even with the thermal sensitive detected in bL34 ts variants, mitoribosome is assembled allowing normal mitochondrial translation (Figure 2) of the mitochondrial products except for Cox1p, whose translation was more evidently lowered in the ts1 cells at both assayed temperatures and in ts3 at the restrictive temperature (Figure 2C).
Figure 2. Translation of newly synthesized mitochondrial products.
Newly synthesized mitochondrial translation products were analyzed from wild-type (wt) cells, Δmrpl34/ts1 (ts1) and Δmrpl34/ts3 (ts3). Cells were grown and assayed at permissive (30° C) and restrictive temperature (37° C) as indicated and described in the Material and Methods section. The extracts of proteins from each culture were separated in two gel systems: (A) 17.5% acrylamide-SDS, (B) 12% acrylamide-6M urea. (C) 17.5% Acrylamide-SDS for ts1 and ts3 comparison. The radiolabeled bands corresponding to the mitochondrial gene products are marked in the margins as indicated: subunits 1 (Cox1), subunit 2 (Cox2), subunit 3 (Cox3) of cytochrome c oxidase; subunit 6 (Atp6), subunit 8 (Atp8) and subunit 9 (Atp9) of ATP synthase; cytochrome b subunit (Cyt b) of ubiquinol cytochrome c reductase; the ribosome Var1 protein. The membranes were also imunodecorated with anti-Porin monoclonal antibody (Invitrogen) as a loading control. The percentages of cells containing fully functional mitochondrial DNA (ρ+) at the end of the experiment are indicated at the bottom. (D) Relative quantification of ts1 products presents in “A” in reference to the respective product present in the wild-type cells at the same growth in the assayed condition (30° C and 37° C). (E) Relative quantification of ts1 products presents in “B” in reference to the respective product present in the wild-type cells at the same growth in the assayed condition (30° C and 37° C). (F) Relative quantification of ts1 and ts3 products presents in “C” in reference to the respective product present in the wild-type assayed at 37° C.
The labeling experiments and gel analyzes were performed at least twice for each assayed condition. Bands intensities in each film were evaluated using software BioRaD Image Lab 6.0. In each measurement the wild-type band was selected as the reference band, and directly compared to the correspondent ts product in order to obtain the relative quantity of the newly synthesized product. Thus, the ts1 products separated in the 17.5% acrylamide-SDS gel (Figure 2A) and in the 12% acrylamide 6M Urea gel (Figure 2B) show a relative variation from 0.5 up to 1.8 times of the wild-type products in the experiments conducted at 30° C. Curiously, Var1p and the Fo-ATPase subunits have shown elevated translation in the ts1 mutant in comparison to the wild-type. In comparison to other products translated at 30° C, only Cox1p intensity showed less than one-third of the wild-type product in both assays (Figures 2D and 2E). Indeed, at the non-permissive temperature Cox1p synthesis was more dramatically reduced to values lower than 0.1 for the ts1 mutant and equal to 0.23 for ts3, which was the lowest relative quantification among the observed ts3 products (Figures 2C and 2F). Nevertheless, in the ts1 cells grown at 37° C Cox3p translation was also reduced to values lower than half of the wild-type reference level. Curiously, Cox1p, Cox3p, and Cob1p, in this order, are the three most hydrophobic mitochondrion-encoded products.
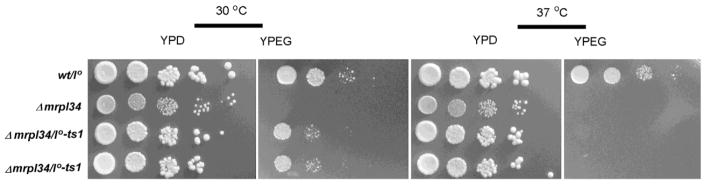
W303-1A and W303-1B strains are known to harbor a considerable number of mitochondrial introns in the COX1 gene. Thus, a possible role of bL34 in COX1 intron processing was assessed through the cross of the wild-type intronless strain aW303/I00 (Barros et al., 2006) with W303ΔMRPL34/TS1 I0 derivative strain, in order to obtain a bl34 null mutant and ts1 mutant in an intronless mitochondria background. The new mutant derivatives, did not present any significant growth difference in comparison with the strains harboring the original W303 mitochondrial DNA background, showing growth defect in respiratory selective media at the non-permissive temperature (Figure 3).
Figure 3. Growth properties of blL34 ts1 mutant in a intronless mitochondrial DNA background.
Growth of the wild-type strain W303/I0 (wt/I0) devoid of mitochondrial introns were compared to mrpl34 null mutant (Δmrpl34) and two independent ts1 mutants also in an mtDNA intronless background. Cultured cells were serially diluted and spotted in rich glucose media (YPD) and rich ethanol-glycerol media (YPEG) plates. Plates were incubated at 30° C and 37° C as indicated and photographed after three days of incubation.
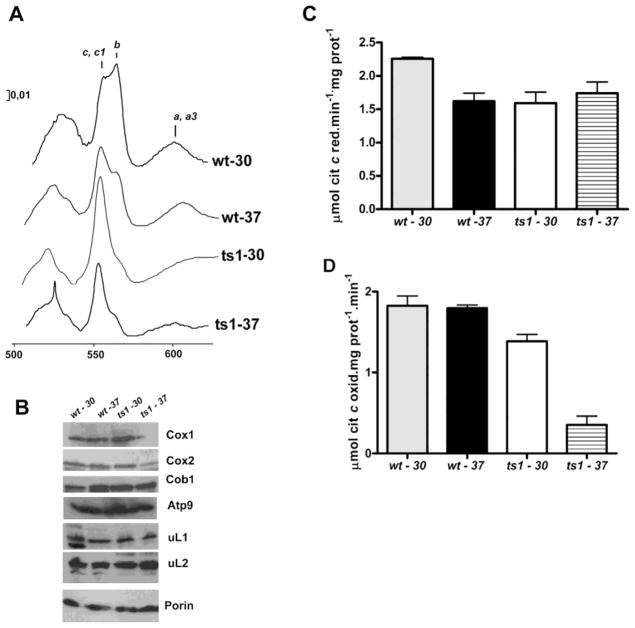
Therefore, the mrpl34/ts1 mutant respiratory deficiency at the non-permissive temperature came from the combined poor translation of Cox1p and Cox3p. Corroborating this hypothesis, the spectrum of mitochondrial cytochromes in the ts1 mutants indicated the presence of a very low level of an “a” type cytochrome (Figure 4A), the cytochrome c oxidase activity was lower in the ts1 mitochondria isolated from cells grown at the restrictive temperature (Figure 4D) and finally we also observed low steady state levels of mitochondrially encoded COX subunits (Cox1p and Cox2p) in the mitochondria isolated from ts1 cells grown at 37° C (Figure 4B). In fact, biogenesis of cytochrome c oxidase was demonstrated to be susceptible to high temperatures (37° C) in cox26 mutants (Strecker et al., 2016). The steady state level of mitoribosomes proteins (uL1, uL2, uL24), Cob1p, and porin did not show significant variation in the tested samples. However, the lower content of “b” type cytochrome in ts1 mitochondria (Figure 4A) is not consistent with the observed steady-state level of cytochrome b protein (Figure 4B) or the NADH cytochrome c reductase activity, which did not show any considerable changes among the tested mitochondria (Figure 4B).
Figure 4. Respiratory properties of bL34 ts1 mutant.
(A) Mitochondria were isolated from the respiratory competent parental strain W303-1B (wt) and from the Δmrpl34/ts1 mutant (ts1) grown in rich 2% galactose media at 30° C and 37° C as indicated. (YPGal). The mitochondria were extracted at a protein concentration of 5 mg/ml in the presence of 1% potassium deoxycholate and 1 M KCl. The spectra of the extracts were obtained at room temperature following oxidation of the sample in the reference cuvette with potassium ferricyanide and reduction of the sample cuvette with sodium dithionite. The α absorption bands corresponding to cytochromes c, c1, b, a, and a3 are indicated. (B) Mitochondrial proteins from wild-type (wt) and Δmrpl34/ts1 (ts1) were also separated by SDS-PAGE, transferred to nitrocellulose, and were reacted with antibodies against the respective antibodies indicated on the right side of the panel. The blots were then treated either with peroxidase-conjugated anti-mouse, or anti-rabbit IgG. The proteins were visualized based on luminol-H2O2 peroxidase chemiluminescence system and intensified with r-coumaric acid. (C) NADH cytochrome c reductase activity in mitochondria isolated as in “A” (D)′ Cytochrome c oxidase activity; the bars in “B” and “C” represent the specific activity obtained in four different measurements from two different mitochondrial preparations.
Another indirect indication that in the ts1 mutants the respiratory growth deficiency at non-permissive temperature (Figure 1A) arose from a combined translational impairment of Cox1p and Cox3p rather than all mitochondrial products comes from the analyses of petite generation in these cells. The number of petites formed in the mrpl34-ts mutant after an overnight growth culture at restrictive temperature (35% for ts1 and 23% for ts3) is much lower in comparison to the mrpl34 null mutant (close to 100%) or other mutants with completely impaired mitochondrial translation (Myers et al., 1985). Even after 48 h of culture at non-permissive temperature all tested mrpl34-ts mutants had considerable amounts of ρ+ cells (60%). Therefore, mitochondrial translation is occurring in these ts mutants at a sufficient rate that allows mtDNA preservation for the translation of Atp6p, Atp8p, and Atp9p that are essential to maintain a minimal of coupled ATP synthase activity necessary for mitochondrial membrane potential maintenance. In the petite obligate hypothesis aberrant expression of the ATPase subunits leads to biased assembly and formation of a defective complex in the inner mitochondrial membrane, which is probably lethal to the cell (Ackerman and Tzagoloff, 2005; Lipinski et al., 2010).
Truncated versions of bL34 and localization studies
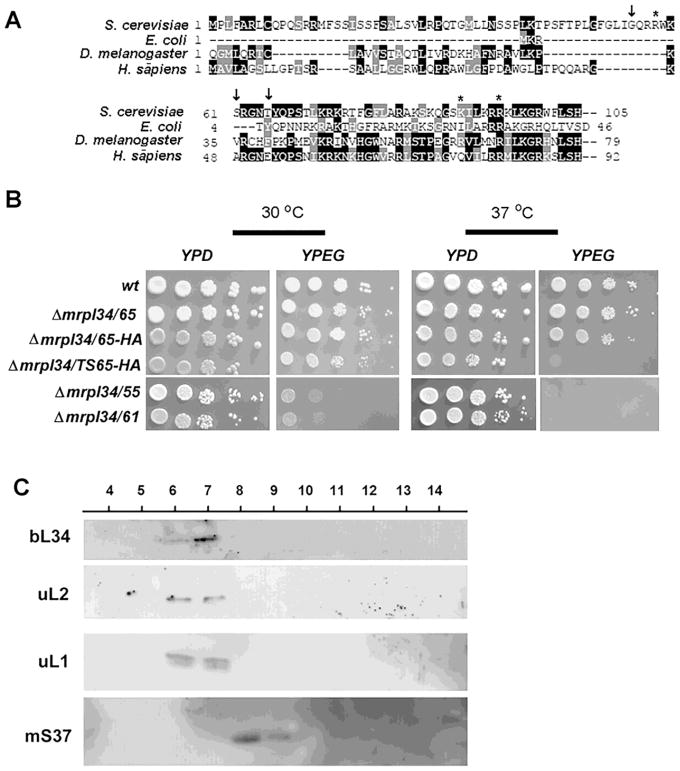
Alignment of bL34 with E. coli L34 also revealed a long N-terminus extension in the protein with no homology between them. According to bL34 primary structure, the mature form of the protein starts at residue 61 (Amunts et al., 2014), which is in the same region we could align the bacterial L34 subunit (Figure 5A). Therefore, it was reasoned that the first 61 residues of bL34 would be important for mitochondrial targeting and processing. In order to test this hypothesis, three truncated forms of bL34 were quimerically fused to the mitochondrial addressing sequence of Neurospora crassa ATP9 (NcATP9) (Barros et al., 2011). The first fusion was performed starting with residue G55 that coincides with the start of identity with other fungi sequences; the second with S61 (starts identity with mammalian sequences), and the third with T65. Curiously only the T65 fusion totally complemented the mrpl34 null mutant growth on ethanol/glycerol (Figure 5B).
Figure 5. Expression, growth and localization of bL34 truncated variants.
(A) Alignment of the N-terminal end of L34 sequences from H. sapiens, S. cerevisiae, E. coli and A. thaliana. The arrows (i) indicate in the Saccharomyces cerevisiae sequence the position of the N-terminal fusions with “HA” tags, and asterisks indicate the position of mutations present in the mrpl34/ts mutants. (B) Growth properties of truncated variants of bL34 starting with T65 (mrpl34/65) G55 (mrpl34/55) and S61 (mrpl34/61) as well as truncated T65 containing the HA fusion (mrpl34/65-HA) and truncated T65 construction obtained from ts2 allele also fused to HA (mrpl34/TS65-HA). Cultured cells were serially diluted and spotted in rich glucose media (YPD) and rich ethanol-glycerol media (YPEG) plates. Plates were incubated at 30° C and 37° C as indicated and photographed after two days of incubation. (C) Mitochondrial ribosomes were purified by sedimentation through a sucrose cushion as described in the Material and Methods section. 20 mL of each fraction was separated by SDS-PAGE in a 12% polyacrylamide gel. Proteins were transferred to nitrocellulose and reacted with rabbit polyclonal antibodies against the large subunits uL1p, uL2p and the small subunit protein mS37. Proteins were visualized with Super Signal (Pierce Biochemicals) following a secondary reaction with peroxidase-coupled anti-rabbit antibody.
In order to have tagged versions of bL34, different tags were added to the C-terminus of the protein. The following tags: His, protein C, biotin and HA were individually added to the C-terminus but none of these constructs was able to rescue the respiratory growth of mrpl34 null mutant, indicating that these fusion proteins were not functional. However, the chimeric construction containing the NcATP9 addressing sequence fused to the HA tag at the N-terminus just before T65 complemented the null mutant. The same tag was also added at the same position of a ts allele and the resultant fusion also maintained the ts phenotype (Figure 5B). The fused protein bL34-HA was detected in mitochondrial fractions from cells cultivated at 37° C indicating its stability as well as the stability of the HA fusion to the mrpl34/ts allele (not shown).
Mitoribosomal subunits extracted from wild-type cells, and purified by sedimentation through a sucrose cushion, were separated in a sucrose gradient. The gradient was collected in 14 equal volume fractions, which were analyzed with antibodies uL1 and uL2 in order to follow the large 54S subunit; and mS37 for the small subunit. Antibodies raised against bL34-trpE fusion protein were used to investigate the sedimentation properties of bL34 in this gradient and a clear co-sedimentation with large subunit proteins was observed (Figure 5C).
bL34 sedimentation properties in sucrose gradients
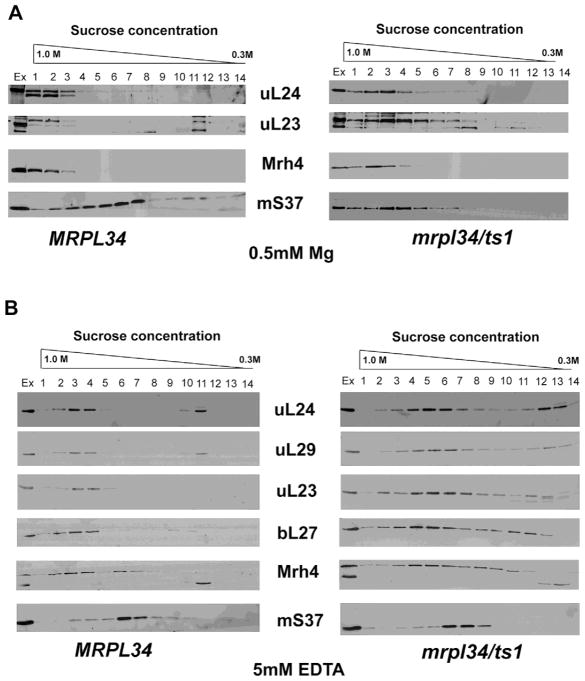
In order to investigate the mitoribosome assembly in the ts mutant under non-permissive temperature, we performed the growth of wild-type and mrpl34/ts1 culture cells in rich-galactose media at 37° C. Mitochondria were isolated and extracted proteins were also subjected to sedimentation in a sucrose gradient as previously described (De Silva et al., 2013), in the presence of EDTA and Mg2+. EDTA promotes the dissociation of mitoribosome into the subunits 54S and 37S, while Mg2+ maintains the structure of mitoribosome 74S. In the presence of Mg2+ the tested mitoribosome associated proteins Mrh4, uL24 and uL23 sediment in fractions corresponding to the assembled 74S and the large 54S subunit (fractions 1–3). Contrasting with the wild-type pattern, in the mrpl34/ts1 extraction uL23 is spread in lighter fractions indicating that its association into the mature 54S is being hampered by the bL34 mutations (Figure 6A). In fact, uL23 interacts with bL34 in the mitoribosome exit tunnel (Amunts et al., 2014). The 37S subunit marker mS37 presents similar pattern of distribution in both strains. At any rate, the proximity of the sedimentation peaks of the wild-type and the ts mutant argues in favor of the presence of mature 54S and putative assembly intermediates with high molecular weight in this mutant. Altogether, these results suggest a late participation of bL34 in the assembly of 54S mitoribosome subunit.
Figure 6. Sedimentation properties of mitoribosome subunits in mrplr34/ts1 mutants.
Cells from wild-type (MRPL34) and mrpl34/ts1 mutants were grown in YPGal at 37° C and used for mitochondrial preparation. Mitochondrial proteins were extracted in the presence of EDTA or Mg2+ as described in the Material and Methods section and sedimented in a linear sucrose gradient 0.3 to 1M for 3h at 40,000 rpm in a 55Ti Beckmann rotor. Fractions were collected from bottom to top, 20 mL of each fraction was separated in a 12% acrylamide-SDS gel system and the western blots assayed for the antibodies indicated in the middle of each panel. The proteins were visualized based on luminol-H2O2 peroxidase chemiluminescence system and intensified with r-coumaric acid. In panel (A) Mitochondrial extracts and the sucrose gradient were prepared with 0.5 mM Mg2+. In panel (B) Mitochondrial extracts and the sucrose gradient were prepared with 5 mM EDTA.
In the presence of EDTA all the tested 54S markers (uL24, uL29, uL23, bL7, and Mrh4) sedimented in lighter and spread fractions in the mrpl34/ts1 than in the wild-type extract (Figure 6B). These results suggest that in mrpl34/ts1 extracts mitoribosome is more susceptible to EDTA (Mg2+ chelation) promoting the dissociation of 54S subunits into intermediates with low molecular weight. Indeed, as discussed later, in bacteria Mg2+ excess can bypass the absence of L34 (Akanuma et al., 2014).
Discussion
S. cerevisiae cultures are known to frequently produce a few percent of ρ–/0 petite cells. Independent of direct factors involved in mtDNA replication and maintenance, many different types of mutations can increase the production of petites (Contamine and Picard, 2000; Lipinski et al., 2010; Gomes et al., 2013). The issue of mitochondrial translation and mtDNA stability has been detailed extensively since the original work of Myers et al. (1985), and without exception, mutants with defective mitochondrial translation result in strains producing ρ–/0 petite cells up to 100%. As a component of mitoribosome subunit 54S, it is expected that bL34 depletion should lead to cultures with a high content of petite cells. In fact, the null mutant mrpl34 could not be studied directly because of the absence of mitochondrial DNA. To circumvent this problem, ts alleles mrpl34-ts1 and mrpl34-ts2 were generated through random PCR mutagenesis. Both ts mutants present slow respiratory growth and the cells were still able to maintain their mitochondrial DNA even after long periods under restrictive conditions (96 h) as well as present reasonable synthesis of newly synthesized mitochondrial products except for Cox1p and Cox3p, whose synthesis were lowered in the ts1 mutant. Cox1p and Cox3p are part of the catalytic core of cytochrome c oxidase enzymatic complex, and probably the modest translation elicits the lower enzymatic activity of the complex. Therefore, the lowered cytochrome c oxidase activity is a reasonable explanation for the respiratory deficiency of the mrpl34-ts cells, sufficient to impair growth on non-fermentable carbon sources, while the translation of Atp6p, Atp8p and Atp9p was sufficient to maintain a minimal of coupled ATP synthase activity, which is a key player in the necessary petite hypothesis for mitochondrial membrane potential maintenance (Ackerman and Tzagoloff, 2005; Lipinski et al., 2010). In fact, Cox24p is another example of a mitoribosome subunits whose absence specifically affects Cox1p translation, mainly in a cox24, cox14 double mutant (Barros et al., 2006; Desai et al., 2017) an intriguing result considering that cox14 deletion characteristically enhances Cox1p translation in different cytochrome c oxidase assembly mutants (Barrientos et al., 2004). However, differently from cox24 mutants, the removal of mitochondrial introns in bL34-ts mutants did not rescue growth at the nonpermissive conditions, indicating that bL34 is not involved in COX1 mRNA processing.
Mitoribosome bL34 interacts with uL23 and uL24, which are components of the exit tunnel, through which newly synthesized peptides pass before emerging from the mitoribosome and seems to be adapted for translating hydrophobic proteins (Amunts et al., 2014). Therefore, an exit tunnel folding problem that prevents the passage of more hydrophobic proteins might be a possible explanation for the poor Cox1p translation in bL34 ts mutants, as well as the loose association with uL23 (Figure 6A).
Indeed, an indication of the partial loss of bL34 function in the ts mutants under restrictive conditions came from analyzes of mitoribosome assembly and subunits formation. In a sucrose gradient containing Mg2+ mitoribosome did not change considerably the sedimentation properties of the tested subunits, except for uL23. On the other hand, Mg2+ chelation with EDTA extraction yielded spread-out patterns for the same subunits. In fact, a suppression of L34 depletion by Mg2+ has already been reported in bacteria indicating that excess of Mg2+ can bypass L34 function in the bacterial 70S ribosome assembly (Akanuma et al., 2014). It would be interesting to investigate whether the same intramitochondrial excess of Mg2+ would provide similar mitoribosome biogenesis rescue in yeast bL34 mutants. These data together with other studies in bacteria suggest that L34 is a late participant and has a limited role in the assembly of the bacterial ribosome (Charollais et al., 2003; Charollais et al., 2004). From our data, based on mrpl34-ts phenotypes, a similar role for bL34 in mitoribosome assembly can also be interpreted but it shall be further investigated with a new batch of anti-bL34 antibodies, or the functional tagged version of bL34.
R95 was identified as a critical residue for protein stability, and its overwhelming presence in the alignment of bL34 family members (in equivalent positions) clearly points its relevance for bL34 function. Indeed, using Pymol modeling software we observed a close association of R95 with 21S rRNA (not shown). It seems reasonable that a thermal instability of an early participant in the biogenesis process of mitoribosome would lead to more severe phenotypes than it was observed for mrpl34-ts. Likewise point mutants with late participation in the process would be more likely able to assemble the mitoribosome and perform translation of most of mitochondrial products.
Mitoribosome biogenesis is still poorly understood, the use of conditional mutants such as bL34 ts used in this work can be of great help to determine the order of events that lead to the assembly of mitoribosome subunits as well as assist in the identification of specific effects of individual subunits in the mitochondrial translation process.
Acknowledgments
We thank Alexander Tzagoloff (Columbia University) for the antibodies used in this work and Ana Carolina Santana Santos (FAPESP 2012/09762-8) for technical assistance. This work was supported by grants and fellowships from Fundaccão de Amparo a Pesquisa de São Paulo (FAPESP - 2013/09482-8; FAPESP 2013/07937-8), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 302935/2014-2). Raquel Monteiro was a fellowship recipient from FAPESP (FAPESP-2011/18892-0).
Abbreviations
- GPD
glycerol 3-phosphate dehydrogenase promoter
- mtLSU
mitochondrial ribosome large subunit
- mtSSU
mitochondrial ribosome small subunit
- NcATP9
Neurospora crassa ATP9 addressing sequence
- PCR
Polymerase Chain Reaction
References
- Ackerman SH, Tzagoloff A. Function, structure, and biogenesis of mitochondrial ATP synthase. Prog Nucleic Acid Res Mol Biol. 2005;80:95–133. doi: 10.1016/S0079-6603(05)80003-0. [DOI] [PubMed] [Google Scholar]
- Akanuma G, Kobayashi A, Suzuki S, Kawamura F, Shiwa Y, Watanabe S, Yoshikawa H, Hanai R, Ishizuka M. Defect in the formation of 70S ribosomes caused by lack of ribosomal protein L34 can be suppressed by magnesium. J Bacteriol. 2014;196:3820–30. doi: 10.1128/JB.01896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Brown A, Bai XC, Llácer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SH, Ramakrishnan V. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–9. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MH, Myers AM, Van Driesche S, Tzagoloff A. COX24 codes for a mitochondrial protein required for processing of the COX1 transcript. J Biol Chem. 2006;281:3743–51. doi: 10.1074/jbc.M510778200. [DOI] [PubMed] [Google Scholar]
- Barros MH, Rak M, Paulela JA, Tzagoloff A. Characterization of Gtf1p, the connector subunit of yeast mitochondrial tRNA-dependent amidotransferase. J Biol Chem. 2011;286:32937–47. doi: 10.1074/jbc.M111.265371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MH, Tzagoloff A. Aep3p-dependent translation of yeast mitochondrial ATP8. Mol Biol Cell. 2017;28:1426–34. doi: 10.1091/mbc.E16-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Korr D, Barwell KJ, Sjulsen C, Gajewski CD, Manfredi G, Ackerman S, Tzagoloff A. MTG1 codes for a conserved protein required for mitochondrial translation. Mol Biol Cell. 2003;14:2292–302. doi: 10.1091/mbc.E02-10-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–82. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Amunts A, Bai XC, Sugimoto Y, Edwards PC, Murshudov G, Scheres SHW, Ramakrishnan V. Structure of the large ribosomal subunit from human mitochondria. Science. 2014;346:718–22. doi: 10.1126/science.1258026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol. 2003;48:1253–65. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–9. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: A plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Fuentes JL, Maddock JR. The yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mrm2. Mol Biol Cell. 2005;16:954–63. doi: 10.1091/mbc.E04-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N, Brown A, Amunts A, Ramakrishnan V. The structure of the yeast mitochondrial ribosome. Science. 2017;355:528–31. doi: 10.1126/science.aal2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva D, Poliquin S, Zeng R, Zamudio-Ochoa A, Marrero N, Perez-Martinez X, Fontanesi F, Barrientos A. The DEAD-box helicase Mss116 plays distinct roles in mitochondrial ribogenesis and mRNA-specific translation. Nucleic Acids Res. 2017;45:6628–43. doi: 10.1093/nar/gkx426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva D, Fontanesi F, Barrientos A. The DEAD box protein Mrh4 functions in the assembly of the mitochondrial large ribosomal subunit. Cell Metab. 2013;18:712–25. doi: 10.1016/j.cmet.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G, Kujawa C, Fukuhara H. Physical and genetic organization of petite and grande yeast mitochondrial DNA. IV. In vivo transcription products of mitochondrial DNA and localization of 23 S ribosomal RNA in petite mutants of Saccharomyces cerevisiae. J Mol Biol. 1974;88:185–203. doi: 10.1016/0022-2836(74)90304-0. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–34. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gomes F, Tahara EB, Busso C, Kowaltowski AJ, Barros MH. Nde1 deletion improves mitochondrial DNA maintenance in Saccharomyces cerevisiae coenzyme Q mutants. Biochem J. 2013;449:595–603. doi: 10.1042/BJ20121432. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;515:283–6. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E.coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–7. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Koerner TJ, Hill JE, Myers AM, Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE-fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–90. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lipinski KA, Kaniak-Golik A, Golik P. Maintenance and expression of the S. cerevisiae mitochondrial genome from genetics to evolution and systems biology. Biochim Biophys Acta - Bioenergetics. 2010;1797:1086–98. doi: 10.1016/j.bbabio.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randal RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- Mears JA, Sharma MR, Gutell RR, McCook AS, Richardson PE, Caulfield TR, Agrawal RK, Harvey SC. A structural model for the large subunit of the mammalian mitochondrial ribosome. J Mol Biol. 2006;358:193–212. doi: 10.1016/j.jmb.2006.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moda BS, Ferreira-Júnior JR, Barros MH. Partial suppression of the respiratory defect of qrs1/her2 glutamyl-tRNA amidotransferase mutants by overexpression of the mitochondrial pentatricopeptide Msc6p. Curr Genet. 2016;62:607–17. doi: 10.1007/s00294-016-0566-6. [DOI] [PubMed] [Google Scholar]
- Myers AM, Pape LK, Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–92. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MF, Alushin GM, Barros MH, Rak M, Tzagoloff A. The putative GTPase encoded by MTG3 functions in a novel pathway for regulating assembly of the small subunit of yeast mitochondrial ribosomes. J Biol Chem. 2012;287:24346–55. doi: 10.1074/jbc.M112.363309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein RJ. Two-step gene disruption in yeast. Methods Enzymol. 1983;101:202–11. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acid as a carrier. Curr Genet. 1989;16:339–46. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–26. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- Strecker V, Kadeer Z, Heidler J, Cruciat CM, Angerer H, Giese H, Pfeiffer K, Stuart RA, Wittig I. Supercomplex-associated Cox26 protein binds to cytochrome c oxidase. Biochim Biophys Acta. 2016;1863:1643–52. doi: 10.1016/j.bbamcr.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpstra P, Zanders E, Butow RA. The association of var1 with the 38 S mitochondrial ribosomal subunit in yeast. J Biol Chem. 1979;254:12653–561. [PubMed] [Google Scholar]
- Tzagoloff A, Akai A, Needleman RB, Zulch G. Assembly of the mitochondrial membrane system. Cytoplasmic mutants of Saccharomyces cerevisiae with lesions in enzymes of the respiratory chain and in the mitochondrial ATPase. J Biol Chem. 1975;250:8236–42. [PubMed] [Google Scholar]
- Tzagoloff A, Myers AM. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–85. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Zampol MA, Busso C, Gomes F, Ferreira-Junior JR, Tzagoloff A, Barros MH. Over-expression of COQ10 in Saccharomyces cerevisiae inhibits mitochondrial respiration. Biochem Biophys Res Comm. 2010;402:82–7. doi: 10.1016/j.bbrc.2010.09.118. [DOI] [PMC free article] [PubMed] [Google Scholar]