Abstract
Identification of all genetic variants associated with complex traits is one of the most important goals in modern human genetics. Genome-wide association studies (GWAS) have been successfully applied to identify common variants, which thus far explain only small portion of heritability. Interests in rare variants have been increasingly growing as an answer for this missing heritability. While next-generation sequencing allows detection of rare variants, its cost is still prohibitively high to sequence a large number of human DNA samples required for rare variant association studies. In this study, we evaluated the sensitivity and specificity of sequencing for pooled DNA samples of multiple individuals (Pool-seq) as a cost-effective and robust approach for rare variant discovery. We comparatively analyzed Pool-seq vs. individual-seq of indexed target capture of up to 960 genes in ~1,000 individuals, followed by independent genotyping validation studies. We found that Pool-seq was as effective and accurate as individual-seq in detecting rare variants and accurately estimating their minor allele frequencies (MAFs). Our results suggest that Pool-seq can be used as an efficient and cost-effective method for discovery of rare variants for population-based sequencing studies in individual laboratories.
Keywords: Genetic variant, Pooled target capture sequencing, Individually indexed target capture sequencing, Rare variant
1. Introduction
Genome-wide association studies (GWAS) have been successful in identifying many novel loci associated with complex traits (http://www.genome.gov/gwastudies). However, GWAS is not optimal for detecting rare variations, and in this regard, genomic markers identified from GWAS on complex traits have not completely accounted for the entire heritability of the trait [1, 2]. To help explain part of this missing heritability, there has been great interest in investigating the role of rare variants (minor allele frequency <0.05) in complex traits with the advent of next-generation DNA sequencing (NGS) technology.
Until recently, the identification of rare genetic variants that might play functional roles in complex traits or diseases has been limited by the resolution of detection technology. The rapid technological advances in NGS, wherein millions of DNA strands are sequenced simultaneously, have enabled the comprehensive characterization of genetic variations including rare variants as well as common variants. Constant increase in throughput of NGS, coinciding with decreasing cost, allowed for the sequencing of the whole human genome in individual laboratories. Still, the cost is often prohibitive for association studies, since a large number of samples are required to gain sufficient statistical power for association studies with rare variants. Thus, it is generally preferable to customize the loci targeted for genomic enrichment using specific regions of interest, such as the coding or regulatory sequences of particular genes. Additionally, nucleotide-based indexing and pre-capture multiplexing of samples in combination further allows sequencing of a larger number of samples, with significantly reduced costs [3].
To make the cost even more affordable for the large number of samples, methods of non-indexed pooling of the equivalent amount of multiple DNA samples together for a library generation can be applied. Since individual information is not assigned in a pool, a number of algorithms have been developed to precisely predict the frequency of called variants in a pool containing multiple samples [4–6]. Several studies have validated the specificity and sensitivity of pooled target capture sequencing (Pool-seq) by comparing the predicted frequency with minor allele frequency (MAF) from other methods, such as genotyping, Sanger sequencing of individual samples, or comparison to an available database [6–8], to demonstrate the accuracy of the variant calls. In addition, an increasing number of studies take advantage of pooled NGS to discover rare variants associated with specific phenotypes [9–11]. Specifically, loci associated from GWAS can be further investigated as candidates for the identification of independent risk variants by re-sequencing of the region in a large number of samples [10]. Other than GWAS, whole-exome sequencing of a small family-based study can also be utilized for the initial candidate loci screening for the further identification of the risk variants by a large sample study [11].
In this study, we used two different sequencing methods, Pool-seq and individually indexed-seq, for the evaluation of efficient variation detection. For both methods, the target regions consisted of 2kb upstream region, exons and exon-intron junctions. For the individually indexed-seq method, we individually indexed 96 samples for target capture sequencing of 960 genes comprised of 5.69 Mb target region. For the Pool-seq method, we used a pooling technique for 56 genes comprised of 410 kb target region for 1,000 samples. Compared to the previously reported Pool-seq studies [6–8], our Pool-seq study is unique in that by far the largest number of individuals (n=1,000) were used for variant discovery in large target regions (410,000 bp) covering both coding and non-coding regions of the genome. Furthermore, a comparative analysis with individually indexed target capture sequencing was performed to comprehensively compare the variant calling and MAF estimation. Independent genotyping studies showed a high concordance with MAF from the Pool-seq variant calls and validated the accuracy of the variant detection by Pool-seq. Here, we demonstrate that Pool-seq is a robust and cost-effective method for variant detection, especially for the rare variants, across genomic regions in a large sample study.
2. Material and Methods
2.1. Samples
The study was approved by the Committee on Clinical Investigations at Albert Einstein College of Medicine. Written informed consent was obtained from all participants. Our study group consisted of 1,029 Ashkenazi Jewish subjects that were previously recruited as part of a longevity study by Dr. Nir Barzilai of the Albert Einstein College of Medicine [12]. All blood samples were rapidly processed to obtain DNA at the General Clinical Research Center of the Albert Einstein College of Medicine. For the individually indexed target capture and next-generation sequencing, we used genomic DNA extracted from immortalized B-lymphocytes from 96 Ashkenazi Jewish samples. For the pooled target capture sequencing and genotyping experiments, we used whole-genome amplified DNA obtained directly from blood samples from 1,000 Ashkenazi Jewish subjects. Whole-genome amplification was performed using illustra GenomiPhi V2 DNA Amplification kits (GE healthcare Life Sciences).
2.2. Target selection and generation of customized target capture for individually indexed next-generation sequencing
We performed individually indexed target capture sequencing of 96 samples with 960 genes relevant to our ongoing research projects. Target genes were selected from pathways of DNA repair and genome maintenance, lipid metabolism, neurodegeneration and cognitive function. At each selected gene locus, we included 2 kb upstream of the transcription start site, all exons, and 20 bp of each exon-intron junction. Bait libraries were designed and assessed for coverage across the target genomic regions using the Agilent eArray website (http://earray.chem.agilent.com/earray/). Design efficiency from eArray was 99%, calculated as the covered fraction of target bases in the oligo design (6,229,389 bp), out of the total submitted target region (6,291,136 bp). Designed bait regions to capture target regions covered 5,692,804 bp.
2.3. Library preparation, target enrichment and sequencing for individually indexed target capture sequencing
We generated 96 libraries which were individually indexed with a unique barcode and 12-plex target capture sequencing was performed for 8-lane sequencing in Illumina HiSeq2000. The library preparation was performed according to the Illumina TruSeq DNA sample preparation version 2 low-throughput (LT) protocol, with the following modifications: (1) genomic DNA was sheared with Covaris system for 300bp; (2) 12 Illumina TruSeq adapters were used; (3) after the ligation of adapters, AMPure XP (Beckman Coulter) beads were used for clean-up steps with 34ul (0.8X volume) for the initial addition and 40ul (0.8X volume) for the second addition; (4) size selection and additional clean-up steps were performed by AMPure XP beads. 0.65X volume of beads were added to each library and after 20 minutes of incubation, separation by magnet was performed and all supernatant was transferred into a new well. 0.85X volume of beads were added to the supernatant and after incubation, it was separated by magnet and supernatant was discarded. Beads were washed with 70% ethanol twice. After complete removal of ethanol, RB was added to suspend the beads. After incubation at room temperature, separation of beads on magnet was performed and eluted products were transferred into the new well; (5) DNA was amplified in 7 cycles for the pre-capture enrichment PCR; (6) 0.85X volumes of AMPure XP beads were added for the clean-up steps after PCR.
The 12-plex target capture was performed according to the Agilent SureSelect Target Enrichment Protocol with the modification based on a high-throughput indexed library preparation and pooled Agilent exome enrichment protocol provided by Evan Geller (personal communication). In brief, 12 post-enriched DNA libraries were combined in equal amounts to a total quantity of 1 μg. In the modified protocol, 6 indexed blocking oligos were supplied by IDT DNA. Each oligo was reconstituted to 300uM with water and equal volumes were combined to make 50uM Indexed Blocking Reagent (IBR). The rest of the protocol was carried out based on Agilent’s protocol. Instead of SureSelect Block #3, IBR was used for the capture. The hybridization mixture was incubated for 24 hours at 65°C to minimize evaporation. Final PCR was performed for a total of 12 cycles. Target-enriched libraries were sent to Axeq Technologies and paired-end sequenced on the Illumina HiSeq2000 with cluster kit version 3 according to Illumina’s protocol. Reads generated were 101 bp in length.
2.4. Data analysis for individually indexed target capture sequencing
Illumina reads were aligned to the human genome, revision hg19 [13], using BWA, version 0.5.9 [14], with setting “-q 17” provided when running the “aln” command. Following alignment, we utilized Picard [http://picard.sourceforge.net] to detect potential PCR duplicates and to calculate on-target statistics. Subsequent processing was performed using the Genome Analysis Toolkit (GATK) [15]. To call variants, we first preprocessed the data, locally realigning the reads around known and suspected indel events, then recalibrated the Illumina base quality scores to more closely reflect actual mismatch rates. We then performed multi-sample genotyping and filtered the results by fitting a Gaussian mixture model to the 7-dimensional point cloud formed by the joint distributions of various statistical annotations calculated for each SNP [16]. The model was trained to assign log-odds likelihood probabilities on the basis of sets of known SNPs from public databases, expanding the set retained until a sensitivity threshold of 99% of accessible, known SNPs was reached. Recommended settings were given during all steps, as detailed in the GATK software manual (<http://www.broadinstitute.org/gsa/wiki/index.php/Best_Practice_Variant_Detection_with_the_GATK_v3>).
The SNP databases used were the 1000 Genomes Project [17], dbSNP build 138 [18], and HapMap3 r3 [19]. Functional annotations were provided by ANNOVAR [20]. Correlation between SNP calls obtained here and those publicly released was determined with the CompOverlap evaluation module of the VariantEval tool from the GATK.
2.5. Target selection and generation of customized target capture for pooled target capture sequencing
We performed 25-sample non-indexed pooling and 20-plex target capture sequencing of 1,000 samples to sequence 56 genes relevant to our ongoing research projects. Among the target genes, 51 genes were also in the gene list of individually indexed-seq. We designed a customized Nimblegen SeqCap EZ Choice library (Roche) target capture to enrich our candidate genes. For each selected gene locus, we included 2 kb upstream region of the transcription start site, all exons, and 20 bp of each exon-intron junction. We used the NimbleDesign (https://design.nimblegen.com/nimbledesign) with ‘Max close match’ set to 5 to design and assess coverage across the target genomic regions of bait libraries. Design efficiency from NimbleDesign was 97.9%, calculated as the covered fraction of target bases in the oligo design (410,497 bp), out of the total submitted target regions (419,327 bp).
2.6. Library preparation, target enrichment and sequencing for pooled target capture sequencing
For each pool, 25 samples were pooled together for equimolar concentration to total 1ug of DNA. A total of 40 pools were generated and 20-plex target capture sequencing was performed for 2-lane sequencing in Illumina HiSeq2000.
Each pooled library was indexed with a unique barcode, which allowed for target capture, enrichment of pooled libraries and multiplex sequencing. The library preparation was performed according to the Illumina TruSeq DNA sample preparation version 2 low-throughput (LT) protocol, with the following modifications: (1) genomic DNA was sheared with Covaris system for 300bp; (2) 20 Illumina TruSeq adapters were used; (3) after the ligation of adapters, AMPure XP (Beckman Coulter) beads were used for clean-up steps with 34ul (0.8X volume) for the initial addition and 40ul (0.8X volume) for the second addition; (4) size selection and additional clean-up step was performed by AMPure XP beads. 0.4X volume of beads were added to each library and after 20 minutes of incubation, separation by magnet was performed and all supernatant was transferred into a new well. 0.65X volume of beads were added to the supernatant and after incubation, separation by magnet was performed and the supernatant was discarded. Beads were washed with 70% ethanol twice. After complete removal of ethanol, RB was added to suspend the beads. After incubation at room temperature, separation of the beads on the magnet was performed and eluted products were transferred into the new well; (5) for the pre-capture enrichment PCR, DNA was amplified in 6 cycles with KAPA HiFi HotStart DNA polymerase with dNTPs (KAPA Biosystems) and PCR primer cocktail from Illumina TruSeq; (5) for the clean-up steps after PCR, 0.85X volumes of AMPure XP beads were added.
The 20-plex target capture was performed according to the Nimblegen SeqCap EZ Choice library target capture protocol, with the following modifications: (1) capture hybridization mixture was incubated for 72 hours at 47°C; (2) post-capture PCR was performed with KAPA HiFi Hotstart Polymerase with the provided PCR primers (KAPA Biosystems) and 11 cycles of PCR reaction was performed; (3) Clean-up of PCR was performed using the Qiagen Qiaquick PCR Purification kit. Target-enriched libraries were sent to Axeq Technologies and paired-end sequenced on the Illumina HiSeq2000 with cluster kit version 3 according to Illumina’s protocol. Reads generated were 101 bp in length.
2.7. Data analysis for pooled targeted capture sequencing
Following sequencing of the targeted DNA, the BWA alignment software, version 0.7.5a [14] was used to align the sequence data to the hg19 human reference genome (GRCh37 assembly, February 2009). The hg19 human reference genome was downloaded from the UCSC Genome Browser [21]. Potential PCR duplicates were removed with the rmdup routine of the samtools software, version 0.1.18 [22]. The Picard tools CollectAlignmentSummaryMetrics and CollectTargetedPcrMetrics, version 1.81, were used to collect statistics about the read coverage in each pool.
Variants were called using the software CRISP, which was specifically developed to call variants in pooled DNA sequence data [6, 23]. CRISP variant calling was performed with a 2,000 bp flanking region surrounding the probe design locations. The CRISP results were postprocessed to calculate the allele counts in the pools.
2.8. Genotyping
To validate variants from pooled target capture sequencing, we designed iPLEX MassArray assays using the web-based assay design suite program on mysequenom website (http://www.mysequenom.com). Variants were successfully assayed based on the high quality of peaks visualized in the MassARRAY® Typer software after performing MassArray. Using the iPLEX assays, genotyping was performed in the Genomics Shared Facility at Albert Einstein College of Medicine for 1,000 samples that were identical with the samples used for pooled target capture sequencing.
3. Results
3.1. Individually indexed target capture and next-generation sequencing of 960 candidate genes in 96 samples
A total of 1,571,149,163 unique reads were aligned to the reference sequence and achieved an average on-target percentage of 90.5% in 96 samples (Table 1). Individual on-target percentage was evenly distributed from 88% to 93% (Supplement Figure S1). Individual fold enrichment of coverage ranged from 384 to 428 in 96 samples with average of 406 (Supplement Figure S1, Table 1). Mean target coverage for individual samples ranged from 60X to 541X with an average of 211X and, as previously reported, this is ample coverage for variant detection [24]. The percentage of all target bases achieving greater than or equal to 20X, 10X, and 2X were 95.2%, 96.8%, and 98%, respectively (Table 1).
Table 1.
Alignment and coverage statistics of 96 samples from individually indexed-seq for 960 genes.
| Target Coverage | %nt on Target | Fold Enrichment | % ≥20x Target Bases | % ≥10x Target Bases | % ≥2x Target Bases | |
|---|---|---|---|---|---|---|
| Mean | 210.8 | 90.5 | 406 | 95.2 | 96.8 | 98.0 |
|
| ||||||
| CVa (95% CI b) |
0.41 (0.35–0.49) |
0.012 (0.011–0.014) |
0.025 (0.022–0.030) |
0.021 (0.018–0.024) |
0.009 (0.008–0.10) |
0.0027 (0.0023–0.0031) |
Coefficient of Variation,
Confidence Interval
Using ANNOVAR [20], we characterized and functionally annotated the variants detected from 96 samples, according to the gene regions in which they resided (Table 2). Overall, novel variants accounted for 16% of the total variants detected in the target gene regions. The 5,200 exonic variants in coding or splicing regions represented about 25% of the total variants found. Intronic variants comprised 5,334 variants (25.7%) of the total variants. Variants in the promoter (2kb upstream) represented 3,914 variants (18.8%). The 3′UTR contained 5,123 variants (24.7%). We further characterized the 5,200 variants in exons (Supplement Table S1), and found that 51.2% were synonymous SNPs and 47.7% were non-synonymous SNPs (nsSNPs). 1.1% of the exonic variants were small insertions or deletions (Indels) which resulted in similar number of frameshift or non-frameshift of the resulting amino acids. Among the 2,482 nsSNPs, 295 (11.8%) were novel SNPs and 1.2% were nonsense SNPs, which prematurely truncate the protein by introducing a stop codon (Supplement Table S1).
Table 2.
Characterization of the variants in 96 samples from individually indexed-seq for 960 genes.
| Databasea | Novel variants | % novel | Total variants | |
|---|---|---|---|---|
| downstreamb | 1 | 1 | 50.00 | 2 |
| intronic | 4676 | 658 | 12.34 | 5334 |
| upstream (2kb)c | 3046 | 868 | 22.18 | 3914 |
| ncRNAd | 9 | 3 | 25.00 | 12 |
| UTR3e | 4030 | 1093 | 21.34 | 5123 |
| UTR5f | 890 | 261 | 22.68 | 1151 |
| Splicingg | 33 | 12 | 26.67 | 45 |
| exonic (coding)h | 4662 | 445 | 8.71 | 5107 |
| exonic;splicing (coding)i | 79 | 14 | 15.05 | 93 |
| total | 17426 | 3355 | 16.14 | 20781 |
Based on dbSNP 138, 1000 Genomes database, and Exome sequencing database
Variant overlaps 1 kb downstream of transcription end site
Variant overlaps 2 kb upstream of transcription start site
Variant overlaps a transcript without coding annotation in the gene definition
Variant overlaps a 3′ untranslated region
Variant overlaps a 5′ untranslated region
Variant is within 3-bp of a splicing junction in intronic region
Variant overlaps a coding exon (but not UTR portion)
Variant is within 3-bp of a splicing junction in exonic region
3.2. Pooling and target capture sequencing of the 56 candidate genes in 1,000 samples
To test the efficiency of detecting variants in a large set of 1,000 genomic DNA samples, we performed targeted and pooled sequencing for 56 genes, of which 51 genes were common to the individually indexed seq. A total of 653,612,106 unique reads aligned with the reference genome and an average on-target percentage of 37.3% was obtained in the 40 pools (Table 3). Individual on-target percentage was evenly distributed between 29% and 45% (Supplement Figure S2). Although the on-target percentage was not high, the average fold enrichment of coverage for the targeted region in 40 pools was 2,020 (Table 3). Individual fold enrichment of coverage ranged from 1,587 to 2,401 (Supplement Figure S2). Mean target coverage for 40 pools was 1,068X. Because each pool has 25 samples, assuming that each sample was captured and sequenced equally, theoretically, mean target coverage for an individual sample for the targeted region would be 43X, which is ample coverage for the detection of variants. A total of 6,551 variants were identified from the 56 genes. The frequency of the variant in Pool-seq was calculated by CRISP [6, 23].
Table 3.
Alignment and coverage statistics of 40 pools in 1,000 samples from pool-seq for 56 genes.
| Target Coverage | %nt on Target | Fold Enrichment | % ≥20x Target Bases | % ≥10x Target Bases | % ≥2x Target Bases | |
|---|---|---|---|---|---|---|
| Mean | 1068 | 37.3 | 2020 | 99.4 | 99.7 | 99.9 |
|
| ||||||
| CVa (95% CIb) |
0.16 (0.13–0.21) |
0.11 (0.09–0.14) |
0.10 (0.09–0.14) |
0.0011 (0.0009–0.0014) |
0.0007 (0.0006–0.0009) |
0.0003 (0.0002–0.0004) |
Coefficient of Variation,
Confidence Interval
We characterized and functionally annotated variants from the 1,000 pooled samples, according to the gene regions in which they reside, using ANNOVAR (Table 4) as we did for the individually indexed target capture and sequencing experiments. Novel variants accounted for 46% of the total variants detected in the target gene region. Exonic variants represented about 12% of the total variants found. Intronic variants represented 3,220 of the total variants (49.2%). The promoter region contained 1,131 variants (17.3%). The 3′UTR contained 1,094 variants (16.7%). We further characterized the variants in exons (Supplement Table S2) and identified 41% as synonymous SNPs and 54% as non-synonymous SNPs. 5% of the exonic variants were small indels which resulted in frameshift or non-frameshift of the resulting amino acids. Among the 420 nsSNPs, 186 (44%) were novel SNPs and 5% were nonsense SNPs (Supplement Table S2).
Table 4.
Characterization of the variants of 40 pools in 1,000 samples from pool-seq for 56 genes.
| Databasea | Novel variants | % novel | Total variants | |
|---|---|---|---|---|
| downstreamb | 57 | 40 | 41.24 | 97 |
| intronic | 1790 | 1430 | 44.41 | 3220 |
| upstream (2kb)c | 543 | 588 | 51.99 | 1131 |
| UTR3d | 539 | 555 | 50.73 | 1094 |
| UTR5e | 109 | 105 | 49.07 | 214 |
| splicingf | 6 | 9 | 60.00 | 15 |
| exonic (coding)g | 453 | 296 | 39.52 | 749 |
| exonic;splicing (coding)h | 17 | 14 | 45.16 | 31 |
| total | 3514 | 3037 | 46.36 | 6551 |
Based on dbSNP 138, 1000 Genomes database, and Exome sequencing database
Variant overlaps 1 kb downstream of transcription end site
Variant overlaps 2 kb upstream of transcription start site
Variant overlaps a 3′ untranslated region
Variant overlaps a 5′ untranslated region
Variant is within 3-bp of a splicing junction in intronic region
Variant overlaps a coding exon (but not UTR portion)
Variant is within 3-bp of a splicing junction in exonic region
3.3. Validation of the accuracy of variant calls from pooling and target capture sequencing by genotyping
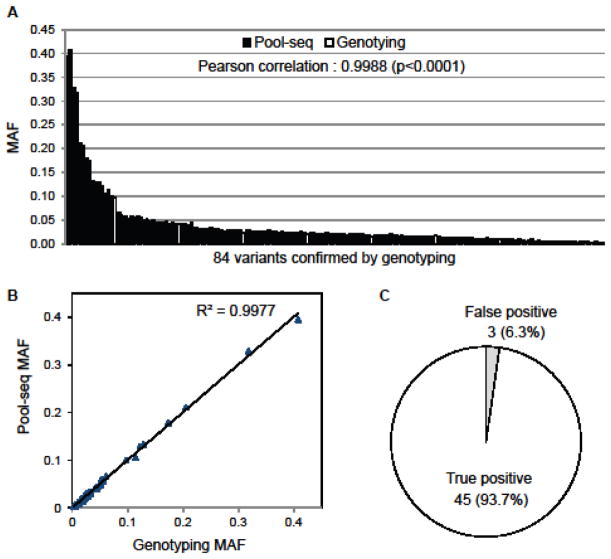
To evaluate the accuracy of the variant calls from Pool-seq, we performed a separate iPLEX MassArray genotyping experiment for 84 randomly-selected variants using the 1000 samples, and compared the MAFs from this genotyping with the Pool-seq variant calls (Figure 1A and 1B). The Pearson correlation of the MAF of 84 variants between Pool-seq and genotyping was 0.9988 (p<0.0001) (Figure 1A) and the correlation coefficient (R2) was 0.9977 (Figure 1B), which clearly demonstrates the accuracy of variant detection by the Pool-seq. Additionally, to evaluate the specificity of the Pool-seq data, we genotyped 48 randomly-selected novel variants and discovered that the false positive rate of Pool-seq is 6.3% which is similar with a previous report that used the CRISP algorithm [23] (Figure 1C).
Figure 1. Evaluation of pool-seq accuracy in detecting variant by genotyping.
A. Randomly selected 84 variants detected from pool-seq were genotyped by iPLEX MassArray and the MAF of each variant from both pool-seq and genotyping is indicated in the bar-graph. Black bar indicates the MAF of variant from pool-seq and white bar indicates the MAF of variant from genotyping. B. MAFs of 84 variants from A are presented as a scattered plot. Triangle dot indicates each 84 variant. C. Randomly selected 48 novel variants detected from pool-seq were genotyped and the proportion of true positive (white) and false positive (gray) are shown in the pie-graph.
3.4. Comparison of the MAF distribution between individually indexed-seq of 96 samples and Pool-seq of 1,000 samples
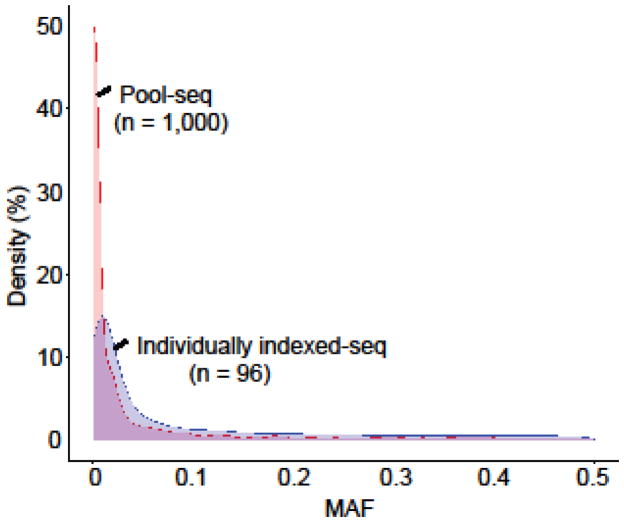
To demonstrate the efficiency of the detection of variants by Pool-seq in large samples, we compared the MAF distribution between individually indexed-seq of 96 samples and Pool-seq of 1,000 samples of same population. Among the total variants discovered from 960 genes by individually indexed-seq, 28.8% (5,980 variants) were discovered as extremely rare variants of MAF less than 0.01 (Table 5). 6,566 (31.6%) were rare variants of MAF greater or equal to 0.01 and less than 0.05 and 8,235 (39.6%) were common variants of MAF greater or equal to 0.05. On the other hand, among total variants discovered from 56 genes by Pool-seq, 58.1% (3,807 variants) were discovered as extremely rare variants of MAF less than 0.01 (Table 5). Similarly, 1,266 (19.3%) were rare variants of MAF greater or equal to 0.01 and less than 0.05 and 1,478 (22.6%) were common variants of MAF greater or equal to 0.05. Additionally, for the rare variants of MAF less than 0.01, 65.4% of detected variants were novel from the Pool-seq while 38.7% of detected variants were novel from the individually indexed-seq (Table 5). We also plotted the density of MAF of the each variant from both Pool-seq and individually indexed-seq in Figure 2, and it clearly displays the high density of extreme rare variants in the Pool-seq.
Table 5.
Characterization of the variants by minor allele frequency (MAF).
| Individually indexed-seq of 96 individuals | Pool-seq of 1,000 individuals | |||||||
|---|---|---|---|---|---|---|---|---|
| Minor allele frequency (MAF) | Databasea | Novel variants | % novel | Total variants (%) | Database | Novel variants | % novel | Total variants (%) |
| 0 < – < 0.01 | 3667 | 2313 | 38.68 | 5980 (28.8) |
1318 | 2489 | 65.38 | 3807 (58.1) |
| 0.01 ≤ – <0.05 | 5586 | 980 | 14.93 | 6566 (31.6) |
848 | 418 | 33.02 | 1266 (19.3) |
| 0.05 ≤ | 7875 | 360 | 4.37 | 8235 (39.6) |
1348 | 130 | 8.80 | 1478 (22.6) |
| Total | 17128 | 3653 | 17.58 | 20781 (100) |
3514 | 3037 | 46.36 | 6551 (100) |
Based on dbSNP 138, 1000 Genomes database, and Exome sequencing database
Figure 2. Comparison of MAF distribution between pool-seq of large samples and individually indexed-seq of small samples.
The plot presents the density of number of variants with each MAF detected in pool-seq of 1,000 samples (red) and individually indexed-seq of 96 samples (blue). X-axis indicates the MAF of variants and y-axis indicates the percentage of density.
3.5. Variant comparison between individually indexed-seq and Pool-seq of 51 overlapping genes
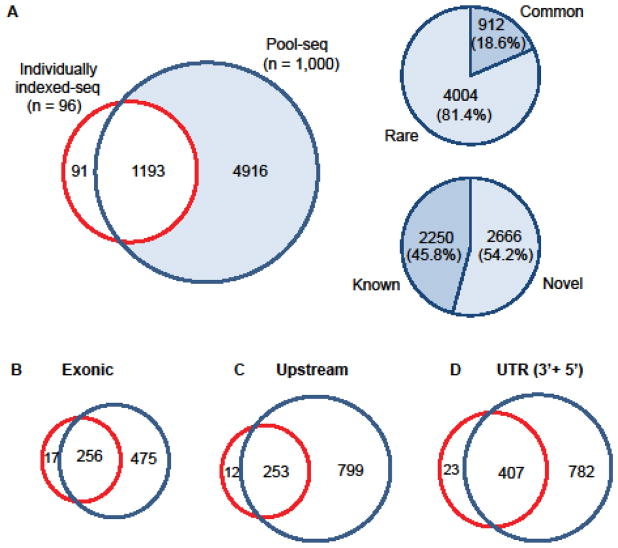
To specifically evaluate the capability to detect variants, we compared variants discovered in 51 candidate genes from both individually indexed-seq and Pool-seq. In the same target region within the 51 genes, Pool-seq of the large sample resulted in detection of more variants compared to individually indexed-seq (Figure 3A). We further characterized the 4,916 variants that were only detected in Pool-seq, and found that 18.6% were common while 81.4% were rare variants with MAF less than 0.05. Additionally, among the 4,916 exclusively detected variants from Pool-seq, more than half of the variants were novel which was also mostly rare. We further grouped the variants according to their genomic region, and found that exonic variants were less likely to be detected than upstream (proximal 2kb promoter region) or UTR variants (Figure 3B, 3C, and 3D). While the number of overlapping variants in both individually indexed-seq and Pool-seq in exonic region was similar to those located upstream, the number of exclusive variants that were only detected in Pool-seq in the upstream region was 1.7 fold higher than the number of exclusive variants in the exonic region (Figure 3B and 3C). On the other hand, the number of exclusive variants that were only detected by Pool-seq in the UTRs was similar to the number of exclusive variants that were only detected by Pool-seq in the upstream region (Figure 3C and 3D). These results indicate that Pool-seq is a robust and effective method to study large samples for detecting rare variants across all genomic regions, including regulatory regions.
Figure 3. Variant comparison between individually indexed-seq and pool-seq of 51 overlapping genes.
A. The Venn diagram indicates the number of variants discovered in individually indexed-seq of 96 samples (red line) and pool-seq of 1,000 samples (blue line) of 51 genes targeted in both methods. Area filled with blue color indicates the exclusive variants only discovered in pool-seq and two pie-graphs on the right side are based on the colored area. Upper-right graph indicates the proportion of rare and common variants and lower-right graph indicates the proportion of known and novel variants. B–D. The Venn diagram indicates the discovered variants from individually indexed-seq and pool-seq in exonic (B), upstream (C) and UTRs (D) of the 51 genes.
4. Discussion
Recent increases in the speed and volume of next generation sequencing technologies have enabled thorough mapping of genetic variations in whole exome or whole genome data. Along with the advance of the technology, there are now many genetic association studies demonstrating the relevance of rare variants with complex phenotypes [25, 26]. Due to the low frequency of rare variants, many statistical methods for rare variant association analysis often consider aggregates of rare variants together [27–29]. Therefore, detection of many rare variants in a large sample size would be required for the association studies. The use of pooling samples together with next-generation sequencing is a reasonable approach for large sample studies because of the affordable cost and reduced labor [7, 30, 31]. In this study, we performed individually indexed-seq for 96 samples and Pool-seq of 1,000 samples to compare the efficiency of variant detection.
For the Pool-seq approach, 25 samples were pooled together into a single pool, and statistical methods were used to calculate the frequency of each allele in the total sample [6, 23]. Current study is different from the other published Pool-seq studies [6–8] for following reasons. First, this study used a large number of individuals (n=1,000) and represents by far the largest study conducted for a Pool-seq analysis, providing enough statistical power required for a human genetic study. Second, in our study, we performed a comparative analysis with individually indexed target capture sequencing and genotyping validation of discovered variants including rare variants to experimentally validate the accurate variant detection of Pool-seq analysis. Whereas the other studies did not perform such comparative analyses. Finally, our study captured large target regions across 410kb, including both coding and non-coding regions, in a large number of 1000 individuals. In contrast, other studies represented either a large target region in a small number of individuals (1.6 Mb capture for up to 50 individuals) or a small target region in a large number of individuals (6.7 Kb capture for 480 individuals) [7, 8]. Thus, our study demonstrated that target capture sequencing of pooled individuals can be successfully performed for large target regions in a large number of individuals, accurately detecting variants and their allele frequencies as experimentally confirmed by individual genotyping analysis. Validation of the variant calls of 84 random variants from the large sample using genotyping, clearly demonstrates the accuracy of the variant calls from Pool-seq (Figure 1A, B). Additionally, we used genotyping to further evaluate 48 novel variants that were detected from Pool-seq to calculate the estimated false positive rate and it resulted in 6.3%, which was in concordance with the previous report [23] (Figure 1C). Considering the false positive rate that was calculated based on the novel variants, we believe that the variants detected by Pool-seq are highly reliable.
In both individually indexed-seq and Pool-seq experiment, we targeted 2kb proximal promoter regions and 20bp exon-intron junctions for the detection of potential splicing variants, along with the coding region. As shown in Table 3.2 and 3.5, we were able to discover many variants in the upstream 2kb region, intronic, UTRs as well as exonic region from both methods. In particular, Pool-seq resulted in a greater number (46.4%) of novel variants which could have potentially higher impact on protein function or expression. As a matter of fact, when we categorized the discovered variants by their MAF in both individually indexed-seq and Pool-seq, we detected many more rare and novel variants by Pool-seq (Table 5). As expected, distribution of variants from Pool-seq showed enriched pattern in low MAF (Figure 2). To evaluate the capability of variant detection in more detail, we compared the variants in 51 genes present in both the individually indexed-seq with small samples and the Pool-seq with large samples. Pool-seq identified more variants which are mostly rare variants (MAF<0.05) in both regulatory and coding region (Figure 3). Not surprisingly, we found almost double the number of the exclusive variants that were only detected in the Pool-seq in upstream or UTRs compared to exonic regions (Figure 3B, 3C, and 3D). Also, we detected many more of the exclusive variants in the Pool-seq in intronic regions compared to all other regions as well (data not shown). This indicates that coding regions are well-conserved compared to the other genomic regions where the alteration of genetic information could be detrimental on the protein function. In addition, the recent ENCODE project demonstrated that many non-coding regions have regulatory functions which can affect the level of expressed coding genes and phenotype [32]. Our results suggest that Pool-seq is an effective way to discover rare variants throughout the genome in large samples.
An unavoidable disadvantage of Pool-seq is the loss of individual sample information. To overcome this, additional genotyping with the same samples could be performed to identify samples harboring specific variants. On the other hand, individually indexed-seq has all of the information for each sample. However, large sample genotyping is further required as well for the case-control association study to have enough association power. In the case of rare variant association studies, since aggregates of rare variants are needed, additional sequencing will be required. Thus, in spite of the tradeoff, Pool-seq is an ideal method for large sample association studies. Pool-seq can be used to select variants for association study using frequency from each case and control group with relatively low false-positive rate. In addition, it can discover many more rare variants which could not be detected in individually indexed-seq with small samples as shown in our study (Figure 3). Furthermore, it can significantly decrease the preparation cost by reducing required number of libraries and target capture [6].
In conclusion, our study demonstrated that Pool-seq is a highly accurate, cost-effective method in identifying variants in human population studies, especially rare variants with minor allele frequency less than 5% in both regulatory as well as coding regions. As compared to the individually indexed-seq, Pool-seq can be utilized to discover the risk variants in a large sample size as a follow up study of candidate loci detected from initial discovery.
Supplementary Material
Highlights.
Evaluation of accuracy of pooled target capture sequencing by genotyping
Analysis of frequency of variants in pooled target capture sequencing
Comparison of variants between pooled sequencing and individually indexed sequencing
Acknowledgments
We would like to thank Dr. Nir Barzilai for generously providing us with the Ashkenazi Jewish DNA samples. Also we would like to thank Archana Tare for critical reading of the manuscript. This work was funded by NIH grants AG024391, AG027734, and AG17242 (Y. S.) and a grant from The Paul F. GlennCenter for the Biology of Human Aging (Y. S.). S. R. is the recipient of a Glenn/AFAR Scholarships for Research in the Biology of Aging.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shearer AE, Hildebrand MS, Ravi H, Joshi S, Guiffre AC, Novak B, Happe S, LeProust EM, Smith RJ. Pre-capture multiplexing improves efficiency and cost-effectiveness of targeted genomic enrichment. BMC Genomics. 2012;13:618. doi: 10.1186/1471-2164-13-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Druley TE, Vallania FL, Wegner DJ, Varley KE, Knowles OL, Bonds JA, Robison SW, Doniger SW, Hamvas A, Cole FS, Fay JC, Mitra RD. Quantification of rare allelic variants from pooled genomic DNA. Nat Methods. 2009;6:263–265. doi: 10.1038/nmeth.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bansal V, Tewhey R, Leproust EM, Schork NJ. Efficient and cost effective population resequencing by pooling and in-solution hybridization. PLoS One. 2011;6:e18353. doi: 10.1371/journal.pone.0018353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day-Williams AG, McLay K, Drury E, Edkins S, Coffey AJ, Palotie A, Zeggini E. An evaluation of different target enrichment methods in pooled sequencing designs for complex disease association studies. PloS one. 2011;6:e26279. doi: 10.1371/journal.pone.0026279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niranjan TS, Adamczyk A, Bravo HC, Taub MA, Wheelan SJ, Irizarry R, Wang T. Effective detection of rare variants in pooled DNA samples using Cross-pool tailcurve analysis. Genome biology. 2011;12:R93. doi: 10.1186/gb-2011-12-9-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, Burtt NP, Rivas M, Guiducci C, Bruno DL, Goldberger OA, Redman MC, Wiltshire E, Wilson CJ, Altshuler D, Gabriel SB, Daly MJ, Thorburn DR, Mootha VK. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet. 2010;42:851–858. doi: 10.1038/ng.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diogo D, Kurreeman F, Stahl EA, Liao KP, Gupta N, Greenberg JD, Rivas MA, Hickey B, Flannick J, Thomson B, Guiducci C, Ripke S, Adzhubey I, Barton A, Kremer JM, Alfredsson L, Sunyaev S, Martin J, Zhernakova A, Bowes J, Eyre S, Siminovitch KA, Gregersen PK, Worthington J, Klareskog L, Padyukov L, Raychaudhuri S, Plenge RM Consortium of Rheumatology Researchers of North A, Rheumatoid Arthritis Consortium I. Rare, low-frequency, and common variants in the protein-coding sequence of biological candidate genes from GWASs contribute to risk of rheumatoid arthritis. Am J Hum Genet. 2013;92:15–27. doi: 10.1016/j.ajhg.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, Harari O, Norton J, Budde J, Bertelsen S, Jeng AT, Cooper B, Skorupa T, Carrell D, Levitch D, Hsu S, Choi J, Ryten M, Hardy J, Ryten M, Trabzuni D, Weale ME, Ramasamy A, Smith C, Sassi C, Bras J, Gibbs JR, Hernandez DG, Lupton MK, Powell J, Forabosco P, Ridge PG, Corcoran CD, Tschanz JT, Norton MC, Munger RG, Schmutz C, Leary M, Demirci FY, Bamne MN, Wang X, Lopez OL, Ganguli M, Medway C, Turton J, Lord J, Braae A, Barber I, Brown K, Passmore P, Craig D, Johnston J, McGuinness B, Todd S, Heun R, Kolsch H, Kehoe PG, Hooper NM, Vardy ER, Mann DM, Pickering-Brown S, Brown K, Kalsheker N, Lowe J, Morgan K, David Smith A, Wilcock G, Warden D, Holmes C, Pastor P, Lorenzo-Betancor O, Brkanac Z, Scott E, Topol E, Morgan K, Rogaeva E, Singleton AB, Hardy J, Kamboh MI, St George-Hyslop P, Cairns N, Morris JC, Kauwe JS, Goate AM UKBE Consortium, UKC The Alzheimer’s Research. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2013 doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, Lipton R, Cheng S, Shuldiner AR. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- 13.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ C. International Human Genome Sequencing. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genomes Project C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International HapMap C. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, Raney BJ, Pohl A, Malladi VS, Li CH, Lee BT, Learned K, Kirkup V, Hsu F, Heitner S, Harte RA, Haeussler M, Guruvadoo L, Goldman M, Giardine BM, Fujita PA, Dreszer TR, Diekhans M, Cline MS, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin RS. Genome Project Data Processing, The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bansal V. A statistical method for the detection of variants from next-generation resequencing of DNA pools. Bioinformatics. 2010;26:i318–324. doi: 10.1093/bioinformatics/btq214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genomes Project C. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Liu X, Yang BZ, Gelernter J. The Role and Challenges of Exome Sequencing in Studies of Human Diseases. Front Genet. 2013;4:160. doi: 10.3389/fgene.2013.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ionita-Laza I, Buxbaum JD, Laird NM, Lange C. A new testing strategy to identify rare variants with either risk or protective effect on disease. PLoS Genet. 2011;7:e1001289. doi: 10.1371/journal.pgen.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Futschik A, Schlotterer C. The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics. 2010;186:207–218. doi: 10.1534/genetics.110.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rellstab C, Zoller S, Tedder A, Gugerli F, Fischer MC. Validation of SNP allele frequencies determined by pooled next-generation sequencing in natural populations of a non-model plant species. PLoS One. 2013;8:e80422. doi: 10.1371/journal.pone.0080422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consortium EP. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.