Abstract
The abuse of antibiotics and following rapidly increasing of antibiotic-resistant pathogens is the serious threat to our society. Natural products from microorganism are regarded as the important substitution antimicrobial agents of antibiotics. We isolated a new strain, Bacillus sp. GFP-2, from the Chiloscyllium plagiosum (Whitespotted bamboo shark) intestine, which showed great inhibitory effects on the growth of both Gram-positive and Gram-negative bacteria. Additionally, the growth of salmon was effectively promoted when fed with inactivated strain GFP-2 as the inhibition agent of pathogenic bacteria. The genes encoding antimicrobial peptides like LCI, YFGAP and hGAPDH and gene clusters for secondary metabolites and bacteriocins, such as difficidin, bacillibactin, bacilysin, surfactin, butirosin, macrolactin, bacillaene, fengycin, lanthipeptides and LCI, were predicted in the genome of Bacillus sp. GFP-2, which might be expressed and contribute to the antimicrobial activities of this strain. The gene encoding β-1,3-1,4-glucanase was successfully cloned from the genome and this protein was detected in the culture supernatant of Bacillus sp. GFP-2 by the antibody produced in rabbit immunized with the recombinant β-1,3-1,4-glucanase, indicating that this strain could express β-1,3-1,4-glucanase, which might partially contribute to its antimicrobial activities. This study can enhance a better understanding of the mechanism of antimicrobial activities in genus Bacillus and provide a useful material for the biotechnology study in antimicrobial agent development.
Keywords: Bacillus sp. GFP-2; Complete genome sequencing; Antimicrobial peptides; Secondary metabolites; Bacteriocins; β-1,3-1,4-Glucanases
Introduction
The studies of antibiotics started with the discovery of penicillin in 1928. With the discoveries of different antibiotics, many effective therapeutic strategies toward incurable or obstinate disease have been developed. However, accompany with the widely usage of antibiotics, the emerging of antibiotic-resistant pathogens is increasing rapidly, suggesting that, without urgent actions, we are heading for a “post-antibiotic era” (Mahlapuu et al. 2016). To solve this threat, many new and non-conventional anti-infective therapies have been developed (or identified) (Czaplewski et al. 2016). Over the past 10 years, many marine natural products were isolated and showed important effects in the aforementioned therapies. Due to the structural diversities of these natural products, researchers have paid high attention to expanding their studies from sponges, corals to microorganisms, among which, marine microorganisms are regarded as major sources of antimicrobial agents (Ng et al. 2015).
Genus Bacillus consists of over 300 species. Members of this genus are Gram-positive, aerobic or facultative anaerobic, rod-shaped bacteria with diverse functions. Recently, peptides produced by members of the genus Bacillus were shown to have a broad spectrum of antimicrobial activity against pathogenic microbes. Strains in genus Bacillus can produce structurally diverse secondary metabolites, which exhibit a wide spectrum of antibiotic activity, but the precise mechanism is still unclear (Li et al. 2012; Sabate and Audisio 2013; Sumi et al. 2015). The antimicrobial peptides (AMPs) produced by Bacillus species can be divided into two subgroups based on the synthesis pathway, one of which includes small microbial peptides that are non-ribosomally synthesized by large enzymatic complexes, whereas the second subgroup comprises ribosomally synthesized peptides (Marahiel 1992; Marx et al. 2001; Nakano and Zuber 1990). Besides, some species in Bacillus, like Bacillus macerans, also contain genes encoding bacterial β-1,3-1,4-glucanases, which belong to the glycosyl hydrolase family 16 (GH16) and can specifically cleave the β-1,4-glycosidic linkage adjacent to 3-O-substituted glucopyranose residues. Because of its specific functions in antimicrobial activity, β-1,3-1,4-Glucanases have been explored in several industrial applications.
In this study, we isolated an antimicrobial bacteria strain, Bacillus sp. GFP-2, from the Chiloscyllium plagiosum (Whitespotted bamboo shark) intestine, which showed inhibitory effects against both Gram-positive strains and Gram-negative strains and could also promote the growth of salmon. The genes encoding antimicrobial peptides and gene clusters for secondary metabolites and bacteriocins, which might, at least partially, account for the antimicrobial activities of this strain, were predicted by complete genome sequencing and analyzing. Furthermore, a novel β-1,3-1,4-glucanase gene was successfully cloned from Bacillus sp. GFP-2. The recombinant β-1,3-1,4-glucanase, heterogeneously expressed in Escherichia coli, was further purified and used to immunize rabbits to produce an antibody, which could specifically detect β-1,3-1,4-glucanase in the protein extract of GFP-2 culture supernatant, further confirming that Bacillus sp. GFP-2 could produce β-1,3-1,4-glucanase and might contribute to its antimicrobial activities.
Materials and methods
Culture of Chiloscyllium plagiosum and isolation of bacteria colony from intestine
Experimental protocols were reviewed and approved by the Animal Ethic Committee of Zhejiang Sci-Tech University. Whitespotted bamboo sharks were raised in glass container containing water with the salinity of between 1.020 and 1.023% at 25 °C. The health condition was monitored every day and healthy ones were used for this experiment. The shark was sacrificed after injection of 25% urethane as the experiment anesthetic and the intestinal contents and mucosal fluid was taken out and diluted in normal saline as the raw bacteria solution. The raw bacteria solution was further inoculated and diluted on Luria broth (LB) agar plates (5 g of yeast extract, 10 g of tryptone, 10 g of NaCl, and 15 g of agar in 1 L ddH2O) and cultured at 37 °C for 12 h. Single bacteria colonies were then picked and further cultured in liquid LB culture media (5 g of yeast extract, 10 g of tryptone, and 10 g of NaCl in 1 L ddH2O) at 37 °C for 12 h with shaking at 220 rpm and then were harvested by centrifugation at 12,000 rpm for 1 min.
Identification of strain GFP-2 and phylogenetic analysis
The strain GFP-2 was cultured in liquid LB media at 37 °C for 12 h with shaking at 220 rpm and then was harvested by centrifugation at 12,000 rpm for 1 min. The Genomic DNA was extracted using the method as described (Sun et al. 2016). The 16S rRNA sequence was amplified by PCR using the following primers: (F27) 5′-AGAGTTTGATCATGGCTCAG -3′ and (R1492) 5′-TACGGTTACCTTGTTACGAC-3′, with the extracted DNA as the template. The almost complete 16S rRNA sequence (1515 bp) of strain GFP-2 was identified on EzBiocloud service (Kim et al. 2012). Phylogenetic trees were reconstructed by the neighbor-joining (NJ; Saitou and Nei 1987) methods with the MEGA 5 program package (Tamura et al. 2011). Evolutionary distances were calculated according to the algorithm of the Kimura two-parameter model (Kimura 1980). OrthoANI values and Unweighted Pair Group Method with Arithmetic Mean (UPGMA) trees between strain GFP-2 and closely related strains were analyzed by Orthologous Average Nucleotide Identity Tool (OAT) (Lee et al. 2016).
Complete genome sequencing and gene prediction
The complete genome was obtained by using Pacbio RSII platform (Pacific Biosciences, CA, USA). After filter and quality control, all clean reads were assembled using the PacBio HGAP Analysis 2.0 and one circular chromosome with 0 gap was constructed for further analysis. Gene prediction was performed using Glimmer v. 3.02 (Delcher et al. 2007), and functions of the gene products were annotated by BLAST+ (Camacho et al. 2009) using NCBI-nr protein database (Coordinators 2017). The rRNA and tRNA genes were identified by using RNAmmer (Lagesen et al. 2007), tRNAscan-SE (Lowe and Eddy 1997) and Rfam (Griffiths-Jones et al. 2003) database. Classification of predicted genes and pathways were analyzed by using COGs (Tatusov et al. 2003) and KEGG (Ogata et al. 1999) databases. In addition, the RAST online service (http://rast.nmpdr.org/) (Aziz et al. 2008) was used for verification of gene prediction, annotation and classification. The genome circle was draw by CGView application (Grant and Stothard 2008). The antimicrobial peptides were predicted by BLAST+ using the antimicrobial peptide database (APD3)(Wang et al. 2016). The gene cluster for secondary metabolites and bacteriocins were predicted with online servers antiSMASH 3.0 (https://antismash.secondarymetabolites.org/) and BAGEL3 (http://bagel.molgenrug.nl/) (van Heel et al. 2013a; Weber et al. 2015).
Antimicrobial activity assay
The antimicrobial ability of strain GFP-2 was identified with inhibition zone method. In brief, (1) inoculated activated Bacillus subtilis BS168 (Gram-positive, OD600nm = 1.0) and Escherichia coli TG1 (Gram-negative, OD600nm = 1.0) as target bacteria into LB agar plates with the concentration of 1:100, v/v; (2) put sterilized oxford cup on pre-coating LB agar plates; (3) added 250 μl of supernatant and related thallus precipitate of strains GFP-2 (OD600nm = 2.0), or 250 μl of ampicillin (1 mg/ml) as the positive control, and then cultured the plates at 37 °C for 12 h; (4) measured the inhibition zones.
Glucan hydrolysis assay
The crude proteins in extracellular metabolites of Bacillus sp. GFP-2 were extracted by ammonium sulfate precipitation method from the culture supernatant. Then the crude proteins or recombinant β-1,3-1,4-glucanase was inoculated into the β-1,3-1,4-glucanase identification agar plates (containing 0.2% of glucan, 0.2% of NaNO3, 0.1% of K2HPO4, 0.05% of KCl, 0.05% of MgSO4, 0.001% of FeSO4, 0.005% of Congo red, and 2% agar) and the plates were incubated at 37 °C for 12 h.
Salmon feeding assay
The growth promoting ability of strain GFP-2 to salmon (Oncorhynchus mykiss) was evaluated by the weight increment and survival rate of salmon with the feed of strain GFP-2 (strain: fodder = 3:100, in wet weight) after 60 days.
Growth curve analysis
Overnight cultured bacteria were inoculated into 5 ml of liquid LB culture media at the ratio of 1:100, in the presence of control medium, 25 µg/ml ampicillin, or indicated amount of GFP-2 culture media, and cultured at 37 °C with shaking at 220 rpm. 0, 1, 2, 3, 4, 5, 6, 8, 10 and 12 h after the inoculation, the growth of bacteria was monitored using optical density (O.D) method at 600 nm.
Cloning of β-1,3-1,4-glucanase gene and heterogeneously expression in Escherichia coli (E. coli)
DNA sequence encoding β-1,3-1,4-glucanase was cloned from GFP-2 genome into the expression construct pET-28a(+) by PCR, which was transformed into E. coli BL21 (DE3) to express the recombinant β-1,3-1,4-glucanase. The recombinant protein was further purified through Ni-chelating affinity chromatography and stored at − 80 °C for further use.
Antibody production by immunizing rabbit with the recombinant β-1,3-1,4-glucanase
2.5 kg weight New Zealand rabbits were immunized with 1 mg of recombinant β-1,3-1,4-glucanase, together with 1 ml of Complete Freund’s Adjuvant (CFA), followed by a second immunization with 0.5 mg of recombinant protein and 0.5 ml of Incomplete Freund’s Adjuvant (IFA) after 15 days. 45 days after the first immunization, rabbit serum containing antibody against β-1,3-1,4-glucanase was collected by centrifugation of the blood at 5000 rpm for 15 min.
Detection of β-1,3-1,4-glucanase in the GFP-2 culture supernatant
The supernatant of strain GFP-2 culture was collected were analyzed by SDS-PAGE and western blot using the aforementioned rabbit serum containing antibody against β-1,3-1,4-glucanase.
Nucleotide sequence accession number
The complete genome sequence of Bacillus sp. GFP-2 was deposited in DDBJ/EMBJ/GenBank under the Accession Number CP021011.
The sequence of β-1,3-1,4-glucanase was deposited in GenBank under the Accession Number BankIt2108772 BSeq#1 MH260376.
Results
Isolation of a bacteria strain GFP-2 from shark intestine that could inhibit both Gram-positive and Gram-negative bacteria
To investigate the potential antimicrobial products from marine, we screened the bacteria from intestine contents from Whitespotted bamboo sharks, by performing the antimicrobial activity assay. And one of the isolated strains, which was named as GFP-2, showed great inhibition effects against Bacillus subtilis BS168, a Gram-positive bacterium, and Escherichia coli TG1, a Gram-negative bacterium. As shown in Fig. 1A, both the precipitated bacteria (c) and the culture supernatant (d), especially the latter, formed obvious inhibition zones in the BS168 culture plate, 1.34 cm and 1.70 cm in diameter, respectively, reaching 49.6 and 63.0% of that in the positive control (b, ampicillin). Similarly, both the precipitated bacteria (c) and the culture supernatant (d) of GFP-2 culture inhibited TG1 growth, with 1.28 cm and 1.56 cm inhibition zones in diameter, respectively, 56.4 and 68.7% of that in the positive control (b, ampicillin), as shown in Fig. 1B. Further growth curve analysis showed that, with the presence of GFP-2 culture supernatant, the growth of both BS168 and TG1 were greatly inhibited, similar to that with the presence of 25 µg/ml of ampicillin (Fig. 1C, D). These results indicated that strain GFP-2 can effectively inhibit the growth of Gram-positive and Gram-negative strains, possibly through its secondary metabolites secreted into the supernatant.
Fig. 1.
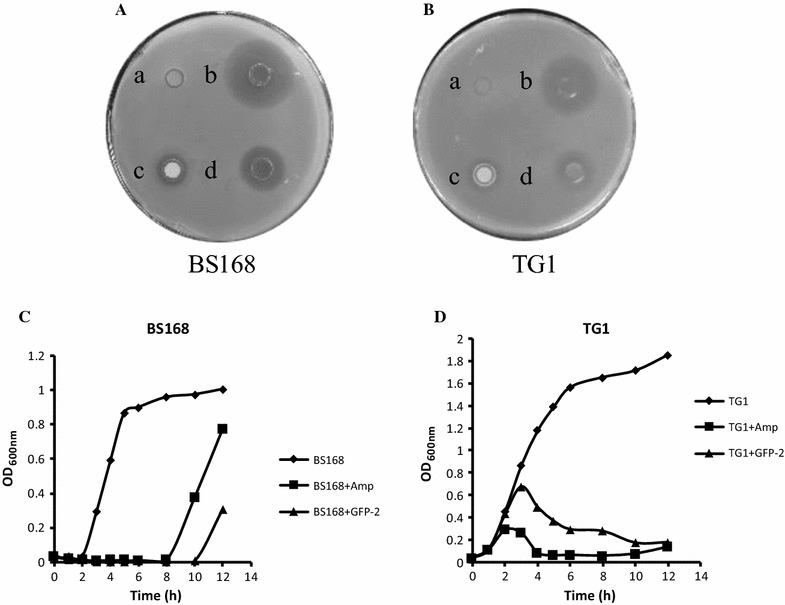
Inhibition of Bacillus subtilis BS168 and Escherichia coli TG1 by Bacillus sp. GFP-2. Activated Bacillus subtilis BS168 (Gram-positive, OD600nm = 1.0) (A) and Escherichia coli TG1 (Gram-negative, OD600nm = 1.0) (B) were inoculated as target bacteria into LB agar plates. Sterilized oxford cups were put on pre-coating LB agar plates and 250 μl of LB medium (a, negative control), supernatant (d) and related thallus precipitate (c) of strains GFP-2 (OD600nm = 2.0), or ampicillin (1 mg/ml) (b, positive control), were added onto the plates, and the plates were incubated at 37 °C for 12 h. Overnight cultured BS168 (OD600nm = 3.28) (C) and TG1 (OD600nm = 1.86) (D) were inoculated, respectively, into 5 ml of liquid LB culture media at the ratio of 1:100, in the presence of control medium (diamond), 25 µg/ml ampicillin (square), or GFP-2 culture media (100 µl for TG1, 500 µl for BS168) (triangle), and cultured at 37 °C with shaking at 220 rpm. 0, 1, 2, 3, 4, 5, 6, 8, 10 and 12 h after the inoculation, the growth of bacteria was monitored using optical density (O.D) method at 600 nm
Strain GFP-2 could facility the growth of salmon (Oncorhynchus mykiss)
Bacterial infection and the following low survival rate and low specific growth rate greatly impaired the pisciculture. To test if the anti-bacteria activities of GFP-2 could benefit the growth of fishes, we fed salmon (Oncorhynchus mykiss) with the Diet 1 (Negative control with normal diet) or Diet 2 (normal diet add with 3% inactivated GFP-2 strain) and estimated the growth performance, such as Gained weight, Specific growth rate (%), Feed conversion ratio and Survival rate (%). As shown in Table 1, after 30 days of feeding with diet 2 containing strain GFP-2, the salmon showed greatly higher gained weight (3.8 fold vs. 1.1 fold) and higher survival rate (79.0% vs. 29.8%) than those with the control diet. These results indicated that the antimicrobial activities of the strain GFP-2 could greatly benefit the growth of fishes and could be used as a new potential antimicrobial agent in the pisciculture.
Table 1.
Growth performances and feed utilization of the Oncorhynchus mykiss fed with different diets: Diet 1 (Negative control with normal diet), Diet 2 (normal diet add with 3% Bacillus sp. GFP-2)
| Diet 1 | Diet 2 | |
|---|---|---|
| Initial weight (g) | 300 | 300 |
| Final weight (g) | 631 | 1442 |
| Gained weight (g) | 331 | 1142 |
| Specific growth rate (%) | 11.03 | 38.07 |
| Feed conversion ratio | 1.27 | 0.37 |
| Survival rate (%) | 29.80 | 79.00 |
Final weight and Initial weight were calculated in average; Gained weight = Final weight (g) − Initial weight (g); Specific growth rate = 100* (ln Final weight − ln Initial weight)/Duration of experiment (60 days); Feed conversion ratio = feed given (dried weight)/Gained weight (wet weight); Survival rate (%) = (final fish number/initial fish number) *100
Strain GFP-2 was identified as a novel strain of Bacillus
To identify the species of strain GFP-2, 16S rRNA Gene Sequencing was performed and MEGA program was used to establish the phylogenetic tree. As shown in Fig. 2a, strain GFP-2 shared 99.9% of 16S rRNA genetic similarities with B. siamensis KCTC 13613T and B. velezensis CR-502T, 99.7% with B. nakamurai NRRL B-41091T, B. amyloliquefaciens DSM 7T and B. subtilis subsp. subtilis NCIB 3610T, but less than 99.5% with the other Bacillus species. And according to this 16S rRNA gene based neighbour-joining tree, strain GFP-2 is located in a separate clade with B. velezensis CR-502T, B. amyloliquefaciens DSM 7T and B. siamensis KCTC 13613T, indicating their closely phylogenetic relationships (Fig. 2a). To further clarify the phylogenetic relationship of strain GFP-2 with these three species, genome-based ANI values were calculated and UPGMA tree were reconstructed (Fig. 2b). The ANI values of strain GFP-2 with B. siamensis KCTC 13613T and B. amyloliquefaciens DSM 7T are 94.1 and 94.4, respectively, lower than the species threshold (95–96%) (Goris et al. 2007), and its ANI values with B. velezensis CR-502T and CBMB205 are 97.8 and 97.7, respectively, showing that strain GFP-2 is a novel strain of B. velezensis. This strain has been uploaded and stored in the China General Microbiological Culture Collection Center (CGMCC No. 13337).
Fig. 2.
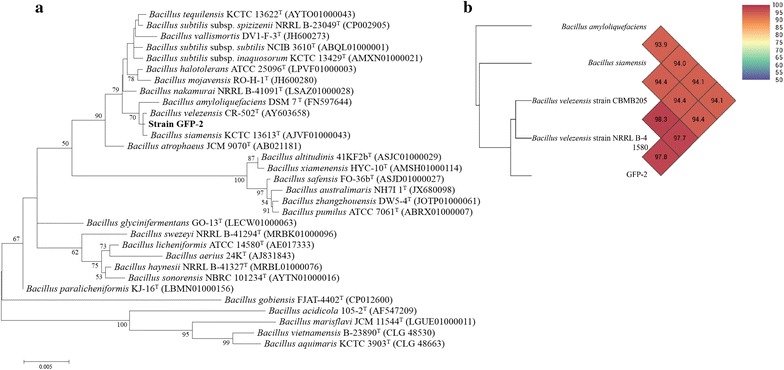
The phylogenetic relationship of Bacillus sp. GFP-2 and related strains. The 16S rRNA sequence of Bacillus sp. GFP-2 was amplified by PCR and was identified using EzBiocloud service. a Phylogenetic trees were reconstructed by the neighbor-joining methods with the MEGA 5 program package. Evolutionary distances were calculated according to the algorithm of the Kimura two-parameter model. b OrthoANI values and Unweighted Pair Group Method with Arithmetic Mean (UPGMA) trees between strain GFP-2 and closely related strains were analyzed by Orthologous Average Nucleotide Identity Tool (OAT)
Genes encoding antimicrobial peptides and gene clusters for secondary metabolites and bacteriocins were predicted in the genome of Bacillus sp. GFP-2
As Bacillus sp. GFP-2, especially its culture supernatant, showed inhibitory effects on both Gram-positive and Gram-negative strains and could benefit the growth of salmon, we proposed that this strain might produce some metabolites which may play important roles in its antimicrobial activities. So complete gene sequencing was performed to analyze the genome of Bacillus sp. GFP-2. The results showed that the genome of strain Bacillus sp. GFP-2 (CGMCC 13337) is one 3975,220 bp circular DNA, with the G + C content of 46.4%, in which, a total of 3711 coding sequences (CDS) were predicted, including 19 rRNA and 86 tRNA (Fig. 3a). The summary of general features and statistics of the genome is listed in Tables 2, 3. Among the CDSs, 2893 were classified into clusters of orthologous groups (COGs) categories, the major of which were related to amino acid transport and metabolism (E), transcription (K), carbohydrate transport and metabolism (G), inorganic ion transport and metabolism (P) and signal transduction mechanisms (T) (Fig. 3b). 2881 CDS were classified into KEGG categories, the major of which were related to pathways of global map (33.1%), including biosynthesis of secondary metabolites and metabolic pathways, amino acid metabolism (16.3%) and carbohydrate metabolism (15.6%) (Fig. 3c).
Fig. 3.
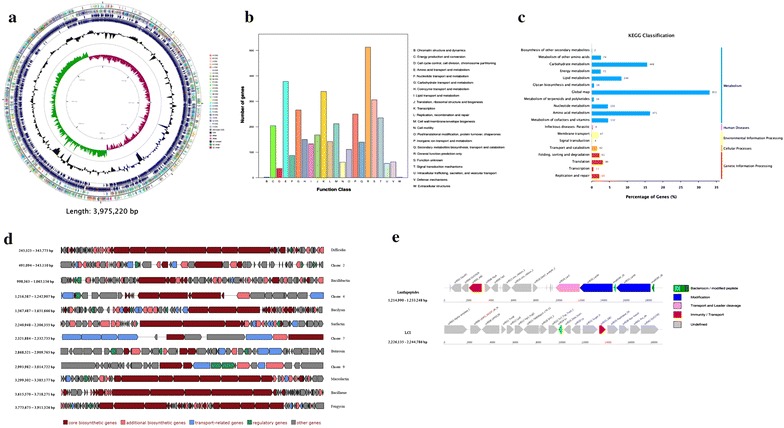
Complete genome sequencing and gene prediction of Bacillus sp. GFP-2. The complete genome was obtained by using Pacbio RSII platform (Pacific Biosciences, CA, USA). After filter and quality control, all clean reads were assembled using the PacBio HGAP Analysis 2.0 and one circular chromosome with 0 gap was constructed for further analysis. The genome of Bacillus sp. GFP-2 was shown as a circular map: from the outer circle inward, each circle displays information about the genome of forward COG-assigned CDS, reverse COG-assigned CDS, forward CDS, reverse CDS, G + C ratio and GC Skew (a). Gene prediction was performed using Glimmer v. 3.02, and functions of the gene products were annotated by BLAST + using NCBI-nr protein database. The rRNA and tRNA genes were identified by using RNAmmer, tRNAscan-SE and Rfam database. Classification of predicted genes and pathways were analyzed by using COGs (b) and KEGG (c) databases. In addition, the RAST online service (http://rast.nmpdr.org/) was used for verification of gene prediction, annotation and classification. The genome circle was draw by CGView application. The antimicrobial peptides were predicted by BLAST+ using the antimicrobial peptide database (APD3). The gene cluster for secondary metabolites (d) and bacteriocins (e) were predicted with online servers antiSMASH 3.0 (https://antismash.secondarymetabolites.org/) and BAGEL3 (http://bagel.molgenrug.nl/) (d)
Table 2.
General features of Bacillus sp. GFP-2 and MIGS mandatory information
| Items | Descriptions |
|---|---|
| General features | |
| Classification | Domain Bacteria |
| Phylum Firmicutes | |
| Class Firmibacteria | |
| Order Bacillales | |
| Family Bacillaceae | |
| Genus Bacillus | |
| Gram strain | Positive |
| Cell shape | Rod |
| Color of colonies | White |
| Temperature | 10–40 °C |
| Optimal NaCl concentration | 1% |
| Optimal pH | 7.0 |
| Observed biotic relationship | Free-living |
| MIGS data | |
| Investigation type | Bacteria_archaea |
| Project name | Genome sequence of Bacillus sp. GFP-2 |
| Latitude and longitude | Not reported |
| Depth | Not reported |
| Geographical location | Not reported |
| Collection date | Not reported |
| Environment (biome) | Shark intestine |
| Number of replicons | 1 |
| Estimated size | 3,975,220 bp |
| Source material identifiers | GFP-2 = CGMCC 13337 |
| Trophic level | Chemoorganotroph |
| Relationship to oxygen | Aerobic |
| Library reads sequenced | The large smrtbell gdna protocol (Pacific Biosciences, USA) |
| Sequencing method | A pacbio RS II platform |
| Assembly data | |
| Assembly method | De novo assembled using the PacBio hierarchical genome assembly process (HGAP)/Quiver software |
| Estimated error rate | 99.99% (average reference consensus) |
| Method of calculation | Estimation from HGAP Assembly quality scores |
| Assembly name | HGAP Assembly version 2 (Pacific Biosciences, USA) |
Table 3.
General features of the genome of Bacillus sp. GFP-2
| Attribute | Value |
|---|---|
| Size (bp) | 3,975,220 |
| DNA G + C content (%) | 46.4 |
| CDSs | 3711 |
| CDSs assigned to COGs (percentages) | 2893 (78.0%) |
| CDSs assigned to KEGG (percentages) | 2881 (77.6%) |
| rRNA operon (16S-5S-23S rRNA) | 1 |
| tRNAs | 86 |
After blasting the genome sequence with the antimicrobial peptide database (APD3), three hypothetical AMPs were predicted. These three predicted AMPs (peg.781, peg.1300 and peg.2297) were closely related to LCI, YFGAP and hGAPDH, respectively (Table 4). In addition, the prediction of gene clusters for secondary metabolites and bacteriocins were performed and 12 clusters for secondary metabolites (Fig. 3d) and two clusters for bacteriocins, including Lanthipeptides (cluster 1) and LCI (cluster 2), (Fig. 3e) were found and identified. Eight clusters of secondary metabolite have specific functions, including the biosynthesis of difficidin (cluster 1), bacillibactin (cluster 3), bacilysin (cluster 5), surfactin (cluster 6), butirosin (cluster 8), macrolactin (cluster 10), bacillaene (cluster 11) and fengycin (cluster 12) (Fig. 3d). The above-mentioned AMPs, secondary metabolites and bacteriocins have been reported having antimicrobial activities against the bacteria and fungi pathogens, indicating that Bacillus sp. GFP-2 may function as antimicrobial agent through producing these molecules.
Table 4.
Sequences of predicted AMPs by Bacillus sp. GFP-2
| Predicted AMPs | Sequences | Related AMPs |
|---|---|---|
| peg.781 | MKFKKVLTGSALSLALLMSAAPAFAASPTASASVENSPISTKADVGINAIKLVQSPDGNFAASFVLNGTKWIFKSKYYDSSKGYWVGIYESVDK | LCI (AP01154) |
| peg.1300 | MKVKVAINGFGRIGRMVFRKAMEDDQIQITAINASYPPETLAHLIKYDTIHGKYDQEVETADDSLIVNGKHIMLFNRRDPRELPWKECGIDIVVEATGKFNSKEKAMSHIEAGAKKVILTAPGKNEDVTIVMGVNEEQFNPDEHVIISNASCTTNCLAPVVKVLDQEFGIESGLMTTVHAYTNDQKNIDNPHKDLRRARACGESIIPTSTGAAKALSLVLPHLKGKLHGLALRVPVPNVSLVDLVCDLKTDVSAEQVNAAFQRAAKTSMYGILDYSDEPLVSSDYNTNSHSAIIDGLTTMVMEDRKVKVLAWYDNEWGYSCRVVDLIRHVAARMKHPSAV | YFGAP (AP02012) |
| peg.2297 | MAVKVGINGFGRIGRNVFRAALNNPEVEVVAVNDLTDANMLAHLLQYDSVHGKLDAEVKVDGSNLVVNGKTIEVSAERDPAKLSWGKQGVEIVVESTGFFTKRADAAKHLEAGAKKVIISAPANEEDITIVMGVNEDKYDAANHHVISNASCTTNCLAPFAKVLNDKFGIKRGMMTTVHSYTNDQQILDLPHKDYRRARAAAENIIPTSTGAAKAVSLVLPELKGKLNGGAMRVPTPNVSLVDLVAELNKDVTAEDVNAALKEAAEGDLKGILGYSEEPLVSGDYNGNANSSTIDALSTMVMEGSMVKVISWYDNESGYSNRVVDLAAYIAKQGL | hGAPDH (AP02017) |
Bacillus sp. GFP-2 could express β-1,3-1,4-glucanase
According to the complete genome sequencing (Fig. 3a), the gene encoding β-1,3-1,4-glucanase was also found in the genome of Bacillus sp. GFP-2. To further confirm if Bacillus sp. GFP-2 could encode and express β-1,3-1,4-glucanase, the crude proteins in extracellular metabolites of Bacillus sp. GFP-2 were extracted by ammonium sulfate precipitation method and the glucan hydrolysis assay was performed. As shown in Fig. 4A, an obvious hydrolysis zone was observed in the plate inoculated with crude proteins (b), indicating that β-1,3-1,4-glucanase might exist in extracellular metabolites of Bacillus sp. GFP-2. Furthermore, the gene sequence encoding β-1,3-1,4-glucanase was successfully cloned from the genome of Bacillus sp. GFP-2 into an expressing construct and heterogeneously expressed in E. coli (Fig. 4B, line 2). And the purified recombinant β-1,3-1,4-glucanase also had the activity of glucan hydrolysis, as shown in Fig. 4C with obvious hydrolysis zones (c and d). Further study suggested that the purified recombinant β-1,3-1,4-glucanase also had inhibitory effects on the growth of Bacillus subtilis WB800N and Escherichia coli BL21 (Fig. 4D), confirming its antimicrobial activities. We further immunized rabbits with the recombinant β-1,3-1,4-glucanase to produce an antibody against β-1,3-1,4-glucanase. And β-1,3-1,4-glucanase was specifically detected by this antibody when the crude proteins from Bacillus sp. GFP-2 culture supernatant were analyzed by SDS-PAGE and western blot (Fig. 4E, lane 1), further confirming that Bacillus sp. GFP-2 indeed could express β-1,3-1,4-glucanase, which may, at least partially, contribute to its antimicrobial activities.
Fig. 4.
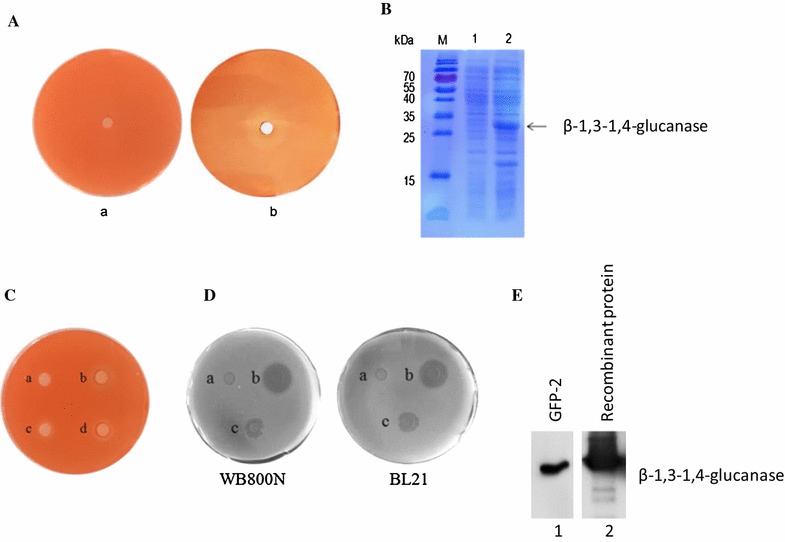
Detection of β-1,3-1,4-glucanase in the culture supernatant of Bacillus sp. GFP-2 and conformation of its antimicrobial activities. A The crude proteins in extracellular metabolites of Bacillus sp. GFP-2 were extracted by ammonium sulfate precipitation method from the culture supernatant. Then 200 μl of Na3PO4 buffer (a), or the crude proteins (b) was inoculated into the β-1,3-1,4-glucanase identification agar plates (see “Materials and methods”) and the plates were incubated at 37 °C for 12 h. B DNA sequence encoding β-1,3-1,4-glucanase was cloned from GFP-2 genome into the expression construct pET-28a(+) by PCR, which was transformed into E. coli BL21(DE3) to express the recombinant β-1,3-1,4-glucanase with (line 2) or without (line 1) the stimulation of IPTG. The recombinant protein was further purified through Ni-chelating affinity chromatography and analyzed by Western blot and Coomassie blue staining. C 200 μl of elution buffer (a), supernatant of engineered bacteria lysate (b), or protein elution solutions (c and d) were inoculated into the β-1,3-1,4-glucanase identification agar plates (see “Materials and methods”) and the plates were incubated at 37 °C for 12 h. D Activated Bacillus subtilis WB800 N (left) and Escherichia coli BL21 (right) were inoculated as target bacteria into LB agar plates. Sterilized oxford cups were put on pre-coating LB agar plates and 250 μl of LB medium (a, negative control), ampicillin (1 mg/ml) (b, positive control), or purified recombinant β-1,3-1,4-glucanase were added onto the plates, and the plates were incubated at 37 °C for 12 h. E GFP-2 culture media (10 μl, line 1) and the recombinant β-1,3-1,4-glucanase (10 μl, 0.913 μg/μl) (line 2) (positive control, see “Materials and methods” for details) were analyzed by SDS-PAGE and western blot using the rabbit serum containing antibody against β-1,3-1,4-glucanase (see “Materials and methods” for details)
Discussion
In this study, a new strain Bacillus sp. GFP-2 was isolated from the intestine of Whitespotted bamboo shark and could effectively inhibit the growth of Gram-positive and Gram-negative strains, showing around 50–70% inhibition ability of ampicillin. In addition, when fed additionally with strain GFP-2, as the inhibition agents of pathogenic bacteria, the growth of salmon (Oncorhynchus mykiss) can be greatly promoted, indicating that strain GFP-2 can effectively inhibit the growth of pathogenic bacteria in the intestine of other fishes besides shark.
After complete genome sequencing, genes encoding AMPs including LCI, YFGAP and hGAPDH-like AMPs and gene clusters for secondary metabolites including difficidin, bacillibactin, bacilysin, surfactin, butirosin, macrolactin, bacillaene and fengycin, and bacteriocins including Lanthipeptides and LCI, were predicted in the genome of strain GFP-2. LCI, an antimicrobial peptide isolated from Bacillus subtilis, shows very strong antagonistic activity against the Gram-negative pathogen Xanthomonas campestris pv Oryzea of rice leaf-blight disease and Gram-negative bacterium Pseudomonas solanacearum PE1 (Gong et al. 2011). YFGAP, a GAPDH-related antimicrobial peptide isolated from the skin of yellowfin tuna, have antimicrobial activity against Gram-positive bacteria, such as Bacillus subtilis, Micrococcus luteus, and Streptococcus iniae. And hGAPDH is a human GAPDH fragment peptide from human placental tissue exhibiting antifungal activity. The existence of LCI, YFGAP, hGAPDH-like AMPs in strain GFP-2 may partially explain its inhibitory effects on Gram negative/positive bacterium. Additionally, YFGAP, hGAPDH-like AMPs are also first predicted in strains of genus Bacillus.
Of the predicted secondary metabolites, difficidin and bacilysin have antibacterial activity against Xanthomonas oryzae rice pathogens (Wu et al. 2015). Surfactins and fengycins are considered as crucial components in control of plant diseases as they act as antifungal and antibacterial metabolites and have been shown to stimulate plant defense system by inducing systemic resistance (Aleti et al. 2016). Butirosin, an aminoglycoside antimicrobial peptide, is known as the important class of agents, especially against Mycobacterium tuberculosis (Kudo and Eguchi 2009). Macrolactin are polyene macrolides containing a 24-membered lactone ring and shows antibiotic effects against vancomycin-resistant Enterococci and methicillin-resistant Staphylococcus aureus (Kim et al. 2011). Bacillaene was first found to inhibit bacterial protein synthesis, such as, it can selectively inhibit antagonistic fungi of Termitomyces and has recently been implicated in interspecies interactions (Muller et al. 2014; Um et al. 2013). Among the two predicted bacteriocins, lanthipeptides are peptides that contain several post-translationally modified amino acid residues and commonly show considerable antimicrobial activity (van Heel et al. 2013b). Meanwhile, the existence of gene cluster for LCI is strong supporting evidence indicating strain GFP-2 can produce LCI. The prediction of these genes in the genome of Bacillus sp. GFP-2 indicated that Bacillus sp. GFP-2 may function as antimicrobial agent through, at least partially, producing these molecules.
The identification of the gene encoding β-1,3-1,4-glucanase in the genome of strain GFP-2 and the detection of β-1,3-1,4-glucanase in the crude proteins from Bacillus sp. GFP-2 culture supernatant confirmed that Bacillus sp. GFP-2 indeed could express β-1,3-1,4-glucanase, which may contribute to its antimicrobial activities.
Our study indicated that the newly isolated strain Bacillus sp. GFP-2 could act as effective antimicrobial agent by producing AMPs, secondary metabolites and bacteriocins. The study of detailed mechanism is still undergoing. These results can enhance a better understanding of the mechanism of inhibition abilities in genus Bacillus and provide a useful tool for biotechnology study in antimicrobial compounds, which may replace abused antibiotics in prevention and clearance of pathogenic bacteria.
Authors’ contributions
KL, ZL, JW and GX conceived and designed the experiments; JW and GX did most of the experiments; YJ confirmed the expression and activity of the recombinant β-1,3-1,4-glucanase; CS, LZ and GL did the gene prediction; RX, LW and HF helped with the antimicrobial activity assay; DW and JC helped with the glucan hydrolysis assay and antibody preparation; KL, CS, JW and GX analyzed the data and wrote this manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Huize Dianze Aquaculture Co. LTD. for their assistance in the feeding of Salmon.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data and material are fully available without restriction. And the dataset supporting the conclusions of this article is included within the article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Experimental protocols were reviewed and approved by the Animal Ethic Committee of Zhejiang Sci-Tech University (No. 20161001).
Funding
This work was funded by National High Technology Research and Development Program (No. 2012ZX09102301-009), Graduate Innovation Research Project of Zhejiang Sci-Tech University (YCX16035), Science Foundation of Zhejiang Sci-Tech University (15042169-Y and16042186-Y).
The funders had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- AMPs
antimicrobial peptides
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- YFGAP
yellowfin tuna glyceraldehyde-3-phosphate dehydrogenase-related antimicrobial peptide
- GH16
glycosyl hydrolase family 16
- UPGMA
unweighted pair group method with arithmetic mean
- OAT
Orthologous Average Nucleotide Identity Tool
- O.D
optical density
- CFA
complete Freund’s adjuvant
- IFA
incomplete Freund’s adjuvant
- CDS
coding sequences
- COGs
clusters of orthologous groups
Footnotes
Jia Wu and Guoqiang Xu contributed equally to this work and are considered as co-first author
Contributor Information
Jia Wu, Email: wujia_719@126.com.
Guoqiang Xu, Email: xuguoqiang022@163.com.
Yangyang Jin, Email: 1456718553@qq.com.
Cong Sun, Email: michael_sc@sina.com.
Li Zhou, Email: 457897197@qq.com.
Guodong Lin, Email: lgd19911128@126.com.
Rong Xu, Email: xurong20172020@163.com.
Ling Wei, Email: 1715857691@qq.com.
Hui Fei, Email: feihui@zju.edu.cn.
Dan Wang, Email: february_dan@aliyun.com.
Jianqing Chen, Email: cjqgqj@126.com.
Zhengbing Lv, Email: zhengbingl@zstu.edu.cn.
Kuancheng Liu, Phone: +86-0571-86843192, Email: kliu@zstu.edu.cn.
References
- Aleti G, Lehner S, Bacher M, Compant S, Nikolic B, Plesko M, Schuhmacher R, Sessitsch A, Brader G. Surfactin variants mediate species-specific biofilm formation and root colonization in Bacillus. Environ Microbiol. 2016;18(8):2634–2645. doi: 10.1111/1462-2920.13405. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017;45(D1):D12–D17. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplewski L, Bax R, Clokie M, Dawson M, Fairhead H, Fischetti VA, Foster S, Gilmore BF, Hancock REW, Harper D, Henderson IR, Hilpert K, Jones BV, Kadioglu A, Knowles D, Olafsdottir S, Payne D, Projan S, Shaunak S, Silverman J, Thomas CM, Trust TJ, Warn P, Rex JH. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect Dis. 2016;16(2):239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong WB, Wang JF, Chen ZL, Xia B, Lu GY. Solution structure of LCI, a novel antimicrobial peptide from Bacillus subtilis. Biochemistry. 2011;50(18):3621–3627. doi: 10.1021/bi200123w. [DOI] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31(1):439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Kim HK, Kim KM, Kim CK, Jeong MH, Ko CY, Moon KH, Kang JS. Antibacterial activities of macrolactin A and 7-O-succinyl macrolactin A from Bacillus polyfermenticus KJS-2 against vancomycin-resistant enterococci and methicillin-resistant Staphylococcus aureus. Arch Pharmacal Res. 2011;34(1):147–152. doi: 10.1007/s12272-011-0117-0. [DOI] [PubMed] [Google Scholar]
- Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide-sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kudo F, Eguchi T. Biosynthetic genes for aminoglycoside antibiotics. J Antibiot. 2009;62(9):471–481. doi: 10.1038/ja.2009.76. [DOI] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66(2):1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- Li GG, Liu BS, Shang YJ, Yu ZQ, Zhang RJ. Novel activity evaluation and subsequent partial purification of antimicrobial peptides produced by Bacillus subtilis LFB112. Ann Microbiol. 2012;62(2):667–674. doi: 10.1007/s13213-011-0303-9. [DOI] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Hakansson J, Ringstad L, Bjorn C. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol. 2016;6:194. doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahiel MA. Molecular biology and regulatory mechanisms of antibiotic production in Bacillus. Die Naturwissenschaften. 1992;79(5):202–212. doi: 10.1007/BF01227129. [DOI] [PubMed] [Google Scholar]
- Marx R, Stein T, Entian KD, Glaser SJ. Structure of the Bacillus subtilis peptide antibiotic subtilosin A determined by 1H-NMR and matrix assisted laser desorption/ionization time-of-flight mass spectrometry. J Protein Chem. 2001;20(6):501–506. doi: 10.1023/A:1012562631268. [DOI] [PubMed] [Google Scholar]
- Muller S, Strack SN, Hoefler BC, Straight PD, Kearns DB, Kirby JR. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus. Appl Environ Microbiol. 2014;80(18):5603–5610. doi: 10.1128/AEM.01621-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Zuber P. Molecular biology of antibiotic production in Bacillus. Crit Rev Biotechnol. 1990;10(3):223–240. doi: 10.3109/07388559009038209. [DOI] [PubMed] [Google Scholar]
- Ng TB, Cheung RC, Wong JH, Bekhit AA, Bekhit Ael D. Antibacterial products of marine organisms. Appl Microbiol Biotechnol. 2015;99(10):4145–4173. doi: 10.1007/s00253-015-6553-x. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabate DC, Audisio MC. Inhibitory activity of surfactin, produced by different Bacillus subtilis subsp. subtilis strains, against Listeria monocytogenes sensitive and bacteriocin-resistant strains. Microbiol Res. 2013;168(3):125–129. doi: 10.1016/j.micres.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sumi CD, Yang BW, Yeo IC, Hahm YT. Antimicrobial peptides of the genus Bacillus: a new era for antibiotics. Can J Microbiol. 2015;61(2):93–103. doi: 10.1139/cjm-2014-0613. [DOI] [PubMed] [Google Scholar]
- Sun C, Fu GY, Zhang CY, Hu J, Xu L, Wang RJ, Su Y, Han SB, Yu XY, Cheng H, Zhang XQ, Huo YY, Xu XW, Wu M. Isolation and complete genome sequence of Algibacter alginolytica sp. nov., a novel seaweed-degrading Bacteroidetes bacterium with diverse putative polysaccharide utilization loci. Appl Environ Microbiol. 2016;82(10):2975–2987. doi: 10.1128/AEM.00204-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um S, Fraimout A, Sapountzis P, Oh DC, Poulsen M. The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp. that inhibit potentially antagonistic fungi. Sci Rep. 2013;3:3250. doi: 10.1038/srep03250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP. BAGEL3: Automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013;41(Web Server issue):W448–W453. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel AJ, Mu D, Montalban-Lopez M, Hendriks D, Kuipers OP. Designing and producing modified, new-to-nature peptides with antimicrobial activity by use of a combination of various lantibiotic modification enzymes. ACS Synth Biol. 2013;2(7):397–404. doi: 10.1021/sb3001084. [DOI] [PubMed] [Google Scholar]
- Wang GS, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Muller R, Wohlleben W, Breitling R, Takano E, Medema MH. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43(W1):W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LM, Wu HJ, Chen L, Yu XF, Borriss R, Gao XW. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci Rep-Uk. 2015;5:12975. doi: 10.1038/srep12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and material are fully available without restriction. And the dataset supporting the conclusions of this article is included within the article.


