Abstract
Prohibitin (PHB) was originally isolated and characterized as an anti-proliferative gene in rat liver. The evolutionarily conserved PHB gene encodes two human protein isoforms with molecular weights of ~33 kDa, PHB1 and PHB2. PHB1 and PHB2 belong to the prohibitin domain family, and both are widely distributed in different cellular compartments such as the mitochondria, nucleus, and cell membrane. Most studies have confirmed differential expression of PHB1 and PHB2 in cancers compared to corresponding normal tissues. Furthermore, studies verified that PHB1 and PHB2 are involved in the biological processes of tumorigenesis, including cancer cell proliferation, apoptosis, and metastasis. Two small molecule inhibitors, Rocaglamide (RocA) and fluorizoline, derived from medicinal plants, were demonstrated to interact directly with PHB1 and thus inhibit the interaction of PHB with Raf-1, impeding Raf-1/ERK signaling cascades and significantly suppressing cancer cell metastasis. In addition, a short peptide ERAP and a natural product xanthohumol were shown to target PHB2 directly and prohibit cancer progression in estrogen-dependent cancers. As more efficient biomarkers and targets are urgently needed for cancer diagnosis and treatment, here we summarize the functional role of prohibitin domain family proteins, focusing on PHB1 and PHB2 in tumorigenesis and cancer development, with the expectation that targeting the prohibitin domain family will offer more clues for cancer therapy.
Facts
PHB1 and PHB2 are widely distributed in cells and correlate with diverse diseases.
PHB1 and PHB2 are involved in multiple biological processes in tumorigenesis like proliferation, metastasis and apoptosis.
PHB1 and PHB2 are regulated by transcriptional regulation, post-transcriptional regulation and protein modification in cancer cells.
Several small molecular inhibitors targeting PHB1 and PHB2 have significant impacts on cancer therapy.
Questions
Why PHB1 exerts controversial impacts on cell proliferation in different cancers?
Which transcription factors regulate PHB1 expression in cancer cells?
What are the mechanisms on regulation of PHB2 in cancer cells?
Can PHB1 or PHB2 inhibitors enhance the treatment efficiency of chemotherapeutic drugs?
Introduction
The prohibitin (PHB) gene was originally isolated and characterized as a candidate anti-proliferative gene in rat liver cells1, and the homologous human gene maps to chromosome 17q21-212–4, encoding two ~33 KD proteins, PHB1 and PHB25. PHB1 and PHB2 belong to the evolutionarily conserved band-7 family, or prohibitin domain family5. Both PHB1 and PHB2 are widely distributed in different cellular compartments including the mitochondria, nucleus and cell membrane, with diverse biological functions. PHB1 located in the inner mitochondrial membrane interacts with PHB2 to stabilize the mitochondria. As mitochondria are the main energy machines for biological activities in cells, expression changes in PHB1 or PHB2 in the mitochondria always induce mitochondria-related processes such as apoptosis6, 7.
PHB1 protein located in the cell membrane functions as viral or bacterial receptors to facilitate the entry of these microorganisms into host cells8. Meanwhile, PHB1 located in the lipid raft of the cell membrane interacts with and activates Raf-1, an evolutionarily conserved oncogene that activates ERK and promotes cancer development9. In addition, membrane-localized PHB2 also promotes cancer cell migration10. Moreover, PHB1 located in the nucleus binds to several transcription factors, such as p53, E2F, and pRb. Early studies claimed that PHB1 accumulates in the nucleus to induce cell cycle arrest and inhibit cell proliferation; however, this theory was challenged in recent years5, 11, 12. Nuclear PHB2 seems to be primarily involved in centromeric cohesion protection and promotion of cell growth13.
PHB1 and PHB2 are also involved in many biological processes related to tumorigenesis. Studies have found that overexpression of PHB1 and PHB2 results in cancer cell metastasis and apoptosis10, 14–16. Interestingly, although PHB1 was first identified as an anti-proliferative protein, the role of PHB1 in cancer cell proliferation remains controversial17, 18. As PHB1 and PHB2 have been shown to be differentially expressed in multiple cancers, PHB1 and PHB2 are potentially useful as new biomarkers and targets in cancer diagnosis and treatment.
Natural products provide an invaluable source of medicinal leads, presenting a significant impact on drug development. RocA is isolated from Aglaia species (Meliaceae), and fluorizoline is synthetized based on natural products from medicinal plants19–21. Both RocA and fluorizoline have been reported to interact with PHB1 directly and disrupt the interaction of PHB1 and Raf-1, therefore inhibits the activation of Raf-1/ERK signaling cascades and suppresss cancer cell growth and metastasis14, 22. RocA was also shown to significantly suppress cancer development in some drug-resistant cells23. Moreover, ERAP, a short synthetic peptide, and xanthohumol, a natural product from medical plants, were demonstrated to suppress cancer cell proliferation by targeting PHB224, 25, indicating that drugs targeting PHB1 and PHB2 may be a promising strategy for cancer treatment.
Although there have been substantial advances in our understanding on the mechanisms of tumorigenesis, efficient remedies for diagnosis and treatment of cancer are still lacking. Considering the special localization and significant roles of prohibitin domain family proteins in cancer, the value of PHB1 and PHB2 in cancer treatment warrants further detailed study. Here, we summarize the current understanding on the functional role of PHB1 and PHB2 in biological processes, particularly tumorigenesis.
Location and function of PHB1 and PHB2
The microenvironment in which proteins reside offers the perfect conditions to exert their function, therefore, localization has a large impact on protein function. According to the literature, both PHB1 and PHB2 are ubiquitously expressed, either in circulating form or in multiple cellular compartments, including the mitochondria, nucleus and plasma membrane6, 11, 26, 27.
PHB1 and PHB2 locate in the inner mitochondrial membrane
PHB1 located in the inner mitochondrial membrane maintains mitochondrial stability by interacting with PHB2 to form a PHB1/PHB2 complex when mitochondria encounter metabolic stress6, 28–30. This process modulates the balance between mitochondrial fusion and fission events31, 32, thus maintaining a healthy mitochondrial network that protects cells from mitochondria-related apoptosis7, 33, 34. Former studies reported that loss of PHB1 and PHB2 in podocytes disrupts the activation of mTORC1 and inhibits kidney filtration31, 35. Levels of mitochondrial PHB1 are significantly decreased in the olfactory bulb, indicating that PHB1 is a driver of olfactory neurodegeneration in intermediate and advanced Alzheimer’s disease stages36. Another study demonstrated that loss of PHB2 from the mitochondrial membrane leads to tau hyperphosphorylation and neurodegeneration37.
Interestingly, experiments performed in transgenic mice illustrated that neuronal expression of mitochondrial PHB1 confers profound neuroprotection38, 39. A proteomics comparison between the substantia nigra (SN) and ventral tegmental area (VTA) dopaminergic neurons also demonstrated neuroprotection of mitochondrial PHB1 in Parkinson’s disease40. Moreover, PHB1 in the mitochondrial membrane is also involved in the regulation of sperm motility as shown by alterations in mitochondrial membrane potential in infertile men with poor sperm quality41. A recent study on PHB2 located in the inner mitochondrial membrane verified that PHB2 acts as a crucial mitophagy receptor involved in targeting mitochondria for autophagic degradation. Briefly, PHB2 was shown to bind the autophagosomal membrane-associated protein LC3 through an LC3-interacting region domain upon mitochondrial depolarization and proteasome-dependent outer membrane rupture, thus inducing eukaryotic mitophagy42.
PHB1 and PHB2 locate in nucleus
Nuclear PHB1 modulates transcriptional activity directly through the interactions with various transcription factors, or indirectly through the interactions with chromatin remodeling proteins5, 11, 12, 43. The level of nuclear PHB1 can be down-regulated upon androgen treatment in cancer cells, indicating that PHB1 has a regulatory role in cell cycle progression44. In prostate cancer cells, PHB1 interacts with and suppresses E2F1 expression, repressing E2F-mediated transcription and inducing cell cycle arrest41, 45. PHB1 in the nucleus also functions as a potent transcriptional corepressor for estrogen receptor α (ERα) to abrogate cell proliferation46. Moreover, the investigation on paclitaxel resistance in cancer cells demonstrated that ERα promotes PHB1 mitochondrial-to-nuclear translocation to regulate estrogen-dependent paclitaxel resistance47.
In leukemic cells, PHB1 is strongly expressed in the nucleus and is a useful biomarker for the identification of leukemia subtypes48. PHB2 in the nucleus is phosphorylated by AKT at Ser-91, acts as a putative nuclear substrate of AKT and induces the differentiation of acute promyelocytic leukemia cells49, 50. Other studies demonstrated that PHB2 is essential for protecting centromeric cohesion in the regulation of sister-chromatid cohesion during mitosis, indicating that PHB2 is necessary for proper mitotic progression13.
PHB1 and PHB2 locate in cell membrane
Some PHB1 proteins have been reported to interact with low density detergent-insoluble lipid raft domains in the plasma membrane26, 51, acting as transmembrane adapters to activate downstream signals14, 52, 53. Previous studies also identified that PHB1 on plasma membrane acts as a viral receptor protein to facilitate virus entry into host cells8, 54, 55. In addition, studies found that expression level of PHB1 on T cell surfaces is significantly up-regulated when T cells are activated56. Interestingly, another study claimed that interaction of PHB1 and Vi capsular polysaccharide (Vi) on T cell plasma membrane is crucial to inhibit T cell activation57, 58. Moreover, PHB1 located on S. typhi-host cell plasma membrane interacts with Vi to down-regulate early inflammatory responses, indicating that PHB1 contributes to the pathogenesis of typhoid fever58. In pancreatic cancer tissue, the expression level of PHB1 on plasma membrane was found to be higher than that in normal tissues, indicating an important role of PHB in tumorigenesis59. In recent studies, PHB1 located on the platelet membrane was demonstrated to be involved in PAR1-mediated human platelet aggregation60, 61, while PHB2 located in rhabdomyosarcoma (RMS) cell membrane was reported to act as a regulator in IGFBP-6-induced RMS cell migration through its interaction with insulin-like growth factor (IGF)-binding protein (IGFBP)-610.
Collectively, PHB1 and PHB2 located in different cellular compartments function in different biological processes, which are involved in multiple diseases (Fig. 1). The functional role of PHB1 and PHB2 in tumorigenesis and the mechanism involved requires further investigation for potential implication of prohibitin domain family proteins in tumor diagnosis and treatment.
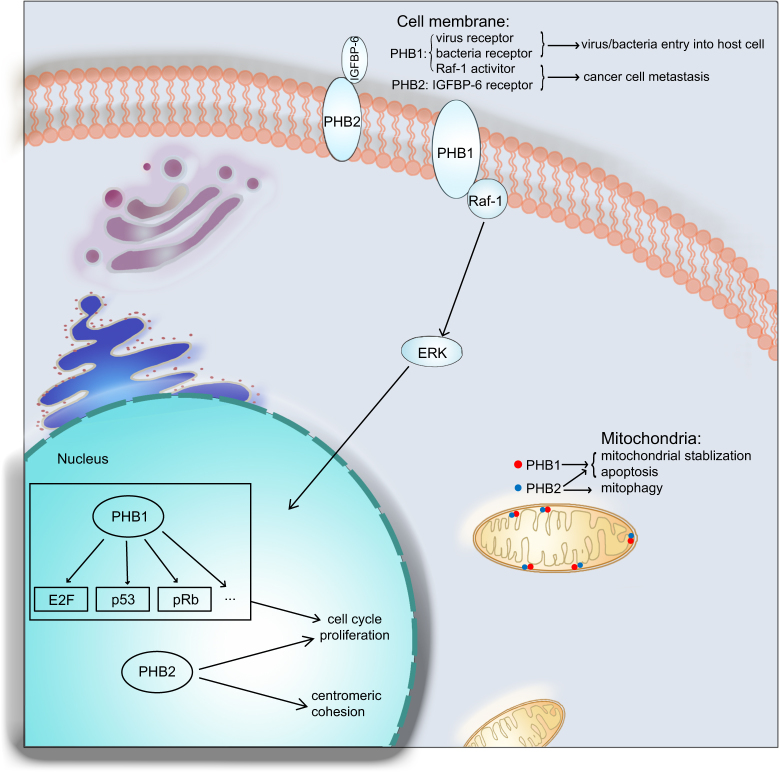
Fig. 1. Location and function of PHB1 and PHB2.
PHB1 and PHB2 locate mainly in mitochondria inner membrane, nucleus and cell membrane. In mitochondria: PHB1 and PHB2 maintain mitochondrial stabilization and cell survival, and PHB2 also functions as a regulator in mitophagy. In nucleus: PHB1 binds to some transcription factors and play an important role in cell cycle process and cell proliferation, and PHB2 protects the centromeric cohesion during mitosis. In cell membrane: PHB1 not only acts as virus receptor to facilitate virus entry into host cells, but also interacts with Raf-1 and activates Raf-1/ERK signaling cascades to promote cancer cell metastasis, whereas PHB2 promotes cancer cell metastasis mainly through the interaction with IGFBP-6
Role of PHB1 and PHB2 in tumorigenesis
Data from The Cancer Genome Atlas (TCGA) and The Human Protein Atlas show that PHB1 and PHB2 are widely expressed in diverse cancers, at both mRNA and protein levels (Fig. 2, Fig. 3)62. A lot of evidence demonstrated that PHB1 and PHB2 are involved in biological processes of cancer development, such as proliferation, apoptosis, and metastasis.
Fig. 2. The mRNA expression levels of PHB1 and PHB2 in multiple cancers.
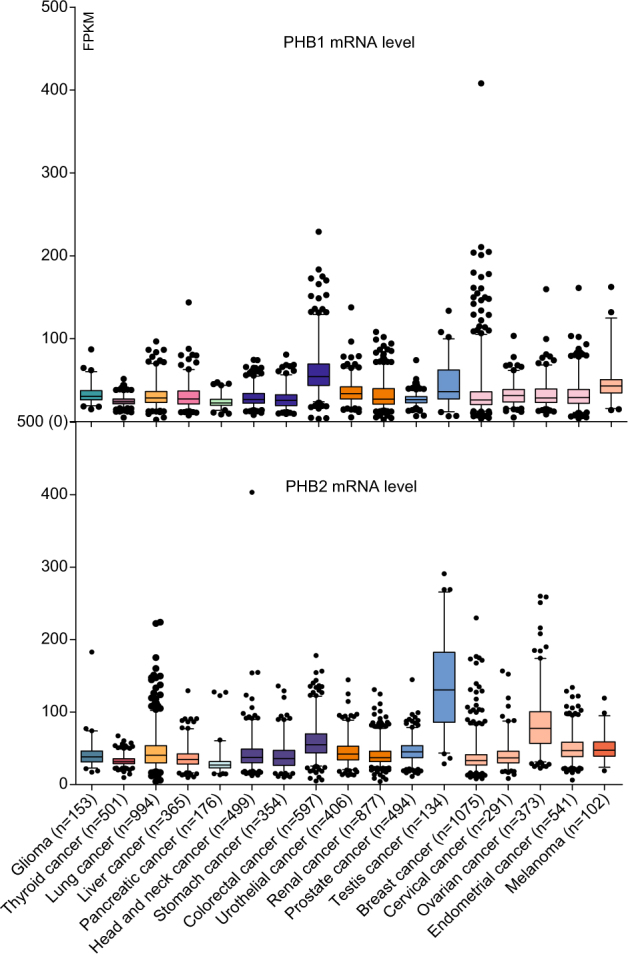
RNA sequencing data were downloaded from The Cancer Genome Atlas (cancergenome.nih.gov). Up: PHB1 mRNA level; down, PHB2 mRNA level. Cancer number = 17; Whiskers: 2.5–97.5 percentile; FPKM Fragments Per Kilobase of exon model per Million mapped fragments
Fig. 3. The protein expression levels of PHB1 and PHB2 in multiple cancers.
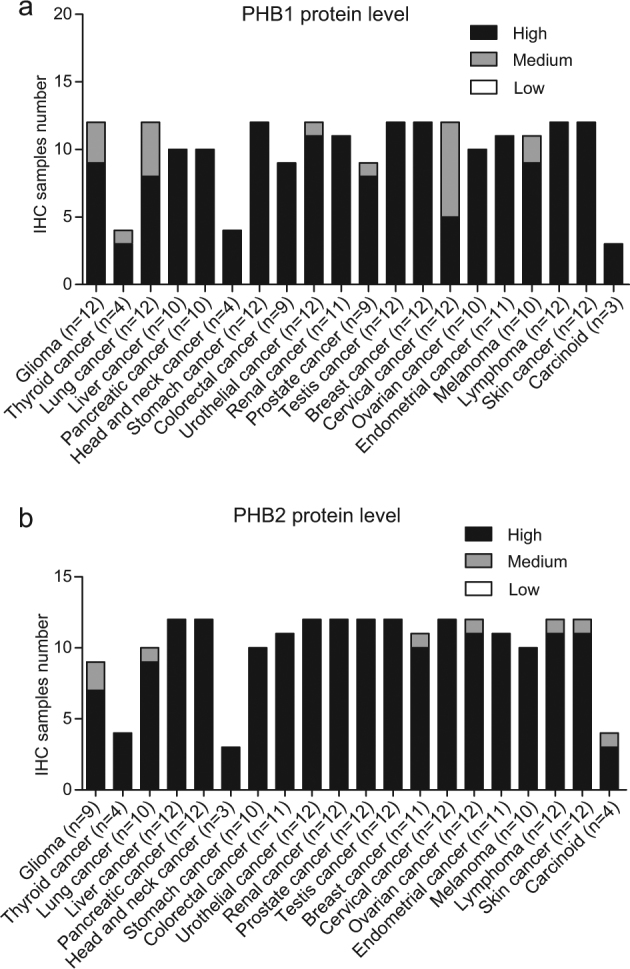
The immunohistochemistry data of clinical samples were downloaded from The Human Protein Atlas (www.proteinatlas.org). a PHB1 protein level. b PHB2 protein level. Cancer number = 20. High, staining score >75%; Medium, 75% > staining score > 25%; Low, staining score < 25%
Roles of PHB1 and PHB2 in cancer cell proliferation
Since the original identification of PHB1 as a proliferation suppressor in rat liver cells1, the effect of PHB1 on cancer proliferation has been studied. Some studies found that PHB1 inhibits breast cancer cell proliferation by up-regulating androgen receptor (AR) expression in estrogen receptor (ER)-positive breast cancer cells15, 63. PHB1 also inhibits proliferation of human osteosarcoma MG-63 cells by interacting with tumor suppressors such as p5364. In glioma cells, PHB1 expression is down regulated by miR-26a, which interferes with the regulation of PHB1 on expression levels of HIF-1 and VEGF, as well as tumor growth65, 66. Moreover, PHB1 is significantly down regulated in nasopharyngeal carcinoma and hepatocellular carcinoma17, 67, indicating that PHB1 may be a promising diagnostic biomarker in these cancers.
However, PHB1 exhibits opposing functions in other cancer types. For example, down-regulation of PHB1 inhibits cancer cell proliferation by inducing G1-G0 arrest in esophageal squamous cell carcinoma (ESCC)18. Overexpression of PHB1 was verified in gallbladder cancer tissues, and further studies identified that PHB1 promotes cancer cell proliferation by activating ERK and downstream signaling pathways68. Similarly, PHB1 was also demonstrated to be overexpressed in bladder cancer tissues; mechanistically, AKT phosphorylates PHB1 at T258, inducing mitochondrial localization of PHB1 and leading to cancer cell proliferation69, 70. Moreover, PHB1 is overexpressed in ovarian epithelial tumors and induces cell proliferation71, 72. In estrogen-dependent cancers such as breast cancer, investigators reported that BIG3, a key molecular regulator in the ER signaling pathway, inhibits the nuclear transportation of the PHB2/REA transcription complex, abolishing the inhibitory effect of the PHB2/REA complex on ERα transcriptional activity73, 74. PHB2 was also shown to promote hepatocellular carcinoma growth and malignancy progression in the hypoxic tumor microenvironment75.
Roles of PHB1 and PHB2 in cancer cell metastasis
Despite many advances in the diagnosis and treatment of cancer, tumor metastasis remains largely incurable and up to 90% of cancer-related deaths are caused by metastatic disease rather than primary tumors76–78. Therefore, efficient biomarkers or targets involved in cancer metastasis are urgently needed to be identified. Interestingly, unlike its controversial role in cancer cell proliferation, PHB1 expression levels are significantly correlated with tumor metastasis and poor prognosis according to the current literature. In many cancers, the Raf-ERK signaling pathway is constitutively activated, promoting cancer cell metastasis. Recent studies have confirmed that PHB1 in lipid rafts is crucial for the activation of Raf-1 and downstream signals. For example, studies in HeLa and CL1-0 cells showed that phosphorylation of PHB1 at T258 and Y259 is necessary for the activation of Raf-ERK signaling cascades supporting cancer cell metastasis14, 79. A similar mechanism was observed in pancreatic ductal adenocarcinoma and gallbladder cancer as well22, 68.
Some researches claimed that a polarized distribution of PHB1 in cells controls the migration direction of colorectal cancer cells. PHB1 can relocate to the luminal side of cells, face extracellular VEGF and indicate the direction of colorectal cancer cell invasion80. In lung cancer cells, studies verified that association of phospho-PHB1 T258 with MEKK1 activates the Snail, the repressor of E-cadherin, enhancing epithelial-mesenchymal transition (EMT) and lung cancer migration/invasion81, 82. PHB1 overexpression is also reported in invasive breast carcinoma, indicating that PHB1 is a potential biomarker in breast cancer83, 84. In prostate cancer, PHB2 was reported to interact with AKT2 and negatively regulate AKT2 expression, inducing cancer cell migration and malignancy49.
Roles of PHB1 and PHB2 in cancer cell apoptosis
Apoptosis is defined as programmed cell death that plays a fundamental role in organism development and tissue homeostasis. Disruption of apoptotic signaling results in multiple diseases85–87. Apoptosis is also important for cancers to eliminate damaged cells that cannot be repaired, maintaining cancer cell vibrancy and aggressiveness88. On the other hand, induction of apoptosis is a promising strategy for cancer treatment to eliminate cancer cells89–91. The role of PHB1 in cancer cell apoptosis has been explored in various studies. In breast and colon cancers, researchers demonstrated that PHB1 binds to the p53 induced gene 3 (PIG3) promoter motif (TGYCC) 15 directly, promoting PIG3-mediated, p53-dependent cancer cell apoptosis92, 93. Another study reported that cholesterol insufficiency in prostate cancer cells causes up-regulation of PHB1, inducing cell cycle arrest and apoptosis94. Furthermore, a series of experiments in colon cancer cells and melanoma cells confirmed that the direct interaction of PHB1 with trifluorothiazoline is necessary for trifluorothiazoline-induced apoptosis19, 32. This phenomenon was also observed in abrin-triggered apoptosis95 and the retinoic acid-resistant leukemia cell line NB4-R196. Interestingly, PHB1 expression significantly decreases, with its localization shifting from cytoplasmic to nuclear, and the co-localization of PHB1 with tumor suppressors such as p53 and Rb promotes apoptosis in cholangiocarcinoma97. Moreover, PHB2 can translocate from the mitochondria to the nucleus during capsaicin-induced apoptosis16. In ER-positive breast cancer, overexpression of PHB1 in MCF-7 and T47D cells significantly induces cell apoptosis15, and the same phenomenon was detected in both gastric and liver cancer cells98, 99.
In fact, PHB1 and PHB2 not only function in biological processes like cancer cell proliferation, metastasis and apoptosis in tumorigenesis and cancer development, but also play a role in modulating other processes such as cell differentiation in hepatocarcinoma, neuroblastoma and acute promyelocytic leukemia50, 100, 101 and mitophagy42. As the functional role of PHB1 and PHB2 in cancer becomes increasingly well defined, it is necessary to evaluate the upstream regulatory mechanisms of PHB1 and PHB2 in tumorigenesis to facilitate our understanding on the prohibitin domain family proteins and to generate clues for targeted drug development.
Regulation of PHB1 and PHB2 in cancers
Proteins in cells are usually regulated in three ways: transcriptional regulation102, 103, post-transcriptional regulation at the mRNA level104, 105 and protein modification106 including degradation107 and structural regulation108. Transcriptional regulation mainly involves the regulation by transcription factors and epigenetic mechanisms109, 110, while post-transcriptional regulation involves RNA interference111, 112. Based on recent studies, all the three approaches of regulation seem to be involved in controlling prohibitin domain family proteins’ effects on cancer cells.
PHB1 is regulated at the transcriptional level
At transcriptional level, researchers found that PHB1 expression level is associated with PHB1 genome copy number and a 3′ untranslated region (UTR) 1630 C > T polymorphism in gastric cancer113. Interestingly, the PHB1 genotype C1703T in the 3′-UTR is correlated with an increase in the risk for melanoma in a high ultraviolet radiation region in Brazil114. The expression of PHB1 was found to significantly increase in the thyroid tumor cells treated with trichostatin A (TSA) and sodium butyrate (NaB), two histone deacetylase inhibitors, demonstrating that HDAC1/2 regulates both PHB1 transcription and alternative splicing115. Unfortunately, it remains unclear which transcription factors are responsible for PHB1 transcription.
PHB1 and PHB2 are regulated at the post-transcriptional level
At the post-transcriptional level, miR-27a targets the 3′-UTR of the PHB1 gene and down-regulates its expression in prostate cancer116. Likewise, the same mechanism has been confirmed in gastric adenocarcinoma cells and glioma cells117, 118. Interestingly, miR-26a also binds directly to the 3′-UTR of PHB1, inhibiting its expression in glioma cells66. In human melanoma cells, studies showed that miR-195 binds to the 3′-UTR of PHB1 as well119. Researchers also demonstrated that miR-539 binds PHB2 and suppresses its expression to induce mitochondrial fission and apoptosis120. Moreover, one study recently confirmed that a long noncoding RNA (lncRNA), prohibitin gene pseudogene 1 (PHBP1), which shares the high level of the nucleotide sequence identity (91.3%) with its cognate gene PHB1, promotes the stabilization of PHB1 mRNA by forming a PHBP1 RNA/PHB1 mRNA heteroduplex in complementary regions, increasing the expression of PHB118.
PHB1 is modified by AKT
Protein modifications such as phosphorylation, glycosylation, ubiquitylation and small ubiquitin-related modifier (SUMO) determine the maturity, stability and activity of proteins121–123. For PHB1, phosphorylation or dephosphorylation at different amino acid sites results in different effects on tumorigenesis. Recent studies have focused on the phosphorylation of PHB1 at Tyr114, Ser121, Thr258, and Tyr 259124, with Thr258 and Tyr 259 phosphorylation well characterized in cancer cells. Phosphorylation of PHB1 at Thr258 in lipid rafts is necessary for the interaction of PHB1 with Raf-1. AKT phosphorylates PHB1 at Thr258, inducing the activation of Raf-1 and the Raf-1/ERK signaling pathway, resulting in cancer metastasis14, 79. Interestingly, Tyr259 phosphorylation induces the activation of Thr258 and the downstream Raf-1/ERK signaling cascades.
Collectively, transcription of PHB1 is regulated by histone acetylation, microRNAs, and some lncRNAs. Moreover, phosphorylation of PHB1 at Thr258 and Tyr259 induced by AKT is important for cancer metastasis (Fig. 4a). Unfortunately, data on the upstream regulation of PHB2 is still lacking. Understanding of the regulation of prohibitin domain family and the related biological processes will offer us clues for further investigations on cancer diagnosis and treatment.
Fig. 4. Regulation of PHB1 and PHB2 in cancer.
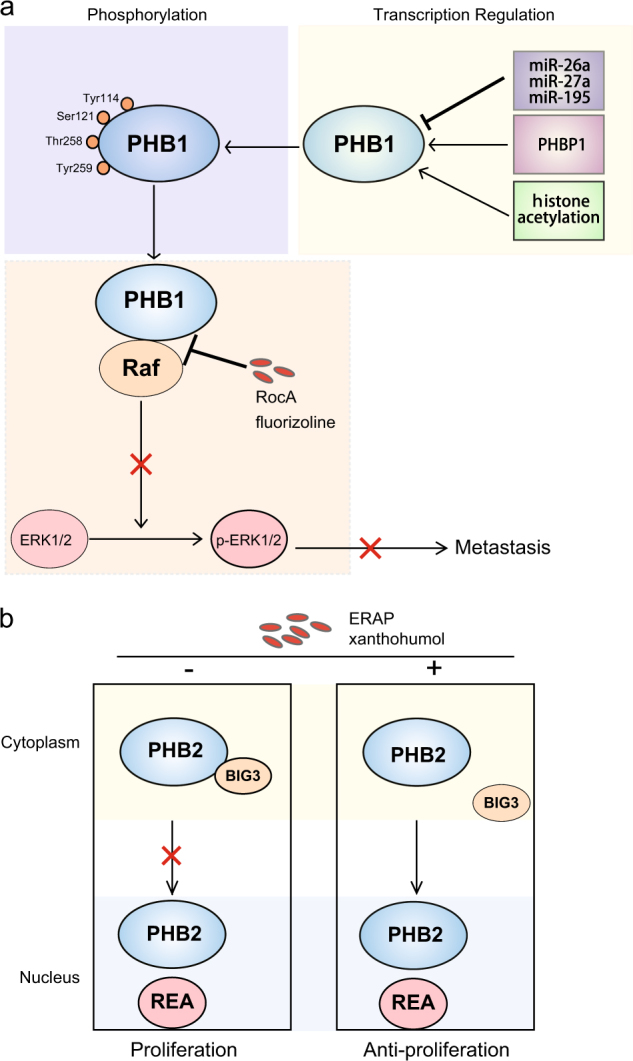
a PHB1 can be down-regulated by miR-26a, miR-27a, and miR-195; the lncRNA named PHBP1 directly binds to and maintain the stabilization of PHB1 mRNA; the acetylation of histone H3 can also increase the expression of PHB1; phosphorylation of PHB1 at different amino acids determines the activation and function of PHB1. Small inhibitors including RcoA and fluorizoline directly bind to PHB1 and interrupt its interaction with Raf, thus blocks Raf-ERK signaling cascades and suppress cancer metastasis. b The short peptide, ERAP, and the natural product derived from medical plants, xanthohumol, could disrupt the PHB2/BIG3 interaction directly and lead to the translocation of PHB2 from cytoplasm to nucleus, thereby, induce cell growth arrest in some estrogen-dependent cancers
Value of PHB1 and PHB2 as therapeutic targets in cancer treatment
Many efficient targeted drugs have been developed for cancer treatment, balanced by increasing instances of clinical drug resistance125, 126. Mechanistic studies found that most target genes or protein mutants develop resistance to targeted drugs over a period of time127–129. Thus, new effective targets are always needed. As summarized above, PHB1 acts as a tumor promoter in most cancer types, and thus is a potential target for novel drug development, especially the PHB1 located in the cell membrane. In fact, several small inhibitors targeting PHB1 or PHB1/Raf-1 or PHB2 interactions have been identified, and tumors were significantly inhibited by these drugs.
Drugs targeting PHB1
Rocaglamide (RocA), a naturally occurring compound isolated from medicinal plants belonging to the genus Aglaia family Meliaceae20, was used for treatment of coughs, injuries, asthma, and inflammatory skin diseases. RocA directly targets PHB1 and prevents its interaction with Raf-1, resulting in impaired ERK1/2 activation in leukemic cells130. The effect of RocA was further displayed in pancreatic ductal adenocarcinoma, where RocA inhibits the interaction of PHB1 and Raf-1, significantly suppressing cancer cell growth and metastasis22. Interestingly, RocA can reverse Raf-1-dependent resistance to vemurafenib, inhibite cell growth and induce apoptosis in melanoma cells23, indicating that RocA has potential value for use in vemurafenib-resistant cancers.
Recently, researchers demonstrated that a small molecular inhibitor fluorizoline, synthetized based on natural products from medical plants, also prevents Ras-Raf interaction in lung cancer cell and inhibits tumor growth and metastasis131. Moreover, fluorizoline induces mitochondrial-dependent apoptosis by targeting at PHB1 in a p53-independent manner in MEF cells, as well as in multiple cancers19, 33, 132. The combined use of RocA and fluorizoline may be a promising strategy for cancer treatment (Fig. 4a).
Drugs targeting PHB2
As PHB2 is involved in estrogen-dependent cancers such as breast cancer, and BIG3 promotes cancer cell proliferation by inhibiting PHB2 nuclear translocation and promoting the activity of ERα, the BIG3/PHB2 interaction is necessary for tumorigenesis73, 74. Therefore, inhibition of the interaction of BIG3-PHB2 is a feasible strategy in estrogen-dependent cancer therapy. In fact, researchers isolated the stable ERα activity-regulator synthetic peptide (ERAP: 165-177 amino acids), a short peptide derived from α-helical BIG3 sequence that specifically binds to PHB2 and competitively prevents the BIG3/PHB2 interaction, and then showed that ERAP promotes PHB2/REA complex nuclear translocation and suppresses cancer cell proliferation in vitro and in vivo133. Moreover, PHB2 released from the BIG3-PHB2 complex by ERAP treatment reduces phosphorylation levels of AKT and MAPK, resulting in significant suppression of proliferation in ERα-positive breast cancer cells. More importantly, the tumor suppressive effects of ERAP are enhanced by combined use with tamoxifen, indicating that ERAP treatment can reverse tamoxifen resistance and enhance tamoxifen responsiveness in ERα-positive breast cancer cells24, 25. The same research team also identified a natural product derived from medical plants named xanthohumol that could interrupt BIG3/PHB2 interaction and suppress ERα-positive breast cancer cell proliferation (Fig. 4b)134.
New target exploration is a continuous effort for new drug development, offering more opportunities to overcome drug resistance in cancer treatment. Based on the special localization and clear mechanism of PHB1 in tumorigenesis, there is a bright future for developing high-efficiency drugs to target PHB1 for cancer treatment. PHB2 inhibitors will also play an important role in estrogen-dependent cancer treatment, especially in premenopausal women.
Conclusion and future perspectives
Since the PHB gene was first isolated and characterized in rat liver in 1989, researchers have been studying this “anti-proliferation” gene to elucidate its role in biological process and diseases.
Biomarkers and genes for targeted drug development are important for cancer diagnosis and treatment, and drugs developed based on proteins such as EGFR have been extensively used for clinical treatment135. However, increasing numbers of patients are presenting with drug resistance when treated with extant drugs for long periods of time128. Thus, exploration of new targets is urgently needed. Differential PHB1 expression has been identified in multiple cancers as compared with normal tissues; hence, the PHB1 protein, especially when located in the cell membrane, may be a perfect choice for targeted drug development, as drugs could target PHB1 directly without transportation into cells. The observations that small molecule inhibitors RocA and fluorizoline target PHB1, and a short peptide ERAP and xanthohumol target PHB2, provide us stimulation in anti-cancer drug design. At least, modifying these existing drugs to increase their efficiency, or screening more potent inhibitors targeting PHB1 and PHB2 may lead to breakthroughs in cancer therapy.
Acknowledgements
We thank Qihua Zhang for the artwork preparation. This work was supported by the National Key Research and Development Program of China (2017YFA0505100/2017YFA0505000), the National Natural Science Foundation of China (31770888, 31570828, 81773085), and Guangdong Natural Science Research Grant (S2013030013315, 2016A030313838).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by P. Pinton
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bin Li, Phone: +86-20-85224372, Email: libin2015@jnu.edu.cn.
Qing-Yu He, Phone: +86-20-85227039, Email: tqyhe@jnu.edu.cn.
References
- 1.McClung JK, et al. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem. Biophys. Res. Commun. 1989;164:1316–1322. doi: 10.1016/0006-291X(89)91813-5. [DOI] [PubMed] [Google Scholar]
- 2.Sato T, et al. The human prohibitin (PHB) gene family and its somatic mutations in human tumors. Genomics. 1993;17:762–764. doi: 10.1006/geno.1993.1402. [DOI] [PubMed] [Google Scholar]
- 3.Sato T, et al. The human prohibitin gene located on chromosome 17q21 is mutated in sporadic breast cancer. Cancer Res. 1992;52:1643–1646. [PubMed] [Google Scholar]
- 4.White JJ, et al. Assignment of the human prohibitin gene (PHB) to chromosome 17 and identification of a DNA polymorphism. Genomics. 1991;11:228–230. doi: 10.1016/0888-7543(91)90126-Y. [DOI] [PubMed] [Google Scholar]
- 5.Mishra S, Murphy LC, Nyomba BL, Murphy LJ. Prohibitin: a potential target for new therapeutics. Trends Mol. Med. 2005;11:192–197. doi: 10.1016/j.molmed.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Nijtmans LG, et al. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27:306–14.14. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wintachai P, et al. Identification of prohibitin as a Chikungunya virus receptor protein. J. Med. Virol. 2012;84:1757–1770. doi: 10.1002/jmv.23403. [DOI] [PubMed] [Google Scholar]
- 9.Desideri E, Cavallo AL, Baccarini M. Alike but different: RAF paralogs and their signaling outputs. Cell. 2015;161:967–970. doi: 10.1016/j.cell.2015.04.045. [DOI] [PubMed] [Google Scholar]
- 10.Fu P, Yang Z, Bach LA. Prohibitin-2 binding modulates insulin-like growth factor-binding protein-6 (IGFBP-6)-induced rhabdomyosarcoma cell migration. J. Biol. Chem. 2013;288:29890–29900. doi: 10.1074/jbc.M113.510826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury I, Thomas K, Thompson WE. Prohibitin(PHB) roles in granulosa cell physiology. Cell Tissue Res. 2016;363:19–29. doi: 10.1007/s00441-015-2302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koushyar S, et al. The prohibitin-repressive interaction with E2F1 is rapidly inhibited by androgen signalling in prostate cancer cells. Oncogenesis. 2017;6:e333. doi: 10.1038/oncsis.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takata H, et al. PHB2 protects sister-chromatid cohesion in mitosis. Curr. Biol. 2007;17:1356–1361. doi: 10.1016/j.cub.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Chiu CF, et al. Raf activation by Ras and promotion of cellular metastasis require phosphorylation of prohibitin in the raft domain of the plasma membrane. Oncogene. 2013;32:777–787. doi: 10.1038/onc.2012.86. [DOI] [PubMed] [Google Scholar]
- 15.Liu P, et al. Prohibitin promotes androgen receptor activation in ER-positive breast cancer. Cell Cycle. 2017;16:776–784. doi: 10.1080/15384101.2017.1295193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuramori C, et al. Capsaicin binds to prohibitin 2 and displaces it from the mitochondria to the nucleus. Biochem. Biophys. Res. Commun. 2009;379:519–525. doi: 10.1016/j.bbrc.2008.12.103. [DOI] [PubMed] [Google Scholar]
- 17.Liao Q, et al. Prohibitin is an important biomarker for nasopharyngeal carcinoma progression and prognosis. Eur. J. Cancer Prev. 2013;22:68–76. doi: 10.1097/CEJ.0b013e328354d351. [DOI] [PubMed] [Google Scholar]
- 18.Feng F, Qiu B, Zang R, Song P, Gao S. Pseudogene PHBP1 promotes esophageal squamous cell carcinoma proliferation by increasing its cognate gene PHB expression. Oncotarget. 2017;8:29091–29100. doi: 10.18632/oncotarget.16196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez-Perarnau A, et al. A trifluorinated thiazoline scaffold leading to pro-apoptotic agents targeting prohibitins. Angew. Chem. Int. Ed. Engl. 2014;53:10150–10154. doi: 10.1002/anie.201405758. [DOI] [PubMed] [Google Scholar]
- 20.Kim S, Salim AA, Swanson SM, Kinghorn AD. Potential of cyclopenta[b]benzofurans from Aglaia species in cancer chemotherapy. Anticancer Agents Med. Chem. 2006;6:319–345. doi: 10.2174/187152006777698123. [DOI] [PubMed] [Google Scholar]
- 21.Ebada SS, Lajkiewicz N, Porco JA, Jr, Li-Weber M, Proksch P. Chemistry and biology of rocaglamides (=flavaglines) and related derivatives from aglaia species (meliaceae) Prog. Chem. Org. Nat. Prod. 2011;94:1–58. doi: 10.1007/978-3-7091-0748-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luan Z, He Y, Alattar M, Chen Z, He F. Targeting the prohibitin scaffold-CRAF kinase interaction in RAS-ERK-driven pancreatic ductal adenocarcinoma. Mol. Cancer. 2014;13:38. doi: 10.1186/1476-4598-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doudican NA, Orlow SJ. Inhibition of the CRAF/prohibitin interaction reverses CRAF-dependent resistance to vemurafenib. Oncogene. 2017;36:423–428. doi: 10.1038/onc.2016.214. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimaru T, et al. Targeting BIG3-PHB2 interaction to overcome tamoxifen resistance in breast cancer cells. Nat. Commun. 2013;4:2443. doi: 10.1038/ncomms3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimaru T, et al. Therapeutic advances in BIG3-PHB2 inhibition targeting the crosstalk between estrogen and growth factors in breast cancer. Cancer Sci. 2015;106:550–558. doi: 10.1111/cas.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrow IC, et al. Flotillin-1/reggie-2 traffics to surface raft domains via a novel golgi-independent pathway. J. Biol. Chem. 2002;277:48834–48841. doi: 10.1074/jbc.M209082200. [DOI] [PubMed] [Google Scholar]
- 27.Bavelloni A, Piazzi M, Raffini M, Faenza I, Blalock WL. Prohibitin 2: At a communications crossroads. IUBMB Life. 2015;67:239–254. doi: 10.1002/iub.1366. [DOI] [PubMed] [Google Scholar]
- 28.Back JW, et al. A structure for the yeast prohibitin complex: structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Sci. 2002;11:2471–2478. doi: 10.1110/ps.0212602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Huertas MR, et al. The presence of HIV-1 Tat protein second exon delays fas protein-mediated apoptosis in CD4 + T lymphocytes: a potential mechanism for persistent viral production. J. Biol. Chem. 2013;288:7626–7644. doi: 10.1074/jbc.M112.408294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Artal-Sanz M, Tavernarakis N. Prohibitin and mitochondrial biology. Trends Endocrinol. Metab. 2009;20:394–401. doi: 10.1016/j.tem.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Ising C, Brinkkoetter PT. Prohibitin signaling at the kidney filtration barrier. Adv. Exp. Med. Biol. 2017;982:563–575. doi: 10.1007/978-3-319-55330-6_29. [DOI] [PubMed] [Google Scholar]
- 32.Bollu LR, et al. Involvement of de novo synthesized palmitate and mitochondrial EGFR in EGF induced mitochondrial fusion of cancer cells. Cell Cycle. 2014;13:2415–2430. doi: 10.4161/cc.29338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moncunill-Massaguer C, et al. A novel prohibitin-binding compound induces the mitochondrial apoptotic pathway through NOXA and BIM upregulation. Oncotarget. 2015;6:41750–41765. doi: 10.18632/oncotarget.6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang HB, et al. Differential expression and regulation of prohibitin during curcumin-induced apoptosis of immortalized human epidermal HaCaT cells. Int. J. Mol. Med. 2014;33:507–514. doi: 10.3892/ijmm.2014.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ising C, et al. Prohibitin-2 depletion unravels extra-mitochondrial functions at the kidney filtration barrier. Am. J. Pathol. 2016;186:1128–1139. doi: 10.1016/j.ajpath.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 36.Lachén-Montes M, et al. Olfactory bulb neuroproteomics reveals a chronological perturbation of survival routes and a disruption of prohibitin complex during Alzheimer’s disease progression. Sci. Rep. 2017;7:9115. doi: 10.1038/s41598-017-09481-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merkwirth C, et al. Loss of prohibitin membrane scaffolds impairs mitochondrial architecture and leads to tau hyperphosphorylation and neurodegeneration. PLoS Genet. 2012;8:e1003021. doi: 10.1371/journal.pgen.1003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kahl, A. et al. Neuronal expression of the mitochondrial protein prohibitin confers profound neuroprotection in a mouse model of focal cerebral ischemia. J. Cereb. Blood Flow Metab. 2017; 10.1177/0271678X17720371. [DOI] [PMC free article] [PubMed]
- 39.Zhou P, et al. Prohibitin reduces mitochondrial free radical production and protects brain cells from different injury modalities. J. Neurosci. 2012;32:583–592. doi: 10.1523/JNEUROSCI.2849-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dutta, D., et al. Low levels of prohibitin in substantia nigra makes dopaminergic neurons vulnerable in Parkinson’s disease. Mol. Neurobiol. 2017; 10.1007/s12035-016-0328-y. [DOI] [PubMed]
- 41.Chai RR, et al. Prohibitin involvement in the generation of mitochondrial superoxide at complex I in human sperm. J. Cell Mol. Med. 2017;21:121–129. doi: 10.1111/jcmm.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei Y, Chiang WC, Sumpter R, Jr, Mishra P, Levine B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168:224–238. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toska E, Shandilya J, Goodfellow SJ, Medler KF, Roberts SG. Prohibitin is required for transcriptional repression by the WT1-BASP1 complex. Oncogene. 2014;33:5100–5108. doi: 10.1038/onc.2013.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gamble SC, et al. Androgens target prohibitin to regulate proliferation of prostate cancer cells. Oncogene. 2004;23:2996–3004. doi: 10.1038/sj.onc.1207444. [DOI] [PubMed] [Google Scholar]
- 45.Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene. 1999;18:3501–3510. doi: 10.1038/sj.onc.1202684. [DOI] [PubMed] [Google Scholar]
- 46.He B, et al. A repressive role for prohibitin in estrogen signaling. Mol. Endocrinol. 2008;22:344–360. doi: 10.1210/me.2007-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong P, et al. Induction of paclitaxel resistance by ERα mediated prohibitin mitochondrial-nuclear shuttling. PLoS One. 2013;8:e83519. doi: 10.1371/journal.pone.0083519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teittinen KJ, et al. Nucleolar proteins with altered expression in leukemic cell lines. Leuk. Res. 2012;36:232–236. doi: 10.1016/j.leukres.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 49.Shen Y, et al. Prohibitin-2 negatively regulates AKT2 expression to promote prostate cancer cell migration. Int. J. Mol. Med. 2018;41:1147–1155. doi: 10.3892/ijmm.2017.3307. [DOI] [PubMed] [Google Scholar]
- 50.Bavelloni A, et al. Prohibitin 2 represents a novel nuclear AKT substrate during all-trans retinoic acid-induced differentiation of acute promyelocytic leukemia cells. FASEB J. 2014;28:2009–2019. doi: 10.1096/fj.13-244368. [DOI] [PubMed] [Google Scholar]
- 51.Knopf JD, et al. The stromal cell-surface protease fibroblast activation protein-α localizes to lipid rafts and is recruited to invadopodia. Biochim. Biophys. Acta. 2015;1853(10 Pt A):2515–2525. doi: 10.1016/j.bbamcr.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 52.Zhou TB, Qin YH. Signaling pathways of prohibitin and its role in diseases. J. Recept. Signal. Transduct. Res. 2013;33:28–36. doi: 10.3109/10799893.2012.752006. [DOI] [PubMed] [Google Scholar]
- 53.Peng YT, Chen P, Ouyang RY, Song L. Multifaceted role of prohibitin in cell survival and apoptosis. Apoptosis. 2015;20:1135–1149. doi: 10.1007/s10495-015-1143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C, et al. Identification of human host proteins contributing to H5N1 influenza virus propagation by membrane proteomics. J. Proteome Res. 2012;11:5396–5405. doi: 10.1021/pr3006342. [DOI] [PubMed] [Google Scholar]
- 55.Paingankar MS, Gokhale MD, Deobagkar DN. Dengue-2-virus-interacting polypeptides involved in mosquito cell infection. Arch. Virol. 2010;155:1453–1461. doi: 10.1007/s00705-010-0728-7. [DOI] [PubMed] [Google Scholar]
- 56.Yurugi H, et al. Expression of prohibitins on the surface of activated T cells. Biochem. Biophys. Res. Commun. 2012;420:275–280. doi: 10.1016/j.bbrc.2012.02.149. [DOI] [PubMed] [Google Scholar]
- 57.Santhanam SK, Dutta D, Parween F, Qadri A. The virulence polysaccharide Vi released by Salmonella Typhi targets membrane prohibitin to inhibit T-cell activation. J. Infect. Dis. 2014;210:79–88. doi: 10.1093/infdis/jiu064. [DOI] [PubMed] [Google Scholar]
- 58.Sharma A, Qadri A. Vi polysaccharide of Salmonella typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc. Natl Acad. Sci. USA. 2004;101:17492–17497. doi: 10.1073/pnas.0407536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong N, Cui Y, Zhou X, Li T, Han J. Identification of prohibitin 1 as a potential prognostic biomarker in human pancreatic carcinoma using modified aqueous two-phase partition system combined with 2D-MALDI-TOF-TOF-MS/MS. Tumour Biol. 2015;36:1221–1231. doi: 10.1007/s13277-014-2742-y. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Wang Y, Xiang Y, Lee W, Zhang Y. Prohibitins are involved in protease-activated receptor 1-mediated platelet aggregation. J. Thromb. Haemost. 2012;10:411–418. doi: 10.1111/j.1538-7836.2011.04607.x. [DOI] [PubMed] [Google Scholar]
- 61.Wang YJ, et al. Prohibitin is involved in the activated internalization and degradation of protease-activated receptor 1. Biochim. Biophys. Acta. 2014;1843:1393–1401. doi: 10.1016/j.bbamcr.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 62.Uhlen, M. et al. A pathology atlas of the human cancer transcriptome. Science357, eaan2507 (2017). [DOI] [PubMed]
- 63.Zhang W, et al. Skp2 is over-expressed in breast cancer and promotes breast cancer cell proliferation. Cell Cycle. 2016;15:1344–1351. doi: 10.1080/15384101.2016.1160986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du MD, He KY, Qin G, Chen J, Li JY. Adriamycin resistance-associated prohibitin gene inhibits proliferation of human osteosarcoma MG63 cells by interacting with oncogenes and tumor suppressor genes. Oncol. Lett. 2016;12:1994–2000. doi: 10.3892/ol.2016.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chumbalkar VC, et al. Differential protein expression in human gliomas and molecular insights. Proteomics. 2005;5:1167–1177. doi: 10.1002/pmic.200401202. [DOI] [PubMed] [Google Scholar]
- 66.Qian X, et al. MicroRNA-26a promotes tumor growth and angiogenesis in glioma by directly targeting prohibitin. CNS Neurosci. Ther. 2013;19:804–812. doi: 10.1111/cns.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu JJ, et al. Proteomic analysis of hepatocellular carcinoma HepG2 cells treated with platycodin D. Chin. J. Nat. Med. 2015;13:673–679. doi: 10.1016/S1875-5364(15)30065-0. [DOI] [PubMed] [Google Scholar]
- 68.Cao Y, et al. Prohibitin overexpression predicts poor prognosis and promotes cell proliferation and invasion through ERK pathway activation in gallbladder cancer. J. Exp. Clin. Cancer Res. 2016;35:68. doi: 10.1186/s13046-016-0346-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang L, et al. Akt phosphorylates Prohibitin 1 to mediate its mitochondrial localization and promote proliferation of bladder cancer cells. Cell Death Dis. 2015;6:e1660. doi: 10.1038/cddis.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu TF, et al. Prohibitin in the pathogenesis of transitional cell bladder cancer. Anticancer Res. 2007;27:895–900. [PubMed] [Google Scholar]
- 71.El-Etreby NM, Ghazy AA, Rashad R. Prohibitin: targeting peptide coupled to ovarian cancer, luteinization and TGF-β pathways. J. Ovarian Res. 2017;10:28. doi: 10.1186/s13048-017-0325-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dai Z, et al. Mitochondrial comparative proteomics of human ovarian cancer cells and their platinum-resistant sublines. Proteomics. 2010;10:3789–3799. doi: 10.1002/pmic.200900685. [DOI] [PubMed] [Google Scholar]
- 73.Nakamura A, Osonoi T, Terauchi Y. Relationship between urinary sodium excretion and pioglitazone-induced edema. J. Diabetes Investig. 2010;1:208–211. doi: 10.1111/j.2040-1124.2010.00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim NH, et al. BIG3 inhibits the estrogen-dependent nuclear translocation of PHB2 via multiple karyopherin-alpha proteins in breast cancer cells. PLoS One. 2015;10:e0127707. doi: 10.1371/journal.pone.0127707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng J, et al. Prohibitin-2 promotes hepatocellular carcinoma malignancy progression in hypoxia based on a label-free quantitative proteomics strategy. Mol. Carcinog. 2014;53:820–832. doi: 10.1002/mc.22040. [DOI] [PubMed] [Google Scholar]
- 76.Turajlic S, Swanton C. Metastasis as an evolutionary process. Science. 2016;352:169–175. doi: 10.1126/science.aaf2784. [DOI] [PubMed] [Google Scholar]
- 77.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chiu CF, Peng JM, Hung SW, Liang CM, Liang SM. Recombinant viral capsid protein VP1 suppresses migration and invasion of human cervical cancer by modulating phosphorylated prohibitin in lipid rafts. Cancer Lett. 2012;320:205–214. doi: 10.1016/j.canlet.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 80.Ma LL, et al. Prohibitin, relocated to the front ends, can control the migration directionality of colorectal cancer cells. Oncotarget. 2017;8:76340–76356. doi: 10.18632/oncotarget.19394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ho MY, Liang CM, Liang SM. MIG-7 and phosphorylated prohibitin coordinately regulate lung cancer invasion/metastasis. Oncotarget. 2015;6:381–393. doi: 10.18632/oncotarget.2804. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Wong PF, et al. Eurycomanone suppresses expression of lung cancer cell tumor markers, prohibitin, annexin 1 and endoplasmic reticulum protein 28. Phytomedicine. 2012;19:138–144. doi: 10.1016/j.phymed.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 83.Canevari RA, et al. Identification of novel biomarkers associated with poor patient outcomes in invasive breast carcinoma. Tumour Biol. 2016;37:13855–13870. doi: 10.1007/s13277-016-5133-8. [DOI] [PubMed] [Google Scholar]
- 84.Koushyar S, Jiang WG, Dart DA. Unveiling the potential of prohibitin in cancer. Cancer Lett. 2015;369:316–322. doi: 10.1016/j.canlet.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 85.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345:1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bredesen DE, Rao RV, Mehlen P. Cell death in the nervous system. Nature. 2006;443:796–802. doi: 10.1038/nature05293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Evan GerardI, Vousden KarenH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 89.Lowe ScottW, Cepero Enrique, Evan Gerard. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 90.Croce CM, Reed JC. Finally, an apoptosis-targeting therapeutic for cancer. Cancer Res. 2016;76:5914–5920. doi: 10.1158/0008-5472.CAN-16-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Igea A, Nebreda AR. The Stress Kinase p38α as a Target for Cancer Therapy. Cancer Res. 2015;75:3997–4002. doi: 10.1158/0008-5472.CAN-15-0173. [DOI] [PubMed] [Google Scholar]
- 92.Guan X, Liu Z, Wang L, Johnson DG, Wei Q. Identification of prohibitin and prohibiton as novel factors binding to the p53 induced gene 3 (PIG3) promoter (TGYCC)(15) motif. Biochem. Biophys. Res. Commun. 2014;443:1239–1244. doi: 10.1016/j.bbrc.2013.12.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang W, et al. BRCA1 regulates PIG3-mediated apoptosis in a p53-dependent manner. Oncotarget. 2015;6:7608–7618. doi: 10.18632/oncotarget.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dong P, Flores J, Pelton K, Solomon KR. Prohibitin is a cholesterol-sensitive regulator of cell cycle transit. J. Cell. Biochem. 2010;111:1367–1374. doi: 10.1002/jcb.22865. [DOI] [PubMed] [Google Scholar]
- 95.Liu YH, Peck K, Lin JY. Involvement of prohibitin upregulation in abrin-triggered apoptosis. Evid. Based Complement. Altern. Med. 2012;2012:605154. doi: 10.1155/2012/605154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Y, He P, Zhang M, Wu D. Lentiviral vector-mediated RNA interference targeted against prohibitin inhibits apoptosis of the retinoic acid-resistant acute promyelocytic leukemia cell line NB4-R1. Mol. Med. Rep. 2012;6:1288–1292. doi: 10.3892/mmr.2012.1105. [DOI] [PubMed] [Google Scholar]
- 97.Song W, Tian L, Li SS, Shen DY, Chen QX. The aberrant expression and localization of prohibitin during apoptosis of human cholangiocarcinoma Mz-ChA-1 cells. FEBS Lett. 2014;588:422–428. doi: 10.1016/j.febslet.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 98.Zhang L, Ji Q, Ni ZH, Sun J. Prohibitin induces apoptosis in BGC823 gastric cancer cells through the mitochondrial pathway. Asian Pac. J. Cancer Prev. 2012;13:3803–3807. doi: 10.7314/APJCP.2012.13.8.3803. [DOI] [PubMed] [Google Scholar]
- 99.Yoo DR, et al. Proteomic identification of anti-cancer proteins in luteolin-treated human hepatoma Huh-7 cells. Cancer Lett. 2009;282:48–54. doi: 10.1016/j.canlet.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 100.Xu DH, et al. Positional and expressive alteration of prohibitin during the induced differentiation of human hepatocarcinoma SMMC-7721 cells. World J. Gastroenterol. 2008;14:5008–5014. doi: 10.3748/wjg.14.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li QF, et al. Localization of prohibitin in the nuclear matrix and alteration of its expression during differentiation of human neuroblastoma SK-N-SH cells induced by retinoic acid. Cell. Mol. Neurobiol. 2011;31:203–211. doi: 10.1007/s10571-010-9608-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Popovitchenko T, Rasin MR. Transcriptional and post-transcriptional mechanisms of the development of neocortical lamination. Front. Neuroanat. 2017;11:102. doi: 10.3389/fnana.2017.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu W, Ma R, Yuan Y. Post-transcriptional Regulation of Genes Related to Biological Behaviors of Gastric Cancer by Long Noncoding RNAs and MicroRNAs. J. Cancer. 2017;8:4141–4154. doi: 10.7150/jca.22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liang Z, et al. SUMOylation of IQGAP1 promotes the development of colorectal cancer. Cancer Lett. 2017;411:90–99. doi: 10.1016/j.canlet.2017.09.046. [DOI] [PubMed] [Google Scholar]
- 107.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 108.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 109.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 110.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 111.Chitwood DH, Timmermans MC. Small RNAs are on the move. Nature. 2010;467:415–419. doi: 10.1038/nature09351. [DOI] [PubMed] [Google Scholar]
- 112.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leal MF, et al. Prohibitin expression deregulation in gastric cancer is associated with the 3’ untranslated region 1630 C T polymorphism and copy number variation. PLoS One. 2014;9:e98583. doi: 10.1371/journal.pone.0098583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Francisco G, et al. Polymorphisms in the p27kip-1 and prohibitin genes denote novel genes associated with melanoma risk in Brazil, a high ultraviolet index region. Melanoma Res. 2013;23:231–236. doi: 10.1097/CMR.0b013e3283612483. [DOI] [PubMed] [Google Scholar]
- 115.Puppin C, Passon N, Franzoni A, Russo D, Damante G. Histone deacetylase inhibitors control the transcription and alternative splicing of prohibitin in thyroid tumor cells. Oncol. Rep. 2011;25:393–397. doi: 10.3892/or.2010.1075. [DOI] [PubMed] [Google Scholar]
- 116.Fletcher CE, et al. Androgen-regulated processing of the oncomir miR-27a, which targets Prohibitin in prostate cancer. Hum. Mol. Genet. 2012;21:3112–3127. doi: 10.1093/hmg/dds139. [DOI] [PubMed] [Google Scholar]
- 117.Liu T, Tang H, Lang Y, Liu M, Li X. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett. 2009;273:233–242. doi: 10.1016/j.canlet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 118.Chen W, et al. Emerging role of microRNA-27a in human malignant glioma cell survival via targeting of prohibitin. Mol. Med. Rep. 2015;12:1515–1523. doi: 10.3892/mmr.2015.3475. [DOI] [PubMed] [Google Scholar]
- 119.Cirilo PDR, et al. MicroRNA-195 acts as an anti-proliferative miRNA in human melanoma cells by targeting Prohibitin 1. BMC Cancer. 2017;17:750. doi: 10.1186/s12885-017-3721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang K, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 121.Frankson R, et al. Therapeutic targeting of oncogenic tyrosine phosphatases. Cancer Res. 2017;77:5701–5705. doi: 10.1158/0008-5472.CAN-17-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 123.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 124.Ande SR, Moulik S, Mishra S. Interaction between O-GlcNAc modification and tyrosine phosphorylation of prohibitin: implication for a novel binary switch. PLoS One. 2009;4:e4586. doi: 10.1371/journal.pone.0004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wood KC. Mapping the pathways of resistance to targeted therapies. Cancer Res. 2015;75:4247–4251. doi: 10.1158/0008-5472.CAN-15-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ramos P, Bentires-Alj M. Mechanism-based cancer therapy: resistance to therapy, therapy for resistance. Oncogene. 2014;34:3617–3626. doi: 10.1038/onc.2014.314. [DOI] [PubMed] [Google Scholar]
- 128.Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat. Med. 2013;19:1389–1400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat. Med. 2013;19:1381–1388. doi: 10.1038/nm.3369. [DOI] [PubMed] [Google Scholar]
- 130.Polier G, et al. The natural anticancer compounds rocaglamides inhibit the Raf-MEK-ERK pathway by targeting prohibitin 1 and 2. Chem. Biol. 2012;19:1093–1104. doi: 10.1016/j.chembiol.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 131.Yurugi H, et al. Targeting prohibitins with chemical ligands inhibits KRAS-mediated lung tumours. Oncogene. 2017;36:4778–4789. doi: 10.1038/onc.2017.93. [DOI] [PubMed] [Google Scholar]
- 132.Cosialls AM, et al. The prohibitin-binding compound fluorizoline induces apoptosis in chronic lymphocytic leukemia cells through the upregulation of NOXA and synergizes with ibrutinib, 5-aminoimidazole-4-carboxamide riboside or venetoclax. Haematologica. 2017;102:1587–1593. doi: 10.3324/haematol.2016.162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yoshimaru T, et al. Stapled BIG3 helical peptide ERAP potentiates anti-tumour activity for breast cancer therapeutics. Sci. Rep. 2017;7:1821. doi: 10.1038/s41598-017-01951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yoshimaru T, et al. Xanthohumol suppresses oestrogen-signalling in breast cancer through the inhibition of BIG3-PHB2 interactions. Sci. Rep. 2014;4:7355. doi: 10.1038/srep07355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gan HK, Burgess AW, Clayton AH, Scott AM. Targeting of a conformationally exposed, tumor-specific epitope of EGFR as a strategy for cancer therapy. Cancer Res. 2012;72:2924–2930. doi: 10.1158/0008-5472.CAN-11-3898. [DOI] [PubMed] [Google Scholar]