Abstract
Phytophthora capsici is the most devastating pathogen for chile pepper production worldwide and current management strategies are not effective. The population structure of the pathogen is highly variable and few sources of widely applicable host resistance have been identified. Recent genomic advancements in the host and the pathogen provide important insights into the difficulties reported by epidemiological and physiological studies published over the past century. This review highlights important challenges unique to this complex pathosystem and suggests strategies for resistance breeding to help limit losses associated with P. capsici.
Keywords: pepper, root-rot, stem-blight, fruit-blight, oomycete
Introduction
The soil-borne oomycete plant pathogen Phytophthora capsici (Leon.) is the most devastating pathogen to chile pepper production. Chile pepper is an increasingly important crop used as a vegetable, spice, food colorant, and medicinal applications. Over the last 30 years, chile pepper consumption has increased 40-fold (Rehrig et al., 2014). Chile pepper is a high-value crop and has immediate economic benefits for producers. Additionally, chile peppers are important sources of essential nutrients providing long term nutritional benefits for consumers. Globally, P. capsici causes more than $100 million in losses annually (Bosland, 2008). This enormity of damage has stimulated extensive collaborations between plant pathologists and plant breeders to better understand the epidemiology of the pathogen as well as the mechanisms of resistance in the host. This review highlights recent work with the Capsicum–Phytophthora pathosystem and discusses novel approaches to more effectively manage this devastating disease.
Pathogen Identification
Phytophthora capsici was first reported in New Mexico by Fabián García as a “souring of the soil” (García, 1908). In 1922, Leonian systematically described P. capsici isolated from chile pepper in 1918 at the New Mexico Agricultural Research Station in Las Cruces, NM, United States (Leonian, 1922). Synonyms for P. capsici include P. hydrophila (Cruzi, 1927), P. parasitica var. capsici (Sarejanni, 1936), and P. palmivora MF4 (Griffin, 1977).
Phytophthora capsici is part of a species complex with several attempts at resolution over the years, but it has not been fully determined whether it is one species with formae speciales, or multiple species. Studies of P. capsici populations recovered from vegetables at diverse locations reveal a very high level of heterozygosity, typical for an obligately outcrossing diploid organism. Despite individual isolates carrying a large complement of genetic variation, populations can vary dramatically, with some comprised almost entirely of long-lived clonal lineages (e.g., Peru, Argentina, Taiwan, and portions of China) and others displaying a wide array of diverse genotypes that change yearly due to the requirement for sexual recombination and development of thick-walled oospores to survive (e.g., United States and Mexico) (Gobena et al., 2012b; Hu et al., 2013a,b; Castro-Rocha et al., 2016, 2017; Barchenger et al., 2017). The extensive of genetic variation may play a part in attacking such a large host group. Satour and Butler (1967) found 45 species of cultivated plants and weeds susceptible to P. capsici. There is currently a lack of clear delineation between P. capsici, the closely related P. tropicalis and multiple, un-named, but clearly evolutionarily distinct species (Lamour et al., 2012). This is due to the historical use of spore shape (sporangial length/breadth ratio), pedicel length and caducity, and the amphigynous oospore structure – which are poor characters for defining evolutionary relationships, and the difficulty in knowing how much genetic differentiation is sufficient to fully resolve the species. In practice, isolates recovered from woody or perennial hosts are not the same species as isolates recovered from herbaceous annual plants (Lamour et al., 2012).
Disease Symptoms
Phytophthora capsici causes root-rot as well as stem-, leaf-, and fruit-blight. These disease syndromes are dependent on host species, point of infection, and also are influenced by environmental conditions. Furthermore, disease severity is affected by plant maturity, with more mature plants generally being more resistant than seedlings or young fruit (Erwin and Ribeiro, 1996; Lamour et al., 2011; Mansfeld et al., 2017).
In chile pepper, the root-rot syndrome caused by P. capsici is associated with root darkening and small lesions that can quickly expand to girdle and kill the root. In seedlings, damping off associated with root-rot can kill plants two to 5 days after inoculation (Erwin and Ribeiro, 1996). In older plants, root infections can result in stunting, wilting, and eventual plant death in approximately 2 weeks. Root-rot is the most destructive and economically important disease syndrome of chile pepper (Walker and Bosland, 1999; Bosland, 2008).
Foliar-blight symptoms include dark, water soaked areas of the leaves (Walker and Bosland, 1999). The disease starts with a small circular or irregular-shaped lesion on the leaves giving a “scalded” appearance. Later, the lesions enlarge, dry, and bleach to a light tan. The disease progresses to the stem as a dark-green and water-soaked lesion. Finally, the plant is defoliated and stems dry and brown (Weber, 1932). Infected leaves will turn brown or tan and may defoliate as infection spreads to the stem (Alcantara and Bosland, 1994). Stem-blight and crown-rot symptoms of chile pepper are often similar. These symptoms include distinctive black or purple lesions near the soil line (Erwin and Ribeiro, 1996; Ristaino and Johnston, 1999). The lesions rapidly coalesce and girdle the main branches of stem, which results in stem or entire plant death (Erwin and Ribeiro, 1996).
The early symptoms of fruit-blight include small, water-soaked, dull-colored spots that can rapidly elongate under favorable conditions. Fruit-blight symptoms can continue to spread until most of the chile pepper pod is symptomatic, resulting in unmarketable fruit. Lesions generally occur at either the stem end or the blossom tip of the fruit, but can spread quickly toward the center of the fruit (Erwin and Ribeiro, 1996). The infected tissue becomes dry, sunken, and paper-like and will often turn a tan or straw color.
Management Strategies
Phytophthora blight encompasses both below-ground and above-ground symptoms (Leonian, 1922). Conditions conducive to root infection by P. capsici are saturated soil for extended periods and warm soil temperatures (Weber, 1932; Walker and Bosland, 1999). Free water in the soil from rainfall and irrigation has a greater effect on disease severity than the initial concentration of inoculum (Ristaino, 1991). Additionally, Phytophthora outbreaks may be more severe in low or shaded areas of a field, due to slow drying in these areas (Bosland and Lindsey, 1991; Goldberg, 1995; Hausbeck and Lamour, 2004).
Foliar-blight and stem-blight are serious problems in areas with high relative humidity (Gevens et al., 2007) or during the fall rainy period in other regions (Barksdale et al., 1984; Alcantara and Bosland, 1994). Splashing water due to heavy rainfall or overhead irrigation may allow normally soil-borne P. capsici to infect aerial plant parts (Black, 1999). The disease may also result from sporangia and zoospores produced on diseased plant parts when environmental conditions are favorable. In New Mexico, United States, plants are contaminated when fruit pickers spread infested soils onto wet leaves; especially when harvesting early in the morning when dew is on the leaves.
Current management practices for Phytophthora are cultural, chemical and planting resistant hosts. These approaches include irrigation management, crop rotation, soil solarization, fungicide applications (Ristaino and Johnston, 1999; Sanogo, 2003; Hausbeck and Lamour, 2004; Granke et al., 2012; Sanogo and Bosland, 2012), and the planting of cultivars that are resistant to local isolates. Generally, these management strategies aim to limit losses associated with the pathogen because once established, P. capsici is very difficult to eradicate (Lamour et al., 2011). Additionally, P. capsici can readily move from field to field and rapidly establish itself in a given region, as surface water used for irrigation is an important means of disseminating the pathogen (Gevens et al., 2007). Extreme weather events (e.g., flooding, hurricanes, or typhoons) can initiate new and widespread infestations (Sheu et al., 2009; Dunn et al., 2010). Since fully restricting the movement of P. capsici among sites is often impossible, the best approach to prevent P. capsici infection in vegetable crops is the development of resistant cultivars because it is less expensive and a sustainable alternative to fungicide applications and other management practices (Hausbeck and Lamour, 2004).
Host Range
Originally considered to be host specific (Tucker, 1931), it has since been shown that P. capsici can infect many other plant species including cultivated crops, ornamentals, and native plants belonging to more than 15 plant families (Satour and Butler, 1967; Erwin and Ribeiro, 1996; Hausbeck and Lamour, 2004; Tian and Babadoost, 2004; French-Monar et al., 2006; Granke et al., 2012). It is a major threat to the important crop plant families Cucurbitaceae, Fabaceae, and Solanaceae (Hausbeck and Lamour, 2004). Soon after its identification in 1922, P. capsici was reported to infect eggplant (Solanum melongena L.) (Cruzi, 1927). Phytophthora capsici was first reported to infect cucurbits when Kreutzer (1937) isolated P. capsici in a field of cucumber (Cucumis sativus L.). Three years later, the pathogen was reported to infect muskmelon (C. melo L.), summer squash (Cucurbita pepo L.), and tomato (S. lycopersicum L.) (Kreutzer et al., 1940; Wiant and Tucker, 1940).
In addition to members of the families Cucurbitaceae, Fabaceae, and Solanaceae, Satour and Butler (1967) found the annual crops of okra (Abelmoschus esculentus L.), safflower (Carthamus tinctorius L.), and spinach (Chenopodium amaranticolor Coste and Reyn.) as well as onion (Allium cepa L.) (Leu and Kao, 1981) are hosts of P. capsici. Additionally, woody perennial crops such as apple (Malus pumila Mill.) (Wiant and Tucker, 1940), avocado (Persea americana Mill.) (Tompkins and Tucker, 1937), black pepper (Piper nigrum L.) (Tsao et al., 1985), cacao (Theobroma cacao L.) (Zentmyer et al., 1977), fig (Ficus carica L.) (Katsura and Tokura, 1955), Fraser fir (Abies fraseri Pursh.) (Quesada-Ocampo et al., 2009), macadamia (Macadamia integrifolia Maiden & Betche) (Hunter et al., 1971), papaya (Carica papaya L.) (Erwin and Ribeiro, 1996), peach (Prunus persica L.) (Tompkins and Tucker, 1937), and rubber (Hevea brasiliensis Müll. Arg.) (Erwin and Ribeiro, 1996) are also host species. Most of these isolates likely belong to evolutionarily distinct species and can no longer share genetic information via mating with the vegetable strains (Donahoo and Lamour, 2008).
Pathogen Distribution
After its first identification, P. capsici was quickly recognized throughout important production regions in the United States. Following New Mexico, the pathogen was identified in California (Tompkins and Tucker, 1937, 1941) in the 1920s, in Colorado (Bodine, 1935; Sandsten, 1939), Florida (Weber, 1932), Arizona (Brown and Evans, 1933), and New York (Wiant and Tucker, 1940) in the 1930s, Texas in the 1940s (French-Monar et al., 2009, New Jersey in the 1960s (Barksdale et al., 1984; Parra and Ristaino, 1998), Hawaii in the 1970s (Hunter et al., 1971), and in South Carolina (Quesada-Ocampo et al., 2011), Michigan (Hausbeck and Lamour, 2004), and Illinois (Babadoost, 2000) in the 1990s. Today, P. capsici is likely established in every state (Hausbeck and Lamour, 2004; Quesada-Ocampo et al., 2011; Granke et al., 2012).
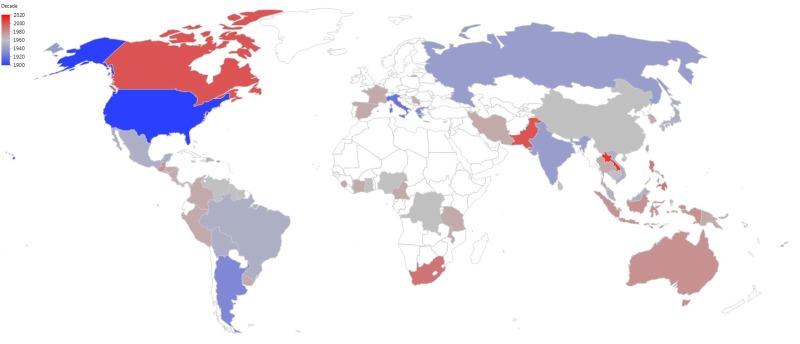
Although it is not known how the pathogen is spread over long distances (Lamour et al., 2011), P. capsici is truly a global disease (Cruzi, 1927; Tucker, 1928; Sarejanni, 1936; Marchionatto, 1938; Godoy, 1940; Thomas et al., 1947; Osnitzkaya, 1949; Malaguti and Pontis-Videla, 1950a,b; Do Amaral, 1952; Katsura and Tokura, 1954; Bell and Alandia, 1957; Turner, 1960, 1961a,b; Holliday and Mowat, 1963; Ravise, 1966; Brasier, 1969; Ershad, 1971; Fernandez-Northcote, 1971; Alfaro Moreno and Vegh, 1972; Clerjeau, 1973; Zentmyer et al., 1973; Aleksić et al., 1975; Kim et al., 1975; Griffin, 1977; Tsao and Tummakate, 1977; Zhou et al., 1984; Alizadeh and Tsao, 1985; Tsao et al., 1985; Carter, 1986; Mu and Tsao, 1987; Tsao and Mu, 1987; Romero-Cova, 1988; Ho, 1990; Thompson et al., 1994; Anderson and Garton, 2000; Gilbert et al., 2001; Guigón-López and González-González, 2001; Velásquez-Valle et al., 2001; Pérez-Moreno et al., 2003; Noveriza and Quimio, 2004; Silvar et al., 2006; Sholberg et al., 2007; Silva-Rojas et al., 2009; Vásquez-López et al., 2009; Zapata-Vázquez et al., 2012; Nguyen, 2015; Callaghan et al., 2016). The chronological spread of P. capsici is presented in Figure 1. However, this is likely not an exhaustive list, as the pathogen could be present in other countries and just not yet reported.
FIGURE 1.
Progressive global spread of Phytophthora capsici from first identification in New Mexico, United States in the early 1900s through to 2017. The countries that are white in color may have P. capsici; however, no reports identifying the pathogen have been published.
Challenges
For nearly a century, researchers around the world have studied the Capsicum–Phytophthora pathosystem, making great strides in understanding this complex interaction. However, even with the greater knowledge gained, the global incidence of the disease is increasing (Hwang and Kim, 1995; Ristaino and Johnston, 1999; Parra and Ristaino, 2001; Kousik and Keinath, 2008; Stam et al., 2013) and most commercial cultivars are either very susceptible or only partially resistant to P. capsici (Ristaino and Johnston, 1999; Hausbeck and Lamour, 2004; Café-Filho and Ristaino, 2008). Progress is slow in limiting losses associated with P. capsici because of the unique challenges presented by this devastating pathogen.
Fungicide Resistance
Although morphologically similar to fungi, oomycetes are genetically and biochemically divergent (Erwin and Ribeiro, 1996) and are generally not sensitive to most broad-spectrum fungicides (Davidse et al., 1991). Therefore, the fungicides growers can rely on to manage oomycetes are limited (Lamour and Hausbeck, 2000). Metalaxyl (Ridomil®; Syngenta) is a phenylalamide fungicide introduced in 1977 that provided systemic protection against oomycetes diseases, including Phytophthora sp. (Cohen et al., 1979; Davidse, 1995; Schwinn and Staub, 1995). Metalaxyl has been used to manage root- and crown-rot of chile pepper (Papavizas and Bowers, 1981; Johnston, 1982; Schlub and Johnston, 1982; Hwang and Sung, 1989; Ristaino et al., 1993; Matheron and Matejka, 1995). Metalaxyl was replaced with mefenoxam (Ridomil Gold®; Syngenta), contains the active enantiomer contained in metalaxyl (Parra and Ristaino, 2001) and has been widely used to manage P. capsici (Lamour and Hausbeck, 2000; Silvar et al., 2006).
The mode of action of phenylamide fungicides is site specific, and fungicide insensitivity was observed in susceptible plant pathogens soon after their introduction in the 1970s (Lamour and Hausbeck, 2000). Insensitivity to mefenoxam and metalaxyl has been widely observed in P. capsici (Bruin and Edington, 1981, 1982; Bower and Coffey, 1985; Abdellaoui-Maane et al., 1988; Lucas et al., 1990; Miller et al., 1994; Hwang and Kim, 1995; Parra and Ristaino, 1998, 2001; Mathis et al., 1999; Ristaino and Johnston, 1999; Agosteo et al., 2000; Lamour and Hausbeck, 2000, 2001b, 2003; Louws et al., 2000; Stevenson et al., 2000, 2001; Babadoost and Islam, 2001; Ploetz R.C. et al., 2001; Tamietti and Valentino, 2001; Matheron and Porchas, 2002; McGovern et al., 2003; Seebold and Horten, 2003; Zhang and Liang, 2003; Hausbeck and Lamour, 2004; McGrath, 2004; Waldenmaier, 2004; French-Monar et al., 2006; Silvar et al., 2006; Gevens et al., 2007; Café-Filho and Ristaino, 2008; Qi et al., 2008; Liu et al., 2009).
Given the global emergence of insensitivity to phenylamide fungicides in P. capsici, alternative fungicides have been evaluated (Cui et al., 2009; Sun et al., 2010; Bi et al., 2014). Some of these compounds include azoxystrobin, cyazofamid, cymoxanil, dimethomorph, fluazinam, fosetyl-A1, oligochitosn, oxathiapiprolin, and zoxmide (Matheron and Porchas, 2000; Ivors et al., 2006; Keinath, 2007; Xu et al., 2007a,b; Ji and Csinos, 2015). However, soon after their first use to manage the disease, insensitivity is often observed (Kousik and Keinath, 2008; Cui et al., 2009; Sun et al., 2010; Bi et al., 2014; Miao et al., 2016).
The unusually rapid and high preponderance of fungicide insensitivity in P. capsici is likely due to the pathogen’s ability to sexually reproduce resulting in high rates of genetic recombination in addition to the production of oospores that can persist in the soil for many years (Lamour and Hausbeck, 2000, 2001b; Bi et al., 2014). Additionally, this is further evidence that resistant cultivars are the best management strategy for P. capsici. In Phytophthora, insensitivity to the phenylamide class of fungicides has been reported to be controlled by a single major effect locus with incomplete dominance that is subject to modifying genes with minor effects (Shattock, 1988; Chang and Ko, 1990; Bhat et al., 1993; Fabritius et al., 1997; Lamour and Hausbeck, 2000). Additionally, once mefenoxam insensitivity has been introduced into a population it is persistent and the frequency of insensitive individuals does not decrease after selection pressure is removed (Bower and Coffey, 1985; Lamour and Hausbeck, 2001b). Sensitivity to dimethomorph was found to be controlled by two dominant genes (Bi et al., 2014) and oxathiapiprlin by a single gene (Miao et al., 2016).
Multiple Disease Syndromes
As previously stated, depending on the point of infection, growing environment, and plant maturity, P. capsici can cause disease on effectively every part of the chile pepper plant (Alcantara and Bosland, 1994; Goldberg, 1995; Ristaino and Johnston, 1999; Walker and Bosland, 1999; Sy et al., 2005). For each P. capsici disease syndrome (root-rot, foliar-blight, stem-blight, and fruit-blight) separate and independent resistant systems have evolved in the host (Monroy-Barbosa and Bosland, 2010), requiring the presence of independent resistance genes for the control of each disease syndrome (Walker and Bosland, 1999; Sy et al., 2005).
The necessity of independent resistance genes for each of the multiple disease syndromes caused by P. capsici in chile pepper increases the complexity of resistance breeding. For host resistance, plant breeders have to pyramid multiple resistance genes in a cultivar to a single race of P. capsici. A similar phenomenon has been observed in the closely related pathosystem of potato (S. tuberosum L.) and P. infestans ([Mont.] de Bary) (Bonde et al., 1940; Rudorf et al., 1950).
Multitude of Races
Within the Phytophthora root-rot and foliar-blight disease syndromes, more than 45 physiological races for have been identified (Hwang et al., 1995; Oelke et al., 2003; Glosier et al., 2008; Sy et al., 2008; Lee et al., 2010; Monroy-Barbosa and Bosland, 2011; da Costa Ribeiro and Bosland, 2012; Jiang et al., 2015; Barchenger, 2017) with different R genes controlling the resistant phenotype against each physiological race of P. capsici within each disease syndrome (Monroy-Barbosa and Bosland, 2008). Screening for resistance has been accomplished on a wide range of genetic material (Kimble and Grogan, 1960; Barksdale et al., 1984; Peter et al., 1984; Reifschneider et al., 1986; Ortega et al., 1991; Candole et al., 2010), and sources for P. capsici resistance have been identified in C. annuum such as Criollo de Morelos 334 (CM334), PI 201232, PI 201234, PI 201237, and PI 640532 (McGregor et al., 2011) from southern Mexico, AC2258 from Central America, and ‘Perennial’ from India. Among the sources of resistance, CM334 has the highest resistance level (Quirin et al., 2005). It is proposed that the center of origin for P. capsici is Mexico, Central or South America (Zentmyer, 1988). The reason the majority of P. capsici resistant chile peppers are from this region can be explained by an evolutionary arms race. This coevolution results in plant specificity and pathogen virulence continually adapting in response to each other. For this reason, breeding for P. capsici resistance in chile pepper is challenging, because new races are continually evolving to overcome the host resistance.
Several P. capsici race identification systems have been proposed (Black, 1999; Oelke et al., 2003; Glosier et al., 2008; Lee et al., 2010); however, these relied on the use of cultivars as the host differential. Using chile pepper cultivars for race detection has limitations because cultivars can vary among seed companies and can segregate (Votava and Bosland, 2002; Candole et al., 2012). Additionally, cultivars can become unavailable, and not all accessions are available to scientists in different countries. Sy et al. (2008) developed a differential set of New Mexico Recombinant Inbred Lines (NMRIL) for P. capsici race characterization that been used for large scale race detection. Recombinant inbred lines (RILs) are often used as host differentials to identify races of pathogens (Lister and Dean, 1993). The RILs allow the maximum genetic variability within a population with homozygous genotypes that can be replicated permanently without the risk of segregation occurring. The NMRILs have been used for race detection in the United States (Monroy-Barbosa and Bosland, 2008, 2010, 2011; Sy et al., 2008; Jiang et al., 2015), Brazil (da Costa Ribeiro and Bosland, 2012), and Taiwan (Barchenger, 2017). The NMRILs have the potential to differentiate thousands of races of P. capsici based on the formula 2n, where 2 is the number of possible reactions (resistant and susceptible) and n is the number of host differentials used.
Mating Type and Genetic Recombination
Phytophthora capsici is a heterothallic species that can reproduce both asexually and sexually (Erwin and Ribeiro, 1996). Once the pathogen is introduced into a field and exposed to water (such as rainfall or irrigation), P. capsici rapidly reproduces asexually through the production of sporangia and motile zoospores (Hausbeck and Lamour, 2004; Lamour and Kamoun, 2009). Each sporangium can produce 20–40 zoospores that can travel in standing water and infect nearby plants (Hausbeck and Lamour, 2004). This swift spread throughout a field can result in losses up to 100% within days. For isolates recovered from the middle and eastern United States, there is no evidence to suggest host specialization (Castro-Rocha et al., 2017). Infection on a single cucumber or pumpkin easily leads to 100’s of millions of spores being released during a rain or irrigation event. Recent studies indicate zoospore progeny can have genomes markedly different, at the chromosome level, from the isolate (or isolates) that initiated the infection and that an impressive, and potentially highly significant, amount of asexual evolution is occurring during spore production (Barchenger et al., 2017; Castro-Rocha et al., 2017; Shrestha et al., 2017).
Sexual reproduction occurs when the two mating types that have been designated as A1 and A2 (Erwin and Ribeiro, 1996), are in close proximity. Exposure to mating type specific hormones α1 and α2 stimulates production of gametangia, outcrossing, and recombinant oospore formation (Ko, 1988). Interestingly, both mating types also produce male and female gametangia and are capable of self-fertilization (Ko, 1988). However, self-fertilization is not likely to occur as often as outcrossing (Uchida and Aragaki, 1980; Dunn et al., 2014a). These recombinant oospores can survive extended periods of cold temperatures (Hausbeck and Lamour, 2004; Babadoost and Pavon, 2013) and are the source of overwintering inoculum in regions with cold winter conditions (Bowers, 1990; Lamour and Hausbeck, 2003; Granke et al., 2012). Regardless of host availability, oospores have been observed to remain viable in diverse soil textures for several years (Babadoost and Pavon, 2009). When the oospores are exposed to a susceptible host and favorable conditions, they rapidly initiate the repeating asexual reproductive cycle and begin their progression throughout the field (Hausbeck and Lamour, 2004; Granke et al., 2012).
Recently, Carlson et al. (2017) identified a 1.6 Mbp region associated with mating type determination, designated the “mating type region” (MTR) in a closed bi-parental field population in New York, United States. This population started with diploid parents and the authors report elevated heterozygosity across the MTR for the A2 mating type relative to the A1 mating type followed intensive inbreeding. This finding was supported by Barchenger et al. (2017), who found that the A1 isolates collected in Taiwan were largely diploid and the A2 isolates were generally triploid or higher ploidy.
Both A1 and A2 mating types of P. capsici have been widely identified within the same field (Ristaino, 1990; Pan, 1997; Parra and Ristaino, 1998, 2001; Lamour and Hausbeck, 2000, 2001a, 2002; Ploetz R. et al., 2001; Fernandez-Pavia et al., 2004; Islam et al., 2004; Ann et al., 2008; Donahoo and Lamour, 2008; Glosier et al., 2008; French-Monar et al., 2009; Sheu et al., 2009; Dunn et al., 2010; Gobena et al., 2012a; Yin et al., 2012; Jiang et al., 2015; Barchenger et al., 2017), increasing the probability of sexual reproduction leading to new races and recombinant oospores resulting in persistence across growing seasons.
Where both mating types exist, sexual reproduction is associated with genetic diversity, persistent pathogens, and often an A1:A2 ratio of ∼1:1 (Lamour and Hausbeck, 2001a; Dunn et al., 2010). Additionally, it has been proposed that there is a climatic influence on mating type distribution. In tropical regions, it is not completely necessary for the pathogen to produce oospores to survive dormantly and clonal lineages may persist for years (Hulvey et al., 2011). A predominance of one or the other mating type has been observed in tropical environments (da Costa Ribeiro and Bosland, 2012; Barchenger et al., 2017). In more temperate environments with greater seasonality, low rates of selfing as well as persistent and a more uniform distribution of mating type has been observed (Lamour and Hausbeck, 2001a; Dunn et al., 2010; Hu et al., 2013b; Carlson et al., 2017). Additionally, exceptions in which there are deviations from a 1:1 ratio in mating type have been observed (Glosier et al., 2008; Sun et al., 2008; Sy et al., 2008; Sheu et al., 2009). This is likely due to the rise of particularly virulent clonal lineages within a growing season and may not reflect the full diversity of a population (Lamour and Hausbeck, 2001b).
Mutation and Loss of Heterozygosity
The high level of diversity found in P. capsici in a single field has also been attributed to mutation and loss of heterozygosity (Lamour et al., 2012; Hu et al., 2013a; Dunn et al., 2014a). Although mutations are the primary source of new genetic variation in oomycetes (Goodwin, 1997), these mutations often cause no observable changes (Silvar et al., 2006). However, rapid genetic changes due to mutation at virulence loci have been observed in P. infestans and P. sojae (Drenth et al., 1994, 1996; Förster et al., 1994; Goodwin et al., 1995; Sujkowski et al., 1996).
Recent work with P. capsici in Taiwan and the closely related species P. colocasiae (host specific to taro [Colocasia esculenta (L.) Schott]), recovered from Nepal, Vietnam, China, and Hawaii, are shedding new light on a novel component to diversity with P. capsici and P. colocasiae and very likely, the genus as a whole (Barchenger et al., 2017; Shrestha et al., 2017). Loss of heterozygosity was described in detail in the paper presenting the draft reference genome for P. capsici (Lamour et al., 2012). This phenomenon occurred on a large scale, across a high percentage of sexual progeny produced to make a detailed genetic map, and was not specific to any one region of the genome and in total – impacted more than 30% of the P. capsici genome (Lamour et al., 2012). How it occurs is a mystery but newer sequencing technologies, particularly whole genome sequencing and targeted amplicon sequencing indicate the genomes for P. capsici and P. tropicalis can differ dramatically from the diploid state. The difference is not limited to a situation where all the chromosomes are triploid or some other ploidy level, instead it was found that individual isolates can be a mosaic of aneuploid variation. Current work investigating single zoospore progeny from multiple field isolates indicates chromosome dosage can be highly variable within a single zoospore-derived isolate and there is little fidelity to the chromosomal complement of the parental strain. This has potentially significant implications for rapid evolution where gene dosage may allow an isolate to overcome a novel human-mediated selection pressure, including resistance genes and Phytophthora-toxic chemicals, and clearly can play a role in the rapid evolution of populations to novel resistance incorporated by the plant breeder.
Breeding Approaches
Classical breeding approaches for transferring resistance to P. capsici into adapted chile pepper germplasm has been a goal of many breeding programs. One major challenge to chile pepper breeders is that different inheritance models have been reported among the sources of resistance to P. capsici. Several laboratories studying CM334 report at least two genes; but often more genes confer resistance (Guerrero-Moreno and Laborde, 1980; Ortega et al., 1991, 1992; Reifschneider et al., 1992; Walker and Bosland, 1999; Thabuis et al., 2003; Sy et al., 2005). Other studies report a single dominant gene (Saini and Sharma, 1978; Kim and Hur, 1990) or a single dominant gene with modifying genes (Barksdale et al., 1984) control resistance such as in PI 201234 and bell pepper (Smith et al., 1967). Multiple genes with additive or epistatic effects are involved in resistance in ‘Perennial’ (Lefebvre and Palloix, 1996). However, it is likely that the qualitative gene model reported for resistance in chile pepper is actually race-specific resistance (Sy et al., 2008; Foster and Hausbeck, 2010) as well as syndrome-specific resistance (Sy et al., 2005). Another effect confounding inheritance studies is variation in the screening techniques among the studies, leading to different interpretations of potentially the same results.
Resistance in chile pepper has polygenetic inheritance based on multimodal distributions and higher order epistasis effects (Pochard and Daubeze, 1980; Palloix et al., 1988; Bartual et al., 1991, 1993; Pflieger et al., 2001; Lefebvre et al., 2002; Ogundiwin et al., 2005; Bonnet et al., 2007; Minamiyama et al., 2007; Truong et al., 2012; Curtis, 2014). Efforts have been made to identify quantitative trait loci (QTL) linked with P. capsici resistance and transfer these QTLs into elite material (Thabuis et al., 2003, 2004b; Ogundiwin et al., 2005; Sugita et al., 2006; Jin et al., 2007; Minamiyama et al., 2007; Kim et al., 2008; Truong et al., 2012; Liu et al., 2014; Naegele et al., 2014). Although, these QTLs are also often associated with race-specific resistance.
Several molecular markers associated with resistance to P. capsici have been reported in chile pepper for more rapid selection (Quirin et al., 2005; Kim et al., 2008; Truong et al., 2012; Chomkaeo et al., 2014; Liu et al., 2014; Wang et al., 2016; Xu et al., 2016). However, to date, these publically available molecular markers are generally not widely applicable, and some level of phenotype and genotype mismatch has been observed when they are used in diverse germplasm. This phenotype–genotype mismatch limits selection efficiency for marker assisted selection and also further highlights the high level of plasticity in the pathogen.
Historically, it is difficult to introduce P. capsici resistance into well-adapted susceptible cultivars. When using classic backcross methods, resistance is lower than the donor parent with threshold effects, which is likely due to the loss of secondary resistance genes (Palloix et al., 1990). Recurrent selection has been used to move polygenic resistance into elite material (Thabuis et al., 2004a). However, linkage drag associated with low yield, small and undesirable fruit, and less vigorous plants is a major limitation to wide adoption of resistant cultivars. Growers would rather plant high yielding, high quality, more uniform cultivars that are susceptible to P. capsici and risk losing a portion of their crop, than plant less adapted but resistant cultivars. Even cultivars that had field resistance to P. capsici, e.g., Paladin (Dunn et al., 2014b), became susceptible within a decade as the pathogen evolved new virulence in New Jersey (Krasnow et al., 2017). An excellent example of the boom-and-bust cycle of disease resistance.
Additionally, Reeves et al. (2013) identified an inhibitor to P. capsici resistance gene (Ipcr) in New Mexico Capsicum Accession 10399 (NMCA10399). Their results indicate that a single dominant gene inhibited polygenic host resistance to multiple isolates of P. capsici. The single dominant gene inhibited resistance to all disease syndromes. The genetic mechanisms of the Ipcr gene is unknown; however, it is hypothesized to interfere with upstream recognition sites in the host. Additionally, the frequency of the Ipcr gene in commercial cultivars is not known. The authors proved that a chile pepper can be susceptible to P. capsici for two reasons: lack of R genes or presence of an inhibitor gene. These findings further complicate a difficult pathosystem and highlight the complexity of breeding for resistance to P. capsici.
Transgenic Issues
Genetic engineering using Agrobacterium-mediated transgenic approaches has long been used to increased plant resistance to biotic stresses. Members of Solanaceae such as eggplant, petunia (Petunia x hybrid Juss.), potato, tobacco (Nicotiana tabacum L.), and tomato are readily transformed and are considered model organisms for this technology. However, chile pepper is extremely recalcitrant to in vitro regeneration and genetic transformation (Li et al., 2003). Regeneration and transformation of chile pepper has been widely reported (Wang et al., 1991; Zhou et al., 1991; Dong et al., 1992; Lee et al., 1993; Ye et al., 1993; Fari and Andrasfalvy, 1994; Zhu et al., 1996; Jayashankar et al., 1997; Kim et al., 1997; Subhash and Christopher, 1997; Manoharan et al., 1998; Steinitz et al., 1999; Wolf et al., 2001; Li et al., 2003; Lopez-Puc et al., 2006; Arcos-Ortega et al., 2010). However, the problem is an overall lack of reproducibility in these published techniques. One reason for the lack of reproducibility is that regeneration and transformation techniques in chile pepper are genotype-specific (Manoharan et al., 1998). Therefore, different protocols are required depending on the accession being transformed. Furthermore, successfully introduced transgenes in plants regenerated in vitro are often not inherited through subsequent generations of self- or cross-pollination. It is hypothesized the transgenes are quickly lost via transposition. More than 81% of the Capsicum genome consists of transposons, which is high compared to closely related tomato (50%) and potato (47%) (Qin et al., 2014).
Strategies
Despite decades of research to better understand the Capsicum–P. capsici pathosystem, P. capsici is still a major limiting factor for pepper production. Some of the strategies developed in other pathosystems with efficacy in limiting losses associated with infection and disease are not practical for P. capsici. There is a need to identify strategies that can be adopted to better breed for resistance to P. capsici in chile pepper.
Screening Methodology
Breeding for resistance to P. capsici is heavily dependent on the accuracy and precision of the disease screening method used (Chavez and Kabelka, 2009). Several disease screens have been developed for P. capsici. For foliar blight screening, using 1,000 zoospores per plant (Alcantara and Bosland, 1994) and 2,000 zoospores per plant using soaked germination paper (Monroy-Barbosa and Bosland, 2010) have been proposed. Additionally, a foliar spray using inoculum has been used. For root-rot screening, 10,000 zoospores per plant (Bosland and Lindsey, 1991) and 100,000 zoospores per plant (Black, 1999) have been used. In addition, a dose of 5,000 zoospores per plant has been used for screening fruit-rot/blight resistance (Biles et al., 1995). Inoculum concentration and plant age play a major role in the level of resistance displayed in the host (da Costa Ribeiro and Bosland, 2012; Barchenger, 2017; Mansfeld et al., 2017). In order to effectively breed for resistance and correctly identify races of P. capsici, standardized screening protocols should be developed and followed by scientists worldwide.
Race Nomenclature
As described above, several studies identified physiological races of P. capsici (Black, 1999; Oelke et al., 2003; Glosier et al., 2008; Monroy-Barbosa and Bosland, 2008, 2010, 2011; Sy et al., 2008; Lee et al., 2010; da Costa Ribeiro and Bosland, 2012; Jiang et al., 2015). Unfortunately, there is a lack of consistency that can limit overall progress among breeders. Some studies use a numerical or alphabetical nomenclature system with the first race designated Race 1 or A (Glosier et al., 2008; Lee et al., 2010). Other studies number races based on virulence with Race 1 being either the most virulent (Sy et al., 2008) or the least virulent (Black, 1999). Despite the inconsistencies in how the races are identified, the most important limitation is they do not provide a naming scheme that allows for more or less virulent races to be described (Barchenger et al., 2017, 2018). Furthermore, there are overlapping names for genetically divergent races.
Over the years many different races for the different disease syndromes of P. capsici have been identified around the world, and the systems used to designate the different races have no real biological meaning. Therefore, the term race is now being supplemented by a new term, virulence phenotype (Barchenger et al., 2018). Virulence phenotype is used to designate the virulence of the P. capsici isolate on the various host resistance genes. Races are identified based on the differential reaction with the NMRIL, which defines isolates by resistance genes and will hopefully contribute to practical advances in breeding.
Global Strategies for Local Gene Deployment
Based on the current knowledge of this complex pathosystem, it may not be possible to develop cultivars with global or even country-wide durable resistance. However, we propose plant breeders utilize global strategies for local gene deployment for P. capsici resistance. The NMRILs have been used globally (Brazil, China, Taiwan, and across the United States) to characterize P. capsici for the past decade (Monroy-Barbosa and Bosland, 2008, 2010, 2011; Sy et al., 2008; da Costa Ribeiro and Bosland, 2012; Hu et al., 2013b; Naegele et al., 2014, 2017; Naegele and Hausbeck, 2014; Rehrig et al., 2014; Jiang et al., 2015; Barchenger et al., 2018). The NMRILs provide a host differential to identify the virulence phenotype in a given region at a particular time (Barchenger et al., 2018). Simultaneously, the NMRILs also provide insights into the resistance gene(s) required in that region.
A recommended strategy to breed for resistance in a particular region is to utilize the NMRILs to identify the virulence phenotypes in a given region and compare these to the virulence phenotypes from other regions (Sy et al., 2008; da Costa Ribeiro and Bosland, 2012; Jiang et al., 2015; Barchenger et al., 2018). The resistant NMRILs can then be utilized to move resistance into elite germplasm for region-targeted resistant cultivars. The NMRILs provide information on both the pathogen and the host that can be utilized in developing a resistance breeding strategy in a particular region. This was recently demonstrated in Taiwan (Barchenger et al., 2017), where we conducted targeted sequencing on P. capsici isolates collected in Taiwan and analyzed the data in terms of the virulence phenotypes developed based on the NMRILs. A clear relationship between polyploidy in the pathogen and fewer susceptible reactions was found among a set of NMRILs. Polyploid isolates were largely present on the East coast of the island and diploid isolates were largely on the West coast, enabling local gene deployment. Utilizing a globally standardized system to characterize resistance on a local scale also allows plant breeders to compare resistance globally and select lines from different countries or regions with similar virulence phenotypes for use in their own breeding program.
Gene Targeted Resistance
The gene-for-gene model (Flor, 1955, 1971) specifies that in race-specific interactions, the host plant inhibits infection through deployment of defense functions via recognition. This is made possible by the presence of dominant resistance genes in the host that enable recognition of effectors in the pathogen. These effectors encode Pathogen Associated Molecular Patterns/Microbial Associated Molecular Patterns (PAMPs/MAMPs) that are recognized by resistant hosts and trigger Pattern Triggered Immunity (PTI). Successful pathogens, such as P. capsici, have evolved a large and diverse set of secreted effectors that can suppress PTI and initiate Effector-Triggered Susceptibility (ETS) (Jones and Dangl, 2006; Hein et al., 2009; Gill et al., 2015).
Using the P. capsici reference genome, Stam et al. (2013) identified pathogen effector proteins. Several effectors produced by P. capsici, such as those in the RXLR, Crn, and PcNpp classes are thought to play important roles in infection of chile pepper. More than 400 candidate RXLP effectors have been identified in the P. capsici genome (Lamour et al., 2012; Stam et al., 2013). Several necrosis-inducing proteins (PcNLP) have been found to play important roles in symptom development in chile pepper (Feng et al., 2011, 2014). Fu et al. (2015) identified several cell-death-inducting members of the pectate lyase gene family (PcPL) that were highly induced during infection, and could be effectors. The ethylene-responsive factor CaPTI1 appears to be involved in defense response to P. capsici (Jin et al., 2015). A single effector, a PcAvr3a-like protein, has been correlated to non-host resistance in several Nicotiana species (Vega-Arreguín et al., 2014). Interestingly, Vega-Arreguín et al. (2017) found the non-host resistance mechanisms to P. capsici are the same as the mechanism for host-resistance. Selections made within the landrace CM334 act like a non-hosts because no isolates, to date, can infect. To be useful for chile pepper breeding, the effector targets in host differentials derived from CM334 need to be identified. The resistant parent of the host differential NMRILs, CM334, has been sequenced (Kim et al., 2014), which is an important step in identifying effector targets. However, to date, no efforts have been made to identify these regions in the NMRILs.
Although detection of effector targets in the host are limited, efforts have been made to identify resistance genes (Silvar et al., 2008; Wang J.E. et al., 2013; Zhang et al., 2013; Rehrig et al., 2014; Xu et al., 2016). Mallard et al. (2013) identified a major QTL, Pc5.1, located on chromosome 5 associated with resistance to 12 isolates of P. capsici from different geographic regions. It has been widely reported that P. capsici resistance genes are clustered on chromosome 5 (Bonnet et al., 2007; Truong et al., 2012; Liu et al., 2014; Rehrig et al., 2014; Wang et al., 2016). The authors conducted a meta-analysis and found this QTL is highly conserved among diverse resistant chile pepper accessions. Several resistance genes are within and very near to Pc5.1, including CaPhyto (Wang et al., 2016), CaDMR1 (Rehrig et al., 2014) and likely others (Liu et al., 2014). The C. annuum Polygalacturonase-inhibiting Protein1 (CaPGIP1) gene has been identified as to reduce susceptibility in GM tobacco plants (Wang X. et al., 2013). The PGIPs are extracellular plant proteins with recognition ability against many PGs produced by fungi (De Lorenzo et al., 2001). Furthermore, ChiIV3 is a positive regulator of plant cell death and triggers defense signaling and upregulation of pathogenesis related genes against P. capsici infection (Liu et al., 2017). Interestingly, there appears to be several different types of R genes in Capsicum. The majority of the R genes are nucleotide-binding and leucine-rich-repeat proteins (NLRs). Work in R gene identification is more extensive in other Solanaceae crops, and several NLRs have been identified with high orthology to those in tomato and potato (Kim et al., 2014). Recent findings suggest massive expansion of NLR genes in Capsicum, largely due to long-terminal-repeat-retrotransposons-mediated retroduplication (Kim et al., 2017). Richins et al. (2010) identified 168 differentially expressed genes under root-rot inoculation of P. capsici and one of these genes, XEGIP, was further characterized by Jones et al. (2015). The XEGIP gene is modeled to inhibit xyloglucan-specific endo β-1,4 glucanase produced by P. capsici and attacks the xyloglucan bonds in plant cell walls (Yoshizawa et al., 2012). However, the capacity of these genes to recognize PAMPs is unknown. Despite the high number and diversity of resistance genes, breeding for resistance to P. capsici is still inadequate and other strategies are needed to more effectively develop durable resistant cultivars. Further genome-wide analysis of the evolution of NLRs and effectors could provide a basis for gene-targeted resistance breeding.
A potential challenge to identifying effector targets in this host is the presence of the Icpr gene as accessions containing the Icpr gene are always completely susceptible (Reeves et al., 2013). The mode of action and the frequency of the Icpr gene in Capsicum populations are unknown.
Double Haploid Development
As previously described, the use of durable sources of P. capsici resistance in traditional chile pepper breeding programs has limitations. However, the use of double haploid (DH) technology could be a way to fix resistance genes in elite material. Hybridizing adapted material with good horticultural traits to accessions with high levels of resistance and developing DH lines from the F1 generation will allow plant breeders to quickly fix resistance without losing the important horticultural traits. However, there are major limitations to using DH lines, including high cost, necessity of expertise in tissue culture, and the development of protocols specific to each laboratory and genotype. Double haploids have been developed in chile pepper in the past; however, most of the time, success rates are generally low (Dumas de Vaulx et al., 1981; Vagera and Havranek, 1985; Morrison et al., 1986; Munyon et al., 1989; Kristiansen and Andersen, 1993; Maheswary and Mak, 1993; Qin and Rotino, 1993; Ltifi and Wenzel, 1994; Mitykó et al., 1995; Dogimont et al., 1996; Dolcet-Sanjuan et al., 1997; Gyulai et al., 2000; Supena et al., 2006). The reason for the high failure rate is unknown, but it is well known that Capsicum is highly recalcitrant to in vitro regeneration and the media required is highly genotype specific.
Omnigenics
Resistance to P. capsici in chile pepper is a highly complex trait. Through genome-wide association studies, the understanding of the genetic basis of complex traits has greatly expanded. Many important loci generally have small effects and complex traits are largely influenced by non-coding variants such as promoters or enhancers (Boyle et al., 2017). Common SNPs distributed throughout the genome with effects below detectable significance levels account for a large portion of the heritability of complex traits (Yang et al., 2010). Therefore, Boyle et al. (2017) proposed the omnigenic model which postulates that most heritability can be explained by effects on genes outside core disease-related pathways. They suggest that essentially any gene with regulatory variants in at least one tissue that contributes to pathogenicity is likely to have non-trivial effects on disease resistance.
While genes conferring resistance in chile pepper have largely been localized on chromosome 5 (Mallard et al., 2013; Kim et al., 2017), no loci have been identified that account for resistance over a wide geographical region or in diverse genetic backgrounds. It is likely that a large number of variants contribute to resistance (Kim et al., 2017). Therefore, the omnigenic model suggests that to understand the whole picture of disease resistance, we should not only study core genes and pathways, but also the multitude of variants throughout the genome that have seemingly small effects on resistance. The omnigenic model has the potential explain why developing molecular markers and breeding for resistance to P. capsici in chile pepper has been limited in the past and should be considered when breeding for resistance in the future.
FAST SNP Markers for Increased Selection Accuracy
The prediction accuracy from genomic selection for most crop species is generally between 40 and 65%, and essentially never 100% (as reviewed by Fu et al., 2017), and this is also true for most molecular markers available for P. capsici resistance in chile pepper. Therefore, Fu et al. (2017) proposed the development of function-associated specific trait (FAST) SNP markers, as an alternative to regular genome selection, for rapid and more accurate trait predictions. The FAST SNPs technology and has not been employed on vegetable crops (including chile pepper) with large and complex genomes; however, FAST SNPs may increase P. capsici resistance prediction accuracy. To facilitate the development of FAST SNP markers, Fu et al. (2017) proposed a procedure based on RNA-seq of 10 or more pairs of individual plants with extreme trait values (resistant vs. susceptible). Ideally, the lines used for FAST SNP marker development are derived from diverse resistance sources. The RNA-seq reads for each pair of lines are then de novo assembled and differential transcripts are identified. Trait-specific markers can then be developed based on consensus SNPs among all the pairs. FAST SNP markers have the potential to offer better marker-based trait prediction; however, more empirical investigations are needed to confirm their true value. Due to the large genome size of the host, the plasticity of the pathogen genome, and the lack of widely applicable molecular markers makes the multifaceted Capsicum–P. capsici pathosystem an attractive candidate for FAST SNP validation in vegetables.
Conclusion
These are exciting times for plant and pathogen research as new tools, particularly at the genomic level, become available and more affordable. Combinations of strategies and collaborative efforts from scientists around the world are required to effectively breed for resistance to P. capsici. Progress in understanding and manipulating the Capsicum–P. capsici system is likely to be useful in other complex host–pathogen systems and increase our odds to develop durable management strategies.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Ariadna Monroy-Barbosa for her valuable suggestions.
References
- Abdellaoui-Maane S., Sing J. M., Sandrenan P., Bonpeix G. doi: 10.1111/j.1365-2338.2000.tb00891.x. [DOI] [Google Scholar]
- Agosteo G. E., Raudino F., Cacciola S. O. (2000). Resistance of Phytophthora capsici to metalaxyl in plastic-house Capsicum crops in southern Italy. 30 257–261. [Google Scholar]
- Alcantara T. P., Bosland P. W. (1994). An inexpensive disease screening technique for foliar blight of chile pepper seedlings. 29 1182–1183. [Google Scholar]
- Aleksić Ž., Aleksić D., Miladinović Ž. (1975). Bolesti paprike i moguænosti njihovog suzbijanja gajenjem otpornih sorti. 37 73–82. [Google Scholar]
- Alfaro Moreno A., Vegh I. (1972). La “tristeza” o “seca” del pimiento producida por Phytophthora capsici (Leonian). 51:3685 10.1016/S0007-1536(85)80155-8 [DOI] [Google Scholar]
- Alizadeh A., Tsao P. H. (1985). Effect of light in sporangium formation, morphology, ontogeny, and caducity of Phytophthora capsici and ‘P. palmivora’ MF4 isolations from black pepper and other hosts. 85 47–69. 10.1094/PDIS.2000.84.6.705B [DOI] [Google Scholar]
- Anderson T. R., Garton R. (2000). First report of blight of field pepper caused by Phytophthora capsici in Ontario. 84 705 10.1094/PDIS.2000.84.6.705B [DOI] [PubMed] [Google Scholar]
- Ann P. J., Wong I. T., Tsai J. N. (2008). Appearance of A2 mating type of Phytophthora capsici in Taiwan. 17:69. [Google Scholar]
- Arcos-Ortega G. F., Chan-kuuk R. A., Gonzalez-Kantun W. A., Souza-Perera R., Nakazawa-Ueji Y. E., Aviles-Berzunza E., et al. (2010). Agrobacterium tumefaciens-transient genetic transformation of habanero pepper (Capsicum chinense Jacq.) leaf explants. 13 7–8. 10.2225/vol13-issue4-fulltext-10 [DOI] [Google Scholar]
- Babadoost M. (2000). Outbreak of Phytophthora foliar blight and fruit rot in processing pumpkin fields in Illinois. 84:1345 10.1094/PDIS.2000.84.12.1345A [DOI] [PubMed] [Google Scholar]
- Babadoost M., Islam S. Z. (2001). Evaluation of fungicides for control of Phytophthora blight of processing pumpkin. 56:V65. [Google Scholar]
- Babadoost M., Pavon C. (2009). “Survival of Phytophthora capsici oospores in soil,” in , Leuven, 11. [DOI] [PubMed] [Google Scholar]
- Babadoost M., Pavon C. (2013). Survival of oospores of Phytophthora capsici in soil. 97 1478–1483. 10.1016/j.riam.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Barchenger D. W. (2017). Ph.D. dissertation, New Mexico State University, Las Cruces, NM. [Google Scholar]
- Barchenger D. W., Lamour K. H., Sheu Z. M., Shrestha S., Kumar S., Lin S. W., et al. (2017). Intra- and intergenomic variation of ploidy and clonality characterize Phytophthora capsici on Capsicum sp. in Taiwan. 16 955–963. 10.1007/s11557-017-1330-0 [DOI] [Google Scholar]
- Barchenger D. W., Sheu Z. M., Kumar S., Lin S. W., Burlakoti R. R., Bosland P. W. (2018). Race characterization of Phytophthora capsici as a basis for global anticipatory resistance breeding in Capsicum. 10.1094/PHYTO-08-17-0289-R [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Barksdale T. H., Papavizas G. C., Johnson S. A. (1984). Resistance to foliar blight and crown rot of pepper caused by Phytophthora capsici. 68 506–509. 10.1094/PD-68-506 [DOI] [Google Scholar]
- Bartual R., Carbonell E. A., Marsal J. I., Tello J. C., Campos T. (1991). Gene action in resistance to peppers (Capsicum annuum) to Phytophthora stem blight (Phytophthora capsici L.). 54 195–200. [Google Scholar]
- Bartual R., Lacasa A., Marsal J. I., Tello J. C. (1993). Epistasis in the resistance of pepper to Phytophthora stem blight (Phytophthora capsici L.) and its significance in the prediction of double cross performances. 72 149–151. 10.1007/BF00023784 [DOI] [Google Scholar]
- Bell F. H., Alandia B. S. (1957). Diseases of temperate climate crops in Bolivia. 41 646–649. [Google Scholar]
- Bhat R. G., McBlain B. A., Schmitthenner A. F. (1993). The inheritance of resistance to metalaxyl and to flurorophenylalnine in matings of homothallic Phytophthora sojae. 97 865–870. 10.1016/S0953-7562(09)81164-7 [DOI] [Google Scholar]
- Bi Y., Hu J., Cui X., Shao J., Lu X., Meng Q., et al. (2014). Sexual reproduction increases the possibility that Phytophthora capsici will develop resistance to dimethomorph in China. 63 1365–1373. 10.1111/ppa.12220 [DOI] [Google Scholar]
- Biles C. L., Bruton B. D., Wall M. M., Rivas M. (1995). Phytophthora capsici zoospore infection of pepper fruit in various physical environments. 75 1–5. [Google Scholar]
- Black L. L. (1999). “Studies on Phytophthora blight in pepper,” in , ed. Talekar N. S. (Shanhua: Asian Vegetable Research and Development Center; ), 25–27. [Google Scholar]
- Bodine E. W. (1935). Blight of peppers. 86:6. 15717130 [Google Scholar]
- Bonde R., Stevenson F. J., Clark C. F. (1940). Resistance of certain potato varieties and seedling progenies to late blight in the tubers. 30 733–748. [Google Scholar]
- Bonnet J., Danan S., Boudet C., Barchi L., Sage-Palloix A. M., Caromel B., et al. (2007). Are the polygenetic architectures of resistance to Phytophthora capsici and P. parasitica independent in pepper?. 115 253–264. 10.1007/s00122-007-0561-x [DOI] [PubMed] [Google Scholar]
- Bosland P. W. (2008). “Think global, breed local: specificity and complexity of Phytophthora capsici,” in , Atlantic City, NJ. [Google Scholar]
- Bosland P. W., Lindsey D. L. (1991). A seedling screen for Phytophthora root rot of pepper, Capsicum annuum. 75 1048–1050. 10.1371/journal.pone.0151401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower L. A., Coffey M. D. (1985). Development of laboratory tolerance to phosphorous acid, and fosetyl-AL and metalaxyl in Phytophthora capsici. 7 1–6. 10.1080/07060668509501507 [DOI] [Google Scholar]
- Bowers J. H. (1990). Effect of soil temperature and soil-water matric potential on the survival of Phytophthora capsici in natural soils. 74 771–777. 10.1094/PD-74-0771 [DOI] [Google Scholar]
- Boyle E. A., Yang I. L., Pritchard J. K. (2017). An expanded view of complex traits: from polygenic to omnigenic. 169 1177–1186. 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasier C. M. (1969). Formation of oospores in vivo by Phytophthora palmivora. 52 273–279. 10.1016/S0007-1536(69)80040-9 [DOI] [Google Scholar]
- Brown J. G., Evans M. N. (1933). A Phytophthora rot of watermelon. 51 45–65. 10.5423/PPJ.OA.02.2012.0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruin G. C. A., Edington L. V. (1981). Adaptive resistance in Peronosporales to metalaxyl. 3 201–206. 10.1080/07060668109501348 [DOI] [Google Scholar]
- Bruin G. C. A., Edington L. V. (1982). Induction of fungal resistance of metalaxyl by ultraviolet irradiation. 72 476–480. 10.1094/Phyto-72-476 [DOI] [Google Scholar]
- Café-Filho A. C., Ristaino J. B. (2008). Fitness of isolates of Phytophthora capsici resistant to mefenoxam from squash and pepper fields in North Carolina. 92 1439–1443. 10.1094/PDIS-92-10-1439 [DOI] [PubMed] [Google Scholar]
- Callaghan S. E., Williams A. P., Burgess T., White D., Keovorlajak T., Phitsanoukane P., et al. (2016). First report of Phytophthora capsici in the Lao PDR. 11 1–4. 10.1007/s13314-016-0210-9 [DOI] [Google Scholar]
- Candole B. L., Conner P. J., Ji P. (2010). Screening Capsicum annuum accessions for resistance to six isolates of Phytophthora capsici. 45 254–259. [Google Scholar]
- Candole B. L., Conner P. J., McGregor C., Waters V., Ji P. (2012). The disease reactions of heirloom bell pepper ‘California Wonder’ to Phytophthora capsici. 3 417–424. [Google Scholar]
- Carlson M. O., Gazave E., Gore M. A., Smart C. D. (2017). Temporal genetic dynamics of an experimental, biparental field population of Phytophthora capsici. 8:26. 10.3389/fgene.2017.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter E. M. (1986). Vegetable disease. 5:15. [Google Scholar]
- Castro-Rocha A., Hulvey J. P., Wick R., Shrestha S. K., Lamour K. (2017). Genetic diversity of Phytophthora capsici recovered from Massachusetts between 1997 and 2014. 16 999–1006. 10.1007/s11557-017-1334-9 [DOI] [Google Scholar]
- Castro-Rocha A., Shrestha A., Lyon B., Grimaldo-Pantoja G. L., Flores-Marges J. P., Valero-Galván J., et al. (2016). An initial assessment of the genetic diversity for Phytophthora capsici in northern and central Mexico. 15:15 10.1007/s11557-016-1157-0 [DOI] [Google Scholar]
- Chang T. T., Ko W. H. (1990). Resistance to fungicides and antibiotics in Phytophthora parasitica. Genetic nature and use in hybrid determination. 80 1414–1421. 10.1094/Phyto-80-1414 [DOI] [Google Scholar]
- Chavez D. J., Kabelka E. A. (2009). “Methods for evaluating foliar response of winter squash to Phytophthora capsici,” in , Duck Key, FL, 15. [Google Scholar]
- Chomkaeo N., Struss D., Sakulsingharoj C., Sangtong V., Pongjareonkit S. (2014). Development of the single nucleotide polymorphism markers for Phytophthora capsici resistance in Capsicum annuum with QTL analysis. 7 192–201. 18795251 [Google Scholar]
- Clerjeau M. (1973). A new collar rot disease of melon caused by Phytophthora nicotianae Br. De H. var. parasitica (Dastur) Waterh. and Phytophthora capsici Leon. 59 54–58. [Google Scholar]
- Cohen Y., Reuveni M., Eyal H. (1979). The systemic antifungal activity of Ridomil against Phytophthora infestans on tomato plants. 69 645–649. 10.1094/Phyto-69-645 [DOI] [Google Scholar]
- Cruzi M. (1927). L’eziologia delta “cangrena peìdale” del Capsicum annuum L. 17 1–19. [Google Scholar]
- Cui X. L., Meng Q. X., Bi Y., Wu P. F., Liu X. L. (2009). Baseline sensitivity and laboratory mutants of Phytophthora capsici resistant to dimethomorph. 39 630–637. [Google Scholar]
- Curtis M. R. (2014). MS thesis, University of California, Davis, CA. [Google Scholar]
- da Costa Ribeiro C. S., Bosland P. W. (2012). Physiological race characterization of Phytophthora capsici isolates from several host plant species in Brazil using New Mexico recombinant inbred lines of Capsicum annuum at two inoculum levels. 137 421–426. [Google Scholar]
- Davidse L. C. (1995). “Phenylamide fungicides. Biochemical action and resistance,” in , ed. Lyr H. (New York, NY: Gustav Fisher Verlag; ), 347–354. [Google Scholar]
- Davidse L. C., van den Berg-Velthuis G. C. M., Mantel B. C., Jespers A. B. K. (1991). “Phenylamides and Phytophthora,” in , eds Lucas J. A., Shattock R. C., Shaw D. S., Cooke L. R. (Cambridge: British Mycological Society; ), 349–360. [Google Scholar]
- De Lorenzo G., D’Ovidio R., Cervone F. (2001). The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. 36 313–335. 10.1146/annurev.phyto.39.1.313 [DOI] [PubMed] [Google Scholar]
- Do Amaral J. F. (1952). Requeima do pimentao. 18 160–161. [Google Scholar]
- Dogimont C., Palloix A., Daubze A. M., Marchou G., Gebre Selassie K., Pochard K. (1996). Genetic analysis of broad spectrum resistance to potyviruses using doubled haploid lines of pepper (Capsicum annuum L.). 88 231–239. 10.1007/BF00023895 [DOI] [Google Scholar]
- Dolcet-Sanjuan R., Claveria E., Huerta A. (1997). Androgenesis in Capsicum annuum L.- Effects of carbohydrate and carbon dioxide enrichment. 122 468–475. [Google Scholar]
- Donahoo R. S., Lamour K. H. (2008). Interspecific hybridization and apomixis between Phytophthora capsici and Phytophthora tropicalis. 100 k911–920. 10.3852/08-028 [DOI] [PubMed] [Google Scholar]
- Dong C., Jiang C. H., Feng L., Guo J. (1992). Transgenic pepper plants (Capsicum annuum L.) containing CMV sat-RNA cDAN. 19 184–186. [Google Scholar]
- Drenth A., Tas I. C. Q., Grovers F. (1994). DNA fingerprinting uncovers a new sexually reproducing population of Phytophthora infestans in the Netherlands. 100 97–107. 10.1007/BF01876244 [DOI] [Google Scholar]
- Drenth A., Whisson S. C., Maclean D. J., Irwin J. A. G., Obst N. R., Ryley M. J. (1996). The evolution of races of Phytophthora sojae in Australia. 86 163–169. 10.1094/Phyto-86-163 [DOI] [Google Scholar]
- Dumas de Vaulx R., Chambonnet D., Pochard E. (1981). Culture in vitro d’anthères de piment (Capsicum annuum L.): amélioration des taux d’obtention de plantes chez différents génotypes par des traitements á + 35°C. 1 859–864. [Google Scholar]
- Dunn A. R., Bruening S. R., Grünwald N. J., Smart C. D. (2014a). Evolution of an experimental population of Phytophthora capsici in the field. 104 1107–1117. 10.1094/PHYTO-12-13-0346-R [DOI] [PubMed] [Google Scholar]
- Dunn A. R., Lange H. W., Smart C. D. (2014b). Evaluation of commercial bell pepper cultivars for resistance to Phytophthora blight (Phytophthora capsici). 15 19–24. [Google Scholar]
- Dunn A. R., Milgroom M. G., Meitz J. C., McLeod A., Fry W. E., McGarth M. T., et al. (2010). Population structure and resistance to mefenoxam of Phytophthora capsici in New York State. 96 1461–1468. 10.1094/PDIS-03-10-0221 [DOI] [PubMed] [Google Scholar]
- Ershad D. (1971). Dahlem: BBA, 140. [Google Scholar]
- Erwin D. C., Ribeiro O. K. (1996). St Paul, MN: The American Phytopathological Society. [Google Scholar]
- Fabritius A., Shattock R. C., Judelson H. S. (1997). Genetic analysis of metalaxyl insensitivity loci in Phytophthora infestans using linked DNA markers. 87 1034–1040. 10.1094/PHYTO.1997.87.10.1034 [DOI] [PubMed] [Google Scholar]
- Fari M., Andrasfalvy A. (1994). Regeneration and cloning of pepper. (Capsicum sp.): a review. 26 9–27. [Google Scholar]
- Feng B. Z., Li P. Q., Fu L., Sun B. B., Zhang X. G. (2011). Identification of 18 genes encoding necrosis-inducing proteins from the plant pathogen Phytophthora capsici (Pythiaceae: Oomycetes). 10 910–922. 10.4238/vol10-2gmr1248 [DOI] [PubMed] [Google Scholar]
- Feng B. Z., Zhu X. P., Fu L., Lv R. F., Storey D., Tooley P., et al. (2014). Characterization of necrosis-inducing NLP proteins in Phytophthora capsici. 14:126. 10.1186/1471-2229-14-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Northcote E. N. (1971). “La marchitez y otras enfermades de Capsicum spp. Cultivades en el Peru,” in , Lima. [Google Scholar]
- Fernandez-Pavia S., Biles C. L., Waugh M., Onsurez-Waugh K., Rodriguez-Alvarado G., Lidell C. M. (2004). Characterization of southern New Mexico Phytophthora capsici Leon. isolates from pepper (Capsicum annuum L.). 22 82–89. [Google Scholar]
- Flor H. H. (1955). Host-parasite interaction in flax rust-its genetics and other implications. 45 680–685. [Google Scholar]
- Flor H. H. (1971). Current status of the gene-for-gene concept. 9 275–296. 10.1146/annurev-phyto-072910-095339 [DOI] [PubMed] [Google Scholar]
- Förster H., Tyler B. M., Coffey M. D. (1994). Phytophthora sojae races have arisen by clonal evolution and by rare outcrosses. 7 780–791. 10.1094/MPMI-7-0780 [DOI] [Google Scholar]
- Foster J. M., Hausbeck M. K. (2010). Resistance of pepper to Phytophthora crown, root, and fruit rot is affected by isolate virulence. 94 24–30. 10.1094/PDIS-94-1-0024 [DOI] [PubMed] [Google Scholar]
- French-Monar R. D., Jones J., Roberts P. D. (2006). Characterization of Phytophthora capsici associated with root and weeds on Florida vegetable farms. 90 345–350. 10.1094/PD-90-0345 [DOI] [PubMed] [Google Scholar]
- French-Monar R. D., Patton A. F., Isakeit T. (2009). “The occurrence and recent spread of Phytophthora capsici in Texas high plains,” in , Duck Key, FL. [Google Scholar]
- Fu L., Zhu C., Ding X., Yang X., Morris P. F., Tyler B. M., et al. (2015). Characterization of cell-death-inducing members of the pectate lyase gene family in Phytophthora capsici and their contributions to infection of pepper. 28 766–775. 10.1094/MPMI-11-14-0352-R [DOI] [PubMed] [Google Scholar]
- Fu Y. B., Yang M. H., Zeng F., Biligetu B. (2017). Searching for an accurate marker-based prediction of an individual quantitative trait in molecular plant breeding. 8:1182 10.3389/fpls.2017.01182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García F. (1908). Las Cruces, NM: New Mexico State University, 67. [Google Scholar]
- Gevens A. J., Donahoo R. S., Lamour K. H., Hausbeck M. K. (2007). Characterization of Phytophthora capsici from Michigan surface irrigation water. 97 421–428. 10.1094/PHYTO-97-4-0421 [DOI] [PubMed] [Google Scholar]
- Gilbert G., Lacroix M., Hamel D. (2001). Disease diagnosed on commercial crops submitted to MAPAQ diagnostic laboratory in 2000. 81 40–58. [Google Scholar]
- Gill U. S., Lee S., Mysore K. S. (2015). Host versus non-host resistance: distinct wars with similar arsenals. 105 580–587. 10.1094/PHYTO-11-14-0298-RVW [DOI] [PubMed] [Google Scholar]
- Glosier B. R., Ogundiwin E. A., Sidhu G. S., Sischo D. R., Prince J. P. (2008). A differential series of pepper (Capsicum annuum) lines delineates fourteen physiological races of Phytophthora capsici. 162 23–30. 10.1007/s10681-007-9532-1 [DOI] [Google Scholar]
- Gobena D., McGarth M. T., Lamour K. H. (2012a). Survival and spread of Phytophthora capsici on Long Island, New York. 11 761–768. 10.1007/s11557-011-0787-5 [DOI] [Google Scholar]
- Gobena D., Roig J., Glamarini C., Hulvey J., Lamour K. (2012b). Genetic diversity of Phytophthora capsici isolates from pepper and pumpkin in Argentina. 104 102–107. 10.3852/11-147 [DOI] [PubMed] [Google Scholar]
- Godoy E. F. (1940). El “mildew” o “tizon” del pimiento producido por la “Phytophthora capsici” in la Republica Argentina. 24 235–280. [Google Scholar]
- Goldberg N. P. (1995). Las Cruces, NM: NMSU Cooperative Extension Service, 549. [Google Scholar]
- Goodwin S. B. (1997). The population genetics of Phytophthora. 87 462–473. 10.1094/PHYTO.1997.87.4.462 [DOI] [PubMed] [Google Scholar]
- Goodwin S. B., Sujkowski L. S., Dyer A. T., Fry B. A., Fry W. E. (1995). Direct detection of gene flow and probable sexual reproduction of Phytophthora infestans in Northern North America. 85 473–479. 10.1094/Phyto-85-473 [DOI] [Google Scholar]
- Granke L. L., Quesada-Ocampo L., Lamour K. H., Hausbeck M. K. (2012). Advances in research on Phytophthora capsici on vegetable crops in the United States. 95 1588–1600. 10.1094/PDIS-02-12-0211-FE [DOI] [PubMed] [Google Scholar]
- Griffin M. J. (1977). Cocoa Phytophthora Workshop. Rothamsted experimental station, England, 24-26 May, 1976. Pest news summaries cent. 23 107–110. 10.1371/journal.pone.0129932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Moreno A., Laborde J. A. (1980). “Current status of pepper breeding for resistance to Phytophthora capsici in Mexico,” in , Wageningen. [Google Scholar]
- Guigón-López C., González-González R. (2001). Estudio regional de las enfermedades del chile (Capsicum annuum, L.) y su comportamiento temporal en el sur de Chihuahua, México. 19 49–56. [Google Scholar]
- Gyulai G., Gemesne J. A., Sagi Z. S., Venczel G., Pinter P., Kristof Z., et al. (2000). Doubled haploid development and PCR-analysis of F1 hybrid derived DH-R2 paprika (Capsicum annuum L.) lines. 156 168–174. 10.1016/S0176-1617(00)80302-8 [DOI] [Google Scholar]
- Hausbeck M. K., Lamour K. H. (2004). Phytophthora capsici on vegetable crops: research progress and management challenges. 88 1292–1303. 10.1094/PDIS.2004.88.12.1292 [DOI] [PubMed] [Google Scholar]
- Hein I., Gilroy E. M., Armstrong M. R., Birch P. R. (2009). The zig-zag-zig in oomycete-plant interactions. 10 547–562. 10.1111/j.1364-3703.2009.00547.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H. H. (1990). Taiwan Phytophthora. 31 89–106. [Google Scholar]
- Holliday P., Mowat W. P. (1963). Foot rot of Piper nigrum L. (Phytophthora palmivora). 5 1–62. [Google Scholar]
- Hu J., Diao Y., Zhou Y., Lin D., Bi Y., Pang Z., et al. (2013a). Loss of heterozygosity drives clonal diversity of Phytophthora capsici in China. 8:e82691. 10.1371/journal.pone.0082691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Pang Z., Bi Y., Shao J., Diao Y., Guo J., et al. (2013b). Genetically diverse long-lived clonal lineages of Phytophthora capsici from pepper in Gansu, China. 103 920–926. 10.1094/PHYTO-01-13-0016-R [DOI] [PubMed] [Google Scholar]
- Hulvey J., Hurtado-Gonzalez O., Aragon-Caballero L., Gobena D., Storey D., Finley L., et al. (2011). Genetic diversity of the pepper pathogen Phytophthora capsici on farms in the Amazonian high jungle of Peru. 2 461–466. 10.4236/ajps.2011.23054 [DOI] [Google Scholar]
- Hunter J. E., Kunimoto R. K., Rohrbach K. G. (1971). Phytophthora blight, a new disease of macadamia. 61 1130–1133. 10.1094/Phyto-61-1130 [DOI] [Google Scholar]
- Hwang B. K., Kim C. H. (1995). Phytophthora blight of pepper and its control in Korea. 79 221–227. 10.1094/PD-79-0221 [DOI] [Google Scholar]
- Hwang B. K., Kim Y. J., Kim C. H. (1995). Differential interactions of Phytophthora capsici isolates with pepper genotypes at various plant growth stages. 102 311–316. 10.1007/BF01878125 [DOI] [Google Scholar]
- Hwang B. K., Sung N. K. (1989). Effect of metalaxyl on capsididol production in stems of pepper plants infected with Phytophthora capsici. 73 748–751. 10.1094/PD-73-0748 [DOI] [Google Scholar]
- Islam S. Z., Babadoost M., Lambert K. N., Ndeme A., Fouly H. M. (2004). Characterization of Phytophthora capsici isolates from processing pumpkin in Illinois. 89 191–197. 10.1094/PD-89-0191 [DOI] [PubMed] [Google Scholar]
- Ivors K. L., Milks D. C., Holmberg C. (2006). Evaluation of fungicides for control of Phytophthora blight of bell pepper. 1:V088. [Google Scholar]
- Jayashankar S., Bagga S., Phillips G. C. (1997). Sweet pepper (Capsicum annuum L.) transformation using Agrobacterium rhizogenes. 32 454–455. [Google Scholar]
- Ji P., Csinos A. S. (2015). Effect of oxathiapiprolin on asexual stages of Phytophthora capsici and disease development on vegetables. 166 229–235. 10.1111/aab.12176 [DOI] [Google Scholar]
- Jiang L., Sanogo S., Bosland P. W. (2015). Using recombinant inbred lines to monitor changes in the race structure of Phytophthora capsici in chile pepper in New Mexico. 16 235–240. 10.1094/PHP-RS-15-0034 [DOI] [Google Scholar]
- Jin A., Yongsheng H., Boaxi Z. (2007). Genetic linkage map construction and QTL analysis for Phytophthora capsici L. in pepper. 10:003. [Google Scholar]
- Jin J. H., Zhang H. X., Tan J. Y., Yan M. J., Li D. W., Khan A., et al. (2015). A new ethylene-responsive factor CaPTI1 gene in pepper (Capsicum annuum L.) involved in the regulation of defense response to Phytophthora capsici. 6:1217. 10.3389/fpls.2015.01217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A. (1982). Control of the crown rot phase of Phytophthora blight of bell pepper with fungicides. 37:73. [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Jones R. F., Bosland P. W., Steiner R. L., Jones R. W., O’Connell M. A. (2015). Detection of gene expression changes in Capsicum annuum L. foliar blight caused by Phytophthora capsici Leon. using qRT-PCR and leaf discs. 50 1342–1348. [Google Scholar]
- Katsura K., Tokura R. (1954). Studies on Phytophthora disease of economic plants (VII). A brown rot of watermelon caused by Phytophthora capsici Leonian. 6 38–48. [Google Scholar]
- Katsura K., Tokura R. (1955). “Studies on Phytophthora disease of economic plants (VIII), a brown rot of eggplants caused by Phytophthora capsici Leonian. Jubilee Pub,” in , Gainesville, FL, 167–172. [Google Scholar]
- Keinath A. P. (2007). Sensitivity of populations of Phytophthora capsici from South Carolina to mefenoxam, dimethomorph, zoxamide, and cymoxanil. 91 743–748. 10.1094/PDIS-91-6-0743 [DOI] [PubMed] [Google Scholar]
- Kim B., Hur J. (1990). Inheritance of resistance to bacterial spot and to Phytophthora blight in pepper. 31 350–357. [Google Scholar]
- Kim B. S., Lee E. K., Chung B. K. (1975). An investigation on the brown rot of eggplant caused by Phytophthora capsici Leonian. 14 77–79. [Google Scholar]
- Kim H. J., Naham S. H., Lee H. R., Yoon G. B., Kim K. T., Kang B. C., et al. (2008). BAC-derived markers converted from RFLP linked to Phytophthora capsici resistance in pepper (Capsicum annuum L.). 118 15–27. 10.1007/s00122-008-0873-5 [DOI] [PubMed] [Google Scholar]
- Kim S., Park J., Yeom S. I, Kim Y. M., Seo E., Kim K. T., et al. (2017). New reference genome sequences of hot pepper reveal the massive evaluation of plant disease-resistance gene by retroduplication. 8 210. 10.1186/s13059-017-1341-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Park M., Yeom S. I., Kim Y. M., Lee J. M., Lee H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. 46 470–478. 10.1038/ng.2877 [DOI] [PubMed] [Google Scholar]
- Kim S. J., Lee S. J., Kim B. D., Park K. H. (1997). Satellite-RNA-mediated resistance to cucumber mosaic virus in transgenic plants of hot pepper. 16 825–830. 10.1007/s002990050328 [DOI] [PubMed] [Google Scholar]
- Kimble K., Grogan G. (1960). Resistance to Phytophthora root rot in pepper. 44 872–873. [Google Scholar]
- Ko W. (1988). Hormonal heterothallism and homothallism in Phytophthora. 26 57–73. 10.1146/annurev.py.26.090188.000421 [DOI] [Google Scholar]
- Kousik C. S., Keinath A. P. (2008). First report of insensitivity to cyazofamid among isolates of Phytophthora capsici from the southeastern United States. 92 979 10.1094/PDIS-92-6-0979A [DOI] [PubMed] [Google Scholar]
- Krasnow C. S., Wyenandt A. A., Kline W. L., Carey J. B., Hausbeck M. K. (2017). Evaluation of pepper root rot resistance in an integrated Phytophthora blight management program. 27 408–415. 10.21273/HORTTECH03697-17 [DOI] [Google Scholar]
- Kreutzer W. A. (1937). A Phytophthora rot of cucumber fruit. 27:955. [Google Scholar]
- Kreutzer W. A., Bodine E. W., Durrell L. W. (1940). Cucurbit disease and rot of tomato fruit caused by Phytophthora capsici. 30 951–957. [Google Scholar]
- Kristiansen K., Andersen S. B. (1993). Effect of donor plant temperature, photoperiod, and age on anther culture response of Capsicum annuum L. 67 105–109. 10.1007/BF00022732 [DOI] [Google Scholar]
- Lamour K. H., Hausbeck M. K. (2000). Mefenozam insensitivity and the sexual stage of Phytophthora capsici in Michigan cucurbit fields. 90 396–400. 10.1094/PHYTO.2000.90.4.396 [DOI] [PubMed] [Google Scholar]
- Lamour K. H., Hausbeck M. K. (2001a). Investigating the spatiotemporal genetic structure of Phytophthora capsici in Michigan. 91 k973–980. 10.1094/PHYTO.2001.91.10.973 [DOI] [PubMed] [Google Scholar]
- Lamour K. H., Hausbeck M. K. (2001b). The dynamics of mefenoxam insensitivity in a recombining population of Phytophthora capsici characterized with amplified fragment length polymorphism markers. 91 553–557. 10.1094/PHYTO.2001.91.6.553 [DOI] [PubMed] [Google Scholar]
- Lamour K. H., Hausbeck M. K. (2002). The spatiotemporal genetic structure of Phytophthora capsici in Michigan and implication for disease management. 92 681–684. 10.1094/PHYTO.2002.92.6.681 [DOI] [PubMed] [Google Scholar]
- Lamour K. H., Hausbeck M. K. (2003). Susceptibility of mefenoxam-treated cucurbits to isolates of Phytophthora capsici sensitive and insensitive to mefenoxam. 87 920–922. 10.1094/PDIS.2003.87.8.920 [DOI] [PubMed] [Google Scholar]
- Lamour K. H., Kamoun S. (2009). Hoboken, NJ: Wiley-Blackwell Publishing; 10.1002/9780470475898 [DOI] [Google Scholar]
- Lamour K. H., Mudge J., Gobena D., Hurtado-Gonzales O. P., Schmutz J., Juo A., et al. (2012). Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. 10 1350–1360. 10.1094/MPMI-02-12-0028-R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour K. H., Stam R., Jupe J., Huitema E. (2011). The oomycete broad-host-range pathogen Phytophthora capsici. 13 329–337. 10.1111/j.1364-3703.2011.00754.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Park K. H., Kim B. D. (1993). In vitro plant regeneration and Agrobacterium-mediated transformation from cotyledon explants of hot pepper (Capsicum annuum cv. Golden Tower). 20 k289–294. [Google Scholar]
- Lee S. J., Park Y. J., Kim H. T., Kim B. S. (2010). The race differentiation of Phytophthora capsici in Korea. 16 153–157. 10.5423/RPD.2010.16.2.153 [DOI] [Google Scholar]
- Lefebvre V., Palloix A. (1996). Both epistatic and additive effects of QTLs are involved in polygenic induced resistance to disease: a case study, the interaction pepper-Phytophthora capsici Leonian. 93 503–511. 10.1007/BF00417941 [DOI] [PubMed] [Google Scholar]
- Lefebvre V., Pflieger S., Thabuis A., Caranta C., Blattes A., Chauvert J. C., et al. (2002). Towards the saturation of the pepper linkage map by alignment of three intraspecific maps including known-function genes. 45 839–845. 10.1139/g02-053 [DOI] [PubMed] [Google Scholar]
- Leonian L. H. (1922). Stem and fruit blight of peppers caused by Phytophthora capsici sp. nov. 12 401–408. [Google Scholar]
- Leu L. S., Kao C. W. (1981). Pepper blight induced by Phytophthora capsici. 23 59–66. [Google Scholar]
- Li D., Zhao K., Xie B., Zhang B., Luo K. (2003). Establishment of a highly efficient transformation system for pepper (Capsicum annuum L.). 21 785–788. [DOI] [PubMed] [Google Scholar]
- Lister C., Dean C. (1993). Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. 4 745–750. 10.1046/j.1365-313X.1993.04040745.x [DOI] [Google Scholar]
- Liu W.-Y., Kang J.-H., Jeong H.-S., Choi H.-J., Yang H.-B., Kim K.-T., et al. (2014). Combined used of bulk-segregated analysis of microarrays reveals SNP markers pinpointing a major QTL for resistance to Phytophthora capsici in pepper. 127 2503–2513. 10.1007/s00122-014-2394-8 [DOI] [PubMed] [Google Scholar]
- Liu Y. G., Zhang H. Y., Guo J. G., Lv H. P. (2009). Study on the resistance of Phytophthora capsici isolates to metalaxyl in Gansu. 7 23–25. [Google Scholar]
- Liu Z., Shi L., Yang S., Lin Y., Weng Y., Li X., et al. (2017). Functional and promoter analysis of ChiIV3, a Chitinase of pepper plant, in response to Phytophthora capsici infection. 18:1661. 10.3390/ijms18081661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Puc G., Canto-Flick A., Barredo-Pool F., Zapata-Castillo P., del C., Motalavo-Peniche M., et al. (2006). Direct somatic embryogenesis: a highly efficient protocol for in vitro regeneration of habanero pepper (Capsicum chinense Jacq.). 41 1645–1650. [Google Scholar]
- Louws F. J., Lanacaster M. E., Holmes G. J., Driver J. G. (2000). Evaluation of fungicides and host resistance for control of Phytophthora crown rot of pepper. 1999 Fungic. 55:188. [Google Scholar]
- Ltifi A., Wenzel G. (1994). Anther culture of hot and sweet pepper (Capsicum annuum L.): Influence of genotype and plant growth temperature. 13 74–77. [Google Scholar]
- Lucas J. A., Greer G., Oudemans P. V., Coffey M. D. (1990). Fungicide sensitivity in somatic hybrids of Phytophthora capsici obtained by protoplast fusion. 36 175–187. 10.1016/0885-5765(90)90105-7 [DOI] [Google Scholar]
- Maheswary V., Mak C. (1993). The influence of genotypes and environments on induction of pollen plants for anther culture of Capsicum annuum L. 1 40–50. [Google Scholar]
- Malaguti G., Pontis-Videla R. E. (1950a). El tizon o ‘mildew’ del pimiento en Venezuela. 14 4–5. [Google Scholar]
- Malaguti G., Pontis-Videla R. E. (1950b). Phytophthora capsici in Venezuela. 44 4–12. [Google Scholar]
- Mallard S., Cantet S., Massire A., Bachellez A., Ewert S., Lefebvre V. (2013). A key QTL cluster is conserved among accessions and exhibits broad-spectrum resistance to Phytophthora capsici: a valuable locus for pepper breeding. 32 349–364. 10.1007/s11032-013-9875-3 [DOI] [Google Scholar]
- Manoharan M., Sree Vidya C. S., Lakshmi Sita G. (1998). Agrobacterium-medicated genetic transformation in hot chilli (Capsicum annuum L. var. Pusa jwala). 131 77–83. 10.1016/S0168-9452(97)00231-8 [DOI] [Google Scholar]
- Mansfeld B. N., Colle M., Kang Y., Jones A. D., Grumet R. (2017). Transcriptomic and metabolomic analyses of cucumber fruit peels reveal a developmental increase in terpenoid glycosides associated with age-related resistance to Phytophthora capsici. 4:17022. 10.1038/hortres.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchionatto J. B. (1938). Argentine republic researches on the pepper diseases at Salta and Jujuy. 12 169–170. [Google Scholar]
- Matheron M. E., Matejka J. C. (1995). Comparative activities of sodium tetrathiocarbonate and metalaxyl on Phytophthora capsici and root and crown rot of chili pepper. 79 56–59. 10.1094/PD-79-0056 [DOI] [Google Scholar]
- Matheron M. E., Porchas M. (2000). Comparison of five fungicides on development of root, crown, and fruit rot of chile pepper and recovery of Phytophthora capsici from soil. 84 1038–1043. 10.1094/PDIS.2000.84.9.1038 [DOI] [PubMed] [Google Scholar]
- Matheron M. E., Porchas M. (2002). Suppression of Phytophthora root and crown rot on pepper plants treated with acibenzolar-S-methyl. 86 292–297. 10.1094/PDIS.2002.86.3.292 [DOI] [PubMed] [Google Scholar]
- Mathis W. L., Williams-Woodward J., Csinos A. S. (1999). Insensitivity of Phytophthora capsici to mefenoxam in Georgia. 89 S49. [Google Scholar]
- McGovern R. J., Davis T. A., Myers D. S., Seijo T. E. (2003). Evaluation of fungicides for control of diseases of tropical pumpkin. 58:V124. [Google Scholar]
- McGrath M. T. (2004). Evaluation of fungicides for managing Phytophthora blight of squash. 59:V054. 10.1002/ps.2308 [DOI] [PubMed] [Google Scholar]
- McGregor C., Waters V., Nambeesan S., MacLean D., Candole B. L., Conner P. (2011). Genotypic and phenotypic variation among pepper accessions resistant to Phytophthora capsici. 46 1235–1240. [Google Scholar]
- Miao J., Cai M., Dong X., Liu L., Lin D., Zhang C., et al. (2016). Resistance assessment for oxathiapoprolin in Phytophthora capsici and the detection of a point mutation (G769W) in PcORP1 that confers resistance. 7:615. 10.3389/fmicb.2016.00615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. A., Bhat R. G., Schmitthenner A. F. (1994). Detection of Phytophthora capsici in pepper and cucurbit crops in Ohio with two commercial immunoassay kits. 78 1042–1046. 10.1094/PD-78-1042 [DOI] [Google Scholar]
- Minamiyama Y., Tsuro M., Kubo T., Hirai M. (2007). QTL analysis for resistance to Phytophthora capsici in pepper using a high density SSR-based map. 57 129–134. 10.1270/jsbbs.57.129 [DOI] [Google Scholar]
- Mitykó J., Andrásfalvy A., Csilléry G., Fári M. (1995). Anther-culture response in different genotypes and F1 hybrids of pepper (Capsicum annuum L.). 114 78–80. 10.1111/j.1439-0523.1995.tb00764.x [DOI] [Google Scholar]
- Monroy-Barbosa A., Bosland P. W. (2008). Genetic analysis of Phytophthora root rot race-specific resistance in chile pepper. 133 825–829. 10.1371/journal.pone.0151401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy-Barbosa A., Bosland P. W. (2010). A rapid technique for multiple-race disease screening for Phytophthora foliar blight on single Capsicum annuum L. plants. 45 1563–1566. [Google Scholar]
- Monroy-Barbosa A., Bosland P. W. (2011). Identification of novel physiological races of Phytophthora capsici causing foliar blight using the New Mexico recombinant inbred pepper lines set as a host differential. 136 205–210. [Google Scholar]
- Morrison R. A., Koning R. E., Evans D. A. (1986). Anther culture of an interspecific hybrid of Capsicum. 126 1–9. 10.1016/S0176-1617(86)80210-3 [DOI] [Google Scholar]
- Mu L., Tsao P. H. (1987). Identities of Phytophthora isolates causing vanilla blight and root rot in French Polynesia. 77:1704. [Google Scholar]
- Munyon I. P., Hubstenberger J. F., Phillips G. C. (1989). Origin of plantlets and callus obtained from chile anther culture. 25 293–296. 10.1007/BF02628469 [DOI] [Google Scholar]
- Naegele R. P., Ashrafi H., Hill T. A., Chin-Wo S. R., van Deynze A. E., Hausbeck M. K. (2014). QTL mapping of fruit rot resistance to the plant pathogen Phytophthora capsici in a recombinant inbred line Capsicum annuum population. 104 479–483. 10.1094/PHYTO-05-13-0143-R [DOI] [PubMed] [Google Scholar]
- Naegele R. P., Granke L. L., Fry T. A., Hill H., Ashrafi A., Van Deynza A., et al. (2017). Disease resistance to multiple fungal and oomycete pathogens evaluated using a recombinant inbred line population in pepper. 107 1522–1531. 10.1094/PHYTO-02-17-0040-R [DOI] [PubMed] [Google Scholar]
- Naegele R. P., Hausbeck M. K. (2014). Evaluation of pepper fruit for resistance to Phytophthora capsici in a recombinant inbred line population, and correlation with fruit shape. 98 885–890. 10.1094/PDIS-03-13-0295-RE [DOI] [PubMed] [Google Scholar]
- Nguyen V. L. (2015). Spread of Phytophthora capsici in black pepper (Piper nigrum) in Vietnam. 7 506–513. 10.4236/eng.2015.78047 [DOI] [Google Scholar]
- Noveriza R., Quimio T. H. (2004). Soil mycoflora of black pepper rhizosphere in the Philippines and their in vitro antagonism against Phytophthora capsici L. 5 1–10. [Google Scholar]
- Oelke L. M., Bosland P. W., Steiner R. (2003). Differentiation of race specific resistance to Phytophthora root rot and foliar blight in Capsicum annuum. 128 213–218. [Google Scholar]
- Ogundiwin E. A., Berke T. F., Massoudi M., Black L. L., Huestis G., Choi D., et al. (2005). Construction of 2 intraspecific linkage maps and identification of resistance QTLs for Phytophthora capsici root-rot and foliar-blight diseases of pepper (Capsicum annuum L.). 48 698–711. 10.1139/g05-028 [DOI] [PubMed] [Google Scholar]
- Ortega R. G., Espanol C. P., Zueco J. C. (1991). Genetics of resistance to Phytophthora capsici in the pepper line ‘SCM-334’. 107 50–55. 10.1111/j.1439-0523.1991.tb00527.x [DOI] [Google Scholar]
- Ortega R. G., Espanol C. P., Zueco J. C. (1992). Genetic relationships among four pepper genotypes resistant to Phytophthora capsici. 108 118–125. 10.1111/j.1439-0523.1992.tb00110.x [DOI] [Google Scholar]
- Osnitzkaya E. A. (1949). Phytophthora of tomatoes in protected soil. 2 62–64. 10.1002/ps.4911 [DOI] [PubMed] [Google Scholar]
- Palloix A., Daubeze A. M., Phaly T., Pochard E. (1990). Breeding transgressive lines of pepper for resistance to Phytophthora capsici in a recurrent selection system. 51 141–150. [Google Scholar]
- Palloix A., Daubeze A. M., Pochard E. (1988).Phytophthora root rot of pepper influence of host genotype and pathogen strain on the inoculum density-disease severity relationships. 123 25–33. 10.1111/j.1439-0434.1988.tb01033.x [DOI] [Google Scholar]
- Pan Z. (1997). Ph.D. dissertation, University of Massachusetts, Amherst, MA. [Google Scholar]
- Papavizas G. H., Bowers J. H. (1981). Comparative fungitoxicity of captafol and metalaxyl to Phytophthora capsici. 71 123–128. 10.1094/Phyto-71-123 [DOI] [Google Scholar]
- Parra G., Ristaino J. B. (1998). Insensitivity to Ridomil Gold (mefenoxam) found among field isolates of Phytophthora capsici causing Phytophthora blight on bell pepper in North Carolina and New Jersey. 82:711 10.1094/PDIS.1998.82.6.711D [DOI] [PubMed] [Google Scholar]
- Parra G., Ristaino J. B. (2001). Resistance to mefenoxam and metalaxyl among field isolates of Phytophthora capsici causing Phytophthora blight of bell pepper. 85 1069–1075. 10.1094/PDIS.2001.85.10.1069 [DOI] [PubMed] [Google Scholar]
- Pérez-Moreno L., Durán-Ortiz L., Ramírez-Malagón R., Sánchez-Palé R., Olalde-Portugal V. (2003). Compatibilidad fisiológica y sensibilidad a fungicidas de aislamientos de Phytophthora capsici Leo. 21 19–25. [Google Scholar]
- Peter K. V., Goth R. W., Webb R. E. (1984). Indian hot peppers as new sources of resistance to bacterial wilt, Phytophthora root rot, and root-knot nematode. 19 337–278. [Google Scholar]
- Pflieger S., Palloix A., Caranta C., Blattes A., Lefebvre V. (2001). Defense response genes co-localize with the quantitative disease resistance loci in pepper. 103 920–929. 10.1007/s001220100726 [DOI] [Google Scholar]
- Ploetz R., Heine G., Haynes J., Watson M. (2001). An investigation of biological attributes that may contribute to the importance of Phytophthora capsici as a vegetable pathogen in Florida. 140 61–67. 10.1111/j.1744-7348.2002.tb00157.x [DOI] [Google Scholar]
- Ploetz R. C., Haynes J., Heine G., Watson M. (2001). Investigating factors that may contribute to the increased prevalence of Phytophthora capsici-induced diseases in South Florida. 91:S72. [Google Scholar]
- Pochard E. D., Daubeze A. M. (1980). Study and evaluation of components of a case of polygenic resistance: the resistance of red pepper to Phytophthora capsici. 30 377–398. [Google Scholar]
- Qi R. D., Ding J. C., Gao Z. M., Ni C. G., Jiang J. J., Li P. (2008). Resistance of Phytophthora capsici isolates to metalaxyl in Anhui Province. 35 245–250. [Google Scholar]
- Qin C., Yub C., Shena Y., Fang X., Chen L., Mind J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. 111 5135–5140. 10.1073/pnas.1400975111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Rotino G. L. (1993). Anther culture of several sweet and hot pepper genotypes. 12 59–62. [Google Scholar]
- Quesada-Ocampo L. M., Fullbright D. W., Hausbeck M. K. (2009). Susceptibility of Fraser fir to Phytophthora capsici. 93 135–141. 10.1094/PDIS-93-2-0135 [DOI] [PubMed] [Google Scholar]
- Quesada-Ocampo L. M., Granke L. L., Mercier M. R., Olsen J., Hausbeck M. K. (2011). Investigating the genetic structure of Phytophthora capsici populations. 101 1061–1073. 10.1094/PHYTO-11-10-0325 [DOI] [PubMed] [Google Scholar]
- Quirin E. A., Ogundiwin E. A., Prince J. P., Mazourek M., Briggs M. O., Chlanda T. S., et al. (2005). Development of sequence characterized amplified region (SCAR) primers for the detection of Phyto.5.2, a major QTL for resistance to Phytophthora capsici Leon. in pepper. 110 605–612. 10.1007/s00122-004-1874-7 [DOI] [PubMed] [Google Scholar]
- Ravise A. (1966). Observations sur la reproduction sexuee de souches du Phytophthora palmivora (Butl.). Bult., parasite de cultures tropicales. 2 91–101. [Google Scholar]
- Reeves G., Monroy-Barbosa A., Bosland P. W. (2013). A novel Capsicum gene inhibits host-specific disease resistance to Phytophthora capsici. 103 472–478. 10.1094/PHYTO-09-12-0242-R [DOI] [PubMed] [Google Scholar]
- Rehrig W. Z., Ashrafi H., Hill T., Prince J., Van Deynze A. (2014). CaDMR1 cosegregates with QTL Pc5.1 for resistance to Phytophthora capsici in pepper (Capsicum annuum). 7 1–12. 10.3835/plantgenome2014.03.0011 [DOI] [Google Scholar]
- Reifschneider F. J. B., Boiteux L. S., Dellavecchia P. T., Poulos J. M., Kuroda N. (1992). Inheritance of adult plant resistance to Phytophthora capsici in pepper. 62 45–49. 10.1007/BF00036086 [DOI] [Google Scholar]
- Reifschneider F. J. B., Cafe’-Filho A. C., Rego A. M. (1986). Factors affecting expression of resistance in pepper (Capsicum annuum) to blight caused by Phytophthora capsici in screening trials. 35 451–456. 10.1111/j.1365-3059.1986.tb02042.x [DOI] [Google Scholar]
- Richins R. D., Micheletto S., O’Connell M. A. (2010). Gene expression profiles unique to chile (Capsicum annuum L.) resistant to Phytophthora root rot. 178 192–201. 10.1016/j.plantsci.2009.11.005 [DOI] [Google Scholar]
- Ristaino J. B. (1990). Intraspecific variation among isolates of Phytophthora capsici from pepper and cucurbit fields in North Carolina. 80 k1253–1259. 10.1094/Phyto-80-1253 [DOI] [Google Scholar]
- Ristaino J. B. (1991). Influence of rainfall, drip irrigation, and inoculum density on the development of Phytophthora root and crown rot epidemics and yield in bell pepper. 81 922–929. 10.1094/Phyto-81-922 [DOI] [Google Scholar]
- Ristaino J. B., Johnston S. A. (1999). Ecologically based approaches to management of Phytophthora blight of bell pepper. 83 1080–1089. 10.1094/PDIS.1999.83.12.1080 [DOI] [PubMed] [Google Scholar]
- Ristaino J. B., Larkin R. P., Campbell C. L. (1993). Spatial and temporal dynamics of Phytophthora epidemics in commercial bell pepper fields. 83 1312–1320. 10.1094/Phyto-83-1312 [DOI] [Google Scholar]
- Romero-Cova S. (1988). Texcoco: Chapingo Autonomous University. [Google Scholar]
- Rudorf W., Schaper P., Ross H., Baerecke M., Torka M. (1950). The breeding of resistant varieties of potatoes. 27 222–235. 10.1007/BF02850189 [DOI] [Google Scholar]
- Saini S. S., Sharma P. P. (1978). Inheritance of resistance to fruit rot (Phytophthora capsici Leon) and induction resistance in bell pepper (Capsicum annuum L.). 27 721–723. 10.1007/BF00023707 [DOI] [Google Scholar]
- Sandsten E. P. (1939). Director’s annual report. Fifty-Second Fiscal Year, 1938–1939. 63. [Google Scholar]
- Sanogo S. (2003). Chile pepper and the threat of wilt diseases. 10.1094/PHP-2003-0430-01-RV 22101927 [DOI] [Google Scholar]
- Sanogo S., Bosland P. W. (2012). “Biology and management of Phytophthora capsici in the southwestern USA,” in , ed. Lamour K. (Wallingford: CABI; ), 87–95. [Google Scholar]
- Sarejanni J. A. (1936). La pourriture du collet des solaneìes cultiveìes et la classification du genre Phytophthora. 2 35–52. [Google Scholar]
- Satour M. M., Butler E. E. (1967). A root and crown rot of tomato caused by Phytophthora capsici and P. parasitica. 57 510–515. [Google Scholar]
- Schlub R. L., Johnston S. A. (1982). Control of Phytophthora on pepper using captafol and metalaxyl. 65 215–219. [Google Scholar]
- Schwinn F., Staub T. (1995). “Oomycete fungicides,” in , ed. Lyr H. (New York, NY: Gustav Fisher Verlag; ), 323–346. [Google Scholar]
- Seebold K. W., Horten T. B. (2003). Evaluation of fungicides for control of Phytophthora crown and fruit rot of summer squash. 58:V098. [Google Scholar]
- Shattock R. C. (1988). Studies on the inheritance of resistance to metalaxyl in Phytophthora infestans. 37 4–11. 10.1111/j.1365-3059.1988.tb02188.x [DOI] [Google Scholar]
- Sheu Z. M., Chen J. R., Wang T. C. (2009). First report of the A2 mating type of Phytophthora capsici infecting peppers (Capsicum annuum) in Taiwan. 93:548 10.1094/PDIS-93-5-0548C [DOI] [PubMed] [Google Scholar]
- Sholberg P. L., Walker M. C., O’Gorman D. T., Jesperson G. D. (2007). First report of Phytophthora capsici on cucurbits and peppers in British Columbia. 29 153–158. 10.1080/07060660709507451 [DOI] [Google Scholar]
- Shrestha S. K., Miyasaka S. C., Shintaku M., Kelly H., Lamour K. (2017). Phytophthora colocasiae from Vietnam, China, Hawaii and Nepal: intra- and inter-genomic variations in ploidy and a long-lived, diploid Hawaiian lineage. 16 893–904. 10.1007/s11557-017-1323-z [DOI] [Google Scholar]
- Silvar C., Merino F., Diaz J. (2006). Diversity of Phytophthora capsici in Northwest Spain: analysis of virulence, metalaxyl response, and molecular characterization. 90 1135–1142. 10.1094/PD-90-1135 [DOI] [PubMed] [Google Scholar]
- Silvar C., Merino F., Diaz J. (2008). Differential activation of defense-related genes in susceptible and resistant pepper cultivars infected with Phytophthora capsici. 165 1120–1124. 10.1016/j.jplph.2007.11.008 [DOI] [PubMed] [Google Scholar]
- Silva-Rojas H. V., Fernández-Pavía S. P., Góngora-Canul C., Macías-López B. C., Ávila-Quezada G. D. (2009). Distribución espacio temporal de la Marchitez del chile (Capsicum annuum L.) en Chihuahua e identificación del agente causal Phytophthora capsici Leo. 27 134–147. [Google Scholar]
- Smith P. G., Kimble K. A., Grogan R. G., Millett A. H. (1967). Inheritance of resistance in pepper to Phytophthora root rot. 57 377–379. [Google Scholar]
- Stam R., Jupe J., Howden A. J. M., Morris J. A., Boevink P. C., Hedley P. E., et al. (2013). Identification and characterization CRN effectors in Phytophthora capsici shows modularity and functional diversity. 8:e59517. 10.1371/journal.pone.0059517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinitz B., Wolf D., Matzevitch-Josef T., Zelcer A. (1999). Regeneration in vitro and genetic transformation of pepper (Capsicum spp.): the current state of the art. 18 9–15. [Google Scholar]
- Stevenson W. R., James R. V., Rand R. E. (2000). Evaluation of selected fungicides to control Phytophthora blight and fruit rot of cucumber. 55:163. [Google Scholar]
- Stevenson W. R., James R. V., Rand R. E. W. (2001). Evaluation of selection fungicides to control Phytophthora blight and fruit rot of cucumber. 56:V16. [Google Scholar]
- Subhash K., Christopher T. (1997). Organogenesis and transformation in Capsicum baccatum. 33:57A. [Google Scholar]
- Sugita T., Yamaguchi K., Kinoshita T., Yuji K., Sugimura Y., Nagata R., et al. (2006). QTL analysis for resistance to Phytophthora blight (Phytophthora capsici Leon.) using an intraspecific doubled-haploid population of Capsicum annuum. 56 137–145. 10.1270/jsbbs.56.137 [DOI] [Google Scholar]
- Sujkowski L. S., Goodwin S. B., Fry W. E. (1996). Changes in specific virulence in Polish populations of Phytophthora infestans: 1985-1991. 102 555–561. 10.1007/BF01877022 [DOI] [Google Scholar]
- Sun H., Wang H., Stammler G., Ma J., Zhou M. (2010). Baseline sensitivity of populations of Phytophthora capsici from China to three carboxylic acid amide (CAA) fungicides and sequence analysis of cholinephosphotransferases from a CAA-sensitive isolate and CAA-resistant laboratory mutants. 158 244–252. 10.1111/j.1439-0434.2009.01606.x [DOI] [Google Scholar]
- Sun W. X., Jia Y. J., O’Neill N. R., Feng B. Z., Zhang X. G. (2008). Genetic diversity of Phytophthora capsici from eastern China. 30 414–424. 10.4238/2012.December.17.4 [DOI] [PubMed] [Google Scholar]
- Supena E. D., Suharsono S., Jacobsen E., Custers J. B. (2006). Successful development of a shed-microspore culture protocol for doubled haploid production in Indonesian hot pepper (Capsicum annuum L.). 25 1–10. 10.1007/s00299-005-0028-y [DOI] [PubMed] [Google Scholar]
- Sy O., Steiner R., Bosland P. W. (2005). Inheritance of Phytophthora stem blight resistance as compared to Phytophthora root rot and Phytophthora foliar blight resistance in Capsicum annuum L. 130 75–78. [Google Scholar]
- Sy O., Steiner R., Bosland P. W. (2008). Recombinant inbred line differential identifies race-specific resistance to Phytophthora root rot in Capsicum annuum. 98 867–870. 10.1094/PHYTO-98-8-0867 [DOI] [PubMed] [Google Scholar]
- Tamietti G., Valentino D. (2001). Physiological characterization of a population of Phytophthora capsici Leon. from northern Italy. 83 199–205. [Google Scholar]
- Thabuis A., Palloix A., Pflieger S., Daubeze A. M., Caranta C., Legebvre V. (2003). Comparative mapping of Phytophthora resistance loci in pepper germplasm: evidence for conserved resistance loci across Solanaceae and for a large genetic diversity. 106 1473–1485. 10.1007/s00122-003-1206-3 [DOI] [PubMed] [Google Scholar]
- Thabuis A., Lefebvre V., Bernard G., Daubeze A. M., Phaly T., Pochard E., et al. (2004a). Phenotypic and molecular evaluation of a recurrent selection program from a polygenic resistance to Phytophthora capsici in pepper. 109 342–351. [DOI] [PubMed] [Google Scholar]
- Thabuis A., Palloix A., Servin B., Daubeze A. M., Signoret P., Hospital F., et al. (2004b). Marker-assisted introgression of 4 Phytophthora capsici resistance QTL alleles into a bell pepper line: validation of additive and epistatic effects. 14 9–20. 10.1023/B:MOLB.0000037991.38278.82 [DOI] [Google Scholar]
- Thomas K. M., Ramakrishnan T. S., Soumini C. K., Balakrishnan M. S. (1947). Studies in the genus Phytophthora. I. Oospore formation and taxonomy of Phytophthora palmivora Butl. 26 147–163. [Google Scholar]
- Thompson A. H., Botha W. J., Uys M. D. R. (1994). Phytophthora capsici (Oomycota:fungi), a first report from South Africa. 60 257–260. 10.1016/S0254-6299(16)30600-7 [DOI] [Google Scholar]
- Tian D., Babadoost M. (2004). Host range of Phytophthora capsici from pumpkin and pathogenicity of isolates. 88 485–489. 10.1094/PDIS.2004.88.5.485 [DOI] [PubMed] [Google Scholar]
- Tompkins C. M., Tucker C. M. (1937). Phytophthora rot of honeydew melon. 54 933–944. [Google Scholar]
- Tompkins C. M., Tucker C. M. (1941). Root rot of pepper and pumpkin caused by Phytophthora capsici. 63 417–426. [Google Scholar]
- Truong H. T. H., Kim K. T., Kim D. W., Kim S., Chase Y., Park J. H., et al. (2012). Identification of isolate-specific resistance QTLs to Phytophthora root rot using an intraspecific recombinant inbred line population of pepper (Capsicum annuum). 61 48–56. 10.1111/j.1365-3059.2011.02483.x [DOI] [Google Scholar]
- Tsao P. H., Kasim R., Mustika I. (1985). Morphology and identity of black pepper Phytophthora isolates in Indonesia. 33 61–66. [Google Scholar]
- Tsao P. H., Mu L. (1987). Involvement of Phytophthora in vanilla root ort in French Polynesia. 77:1704. [Google Scholar]
- Tsao P. H., Tummakate A. (1977). The identity of a Phytophthora species from black pepper in Thailand. 69 631–637. 10.2307/3758568 [DOI] [Google Scholar]
- Tucker C. M. (1928). Report of the plant pathologist. 1928 29–35. [Google Scholar]
- Tucker C. M. (1931). Columbia, MO: University of Missouri. 153. 10.1046/j.1464-6722.2001.00073.x [DOI] [Google Scholar]
- Turner P. D. (1960). Strains of Phytophthora palmivora (Butl.) from Theobroma cacao L. I. Isolates from West Africa. 43 665–672. 10.1016/S0007-1536(60)80057-5 [DOI] [Google Scholar]
- Turner P. D. (1961a). Strains of Phytophthora palmivora (Butl.) from Theobroma cacao L. 44 409–416. 10.1016/S0007-1536(61)80035-1 [DOI] [Google Scholar]
- Turner P. D. (1961b). Complementary isolates on Phytophthora palmivora from cacao and rubber and their taxonomy. 51 161–164. [Google Scholar]
- Uchida J. Y., Aragaki M. (1980). Chemical stimulation of oospore formation in Phytophthora capsici. 72 1103–1108. 10.2307/3759563 [DOI] [Google Scholar]
- Vagera J., Havranek P. (1985). In vitro induction of androgenesis in Capsicum annuum L. and its genetic aspects. 27 10–21. 10.1007/BF02894626 [DOI] [Google Scholar]
- Vásquez-López A., Tlapal-Bolaños B., Yáñez-Morales M., Pérez-Pacheco R., Quintos-Escalante M. (2009). Etiología de la marchitez del “chile de agua” (Capsicum annuum L.) en Oaxaca. 32 k127–134. [Google Scholar]
- Vega-Arreguín J. C., Jalloh A., Bos J. I., Moffett P. (2014). Recognition of an Avr3a homologue plays a major role in mediating nonhost resistance to Phytophthora capsici in Nicotiana species. 27 770–780. 10.1094/MPMI-01-14-0014-R [DOI] [PubMed] [Google Scholar]
- Vega-Arreguín J. C., Shimada-Beltran H., Sevillano-Serrano J., Moffett P. (2017). Non-host plant resistance against Phytophthora capsici is mediated in part by members of the I2 R gene family in Nicotiana spp. 8:205. 10.3389/fpls.2017.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez-Valle R., Medina-Aguilar M., Luna-Ruiz J. (2001). Sintomatología y géneros de patógenos asociados con las pudriciones de la raíz del chile (Capsicum annuum L.) en el norte-centro de México. 19 175–181. 26650553 [Google Scholar]
- Votava E. J., Bosland P. W. (2002). A cultivar by any other name: genetic variability in heirloom bell pepper ‘California Wonder’. 37 k1100–1102. [Google Scholar]
- Waldenmaier C. M. (2004). Evaluation of fungicides for control of pumpkin diseases. 59:V064. [Google Scholar]
- Walker S. J., Bosland P. W. (1999). Inheritance of Phytophthora root rot and foliar blight resistance in pepper. 124 14–18. 10.1094/PHYTO-09-12-0242-R [DOI] [PubMed] [Google Scholar]
- Wang J. E., Li D. W., Zhang Y. L., Zhao Q., He Y. M., Gong Z. H. (2013). Defense responses of pepper (Capsicum annuum L.) infected with incompatible and compatible strains of Phytophthora capsici. 1236 625–638. 10.1007/s10658-013-0193-8 [DOI] [Google Scholar]
- Wang X., Zhu X., Tooley P., Zhang Z. (2013). Cloning and functional analysis of three genes encoding polygalacturonase-inhibiting proteins from Capsicum annuum and transgenic CaPGIP1 in tobacco in relation to increased resistance to two fungal pathogens. 81 379–400. 10.1007/s11103-013-0007-6 [DOI] [PubMed] [Google Scholar]
- Wang P., Wang L., Guo J., Yang W., Shen H. (2016). Molecular mapping of a gene conferring resistance to Phytophthora capsici Leonian race 2 in pepper line PI201234 (Capsicum annuum L.). 36:66 10.1007/s11032-016-0464-0 [DOI] [Google Scholar]
- Wang Y., Yang M., Pan N., Chen Z. H. (1991). Plant regeneration and transformation of sweet pepper (Capsicum frutescens). 33 780–786. [Google Scholar]
- Weber G. F. (1932). Blight of peppers in Florida caused by Phytophthora capsici. 22 775–780. [Google Scholar]
- Wiant J. S., Tucker C. M. (1940). A rot of winter green watermelons caused by Phytophthora capsici. 60 73–88. [Google Scholar]
- Wolf D., Matzevitch T., Steinitz B., Zelcer A. (2001). Why is it difficult to obtain transgenic pepper plants? 560 229–233. 10.17660/ActaHortic.2001.560.44 [DOI] [Google Scholar]
- Xu J., Zhao X., Han X., Du Y. (2007a). Antifungal activity of oligochitosan against Phytophthora capsici and other plant pathogenic fungi in vitro. 87 220–228. 10.1016/j.pestbp.2006.07.013 [DOI] [Google Scholar]
- Xu J., Zhao X., Wang X., Zhao Z., Du Y. (2007b). Oligochitosan inhibits Phytophthora capsici by penetrating the cell membrane and putative binding to intracellular targets. 88 167–175. 10.1016/j.pestbp.2006.10.010 [DOI] [Google Scholar]
- Xu X., Chao J., Cheng X., Wang R., Sun B., Wang H., et al. (2016). Mapping of novel race specific resistance gene to Phytophthora root rot in pepper (Capsicum annuum) using bulked segregant analysis combined with specific length amplified fragment sequencing strategy. 11:e0151401. 10.1371/journal.pone.0151401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., Nyholt D. R., et al. (2010). Common SNPs explain a large proportion of the heritability for human height. 42 565–569. 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Li H., Zhang J., Jing Y. (1993). Genetic transformation and plant regeneration in pepper. 35 88–93. [Google Scholar]
- Yin J., Jackson K. L., Candole B. L., Csinos A. S., Langston D. B., Ji P. (2012). Aggressiveness and diversity of Phytophthora capsici on vegetable crops in Georgia. 160 191–200. 10.1111/j.1744-7348.2012.00532.x [DOI] [Google Scholar]
- Yoshizawa T., Shimizu T., Hirano H., Sato M., Hastmoto H. (2012). Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor. 287 18710–18716. 10.1074/jbc.M112.350520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata-Vázquez A., Sanchez-Sanchez M., del-Rio-Robledo A., Silos-Espino H., Perales-Segovia C., Flores-Benitez S., et al. (2012). Phytophthora capsici epidemic dispersion on commercial pepper fields in Aguascalientes, Mexico. 2012:341764. 10.1100/2012/341764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentmyer G. A. (1988). Origin and distribution of four species of Phytophthora. 91 367–379. 10.1016/S0007-1536(88)80111-6 [DOI] [Google Scholar]
- Zentmyer G. A., Kaosiri T., Idosu G. (1977). Taxonomic variants in the Phytophthora palmivora complex. 69 329–332. 10.1016/S0007-1536(77)80060-0 [DOI] [Google Scholar]
- Zentmyer G. A., Mitchell D. J., Jefferson L., Roheim J., Carnes D. (1973). Distribution of mating types of Phytophthora palmivora. 63 663–667. 10.1094/Phyto-63-663 [DOI] [Google Scholar]
- Zhang G. Z., Liang Y. (2003). Identification of pathogen of Capsicum epidemic diseases and its bio-characteristics and measurements of related germicide. 26 23–30. [Google Scholar]
- Zhang Y. L., Jia Q. L., Li D. W., Wang J. E., Yin Y. X., Gong Z. H. (2013). Characteristic of the pepper CaRGA2 gene in defense responses against Phytophthora capsici Leonian. 14 8985–9004. 10.3390/ijms14058985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q. M., Li L. Y., Yang S. H. (1984). Investigations of Phytophthora blight. 01:1. [Google Scholar]
- Zhou Z. H., Su M., Chen D., Liu R., Song L., Yang J. (1991). Preliminary study of regeneration induction and gene transformation of CMVcp in pepper. 6 69–72. [Google Scholar]
- Zhu Y. X., Ou Yang W. J., Zhang Y. F., Chen Z. L. (1996). Transgenic sweet pepper plants from Agrobacterium mediated transformation. 16 71–75. 10.1007/BF01275453 [DOI] [PubMed] [Google Scholar]