Abstract
The gelatinization temperature of endosperm starch in most japonica rice cultivars is significantly lower than that in most indica rice cultivars. This is because three single nucleotide polymorphisms in the Starch synthase (SS) IIa gene in japonica rice cultivars (SSIIaJ) significantly reduce SSIIa activity, resulting in an increase in amylopectin short chains with degree of polymerization (DP) ≤ 12 compared to indica rice cultivars (SSIIaI). SSIIa forms a trimeric complex with SSI and starch branching enzyme (BE) IIb in maize and japonica rice, which is likely important for the biosynthesis of short and intermediate amylopectin chains (DP ≤ 24) within the amylopectin cluster. It was unknown whether the complete absence of SSIIa further increases amylopectin short chains and reduces gelatinization temperature and/or forms altered protein complexes due to the lack of a suitable mutant. Here, we identify the SSIIa-deficient mutant rice line EM204 (ss2a) from a screen of ca. 1,500 plants of the rice cultivar Kinmaze (japonica) that were subjected to N-methyl-N-nitrosourea mutagenesis. The SSIIa gene in EM204 was mutated at the boundary between intron 5 and exon 6, which generated a guanine to adenine mutation and resulted in deletion of exon 6 in the mRNA transcript. SSIIa activity and SSIIa protein in developing endosperm of EM204 were not detected by native-PAGE/SS activity staining and native-PAGE/immunoblotting, respectively. SSIIa protein was completely absent in mature seeds. Gel filtration chromatography of soluble protein extracted from developing seeds showed that the SSI elution pattern in EM204 was altered and more SSI was eluted around 300 kDa, which corresponds with the molecular weight of trimeric complexes in wild type. The apparent amylose content of EM204 rice grains was higher than that in its parent Kinmaze. EM204 also had higher content of amylopectin short chains (DP ≤ 12) than Kinmaze, which reduced the gelatinization temperature of EM204 starch by 5.6°C compared to Kinmaze. These results indicate that EM204 starch will be suitable for making foods and food additives that easily gelatinize and slowly retrograde.
Keywords: amylopectin, amylose, endosperm starch, low gelatinization temperature, protein complex, rice, starch synthase IIa
Introduction
Starch synthases (SSs) have a central role in starch biosynthesis. SSs elongate α-1,4-linked linear glucan chains of starch by transferring an ADP glucose (ADPG) residue to the non-reducing end of glucans. SSIIa functions to elongate amylopectin short chains with degree of polymerization (DP) ≤12 to 13 ≤ DP ≤ 24, and is highly conserved in rice (Umemoto et al., 2002; Nakamura et al., 2005), maize (Zhang et al., 2004; Liu et al., 2012), wheat (Yamamori et al., 2000), barley (Morell et al., 2003), sweet potato (Katayama et al., 2002; Kitahara et al., 2005), and Arabidopsis (Zhang et al., 2008).
The rice SSIIa gene controls the chain-length distribution of endosperm amylopectin in indica and japonica rice cultivars and corresponds to the alk (alkali disintegration of starch granules) gene (Umemoto et al., 1999, 2002). SSIIa from japonica rice cultivars (Nipponbare and Kinmaze) contains four amino acid replacements compared with indica rice cultivars (IR36 and Kasalath), and three of these are associated with significant reduction in japonica SSIIa activity (Nakamura et al., 2005). The SSIIa activity of recombinant SSIIaJ derived from the japonica rice cultivars Nipponbare and Kinmaze is estimated as ca. 10% of that of recombinant SSIIaI derived from the indica rice cultivar IR36 (Nakamura et al., 2005). Differences in the amylopectin chain-length distribution significantly affect the starch gelatinization temperature. The gelatinization temperature of japonica rice starch is ca. 8°C lower than that of indica rice starch (Nakamura et al., 2002). This is because long parallel chains of amylopectin in indica rice expressing SSIIaI can form a longer double helix; by contrast, short chains with DP ≤ 12 can only form a short double helix in japonica rice expressing SSIIaJ.
Recently, direct evidence of protein–protein interactions among starch biosynthetic enzymes was observed in developing endosperms of wheat (Tetlow et al., 2004) and other cereals (Hennen-Bierwagen et al., 2008; Tetlow et al., 2008; Ahmed et al., 2015; Crofts et al., 2015). It is thought that SSIIa forms trimeric complexes with SSI and branching enzyme IIb (BEIIb) in developing endosperms of maize and rice, and these complexes seem to be important for the biosynthesis of short and intermediate starch chains (DP ≤ 24) within the amylopectin cluster (Liu et al., 2012; Crofts et al., 2015, 2017a). It remains to be determined how these trimeric complexes, higher molecular complexes, and monomers relate to the resulting starch structures. Our double mutant rice lines reveal a relay reaction between BEIIb and SSI, where SSI elongates the branches formed by BEIIb (Abe et al., 2014). SSI and BEIIb associate and synergistically enhance their mutual functions as shown by the in vitro analyses of recombinant enzymes (Nakamura et al., 2014). SSIIa can complement a large part of SSI function (Crofts et al., 2017b). These combined results indicate that the SSI, SSIIa, and BEIIb isozymes closely communicate during starch biosynthesis.
SSIIa deficiency in maize (Takeda and Preiss, 1993), wheat (Yamamori et al., 2000), barley (Morell et al., 2003), and Arabidopsis (Zhang et al., 2008) leads to higher amylose contents than in wild types (Fujita and Nakamura, 2012). In rice, the SSIIa and granule-bound starch synthase I (GBSSI) genes, which primarily synthesize amylose, are closely located on chromosome 6. Therefore, most indica rice cultivars have wild-type SSIIaI and GBSSII alleles. By contrast, most japonica rice cultivars have SSIIaJ and GBSSIJ alleles that generate enzymes with low activities (Sano, 1984; Nakamura et al., 2002). Therefore, the true effects of SSIIaJ on amylose content in japonica rice cultivars remains unknown, although the amylose contents of japonica cultivars are lower than those of indica rice cultivars (Sano, 1984).
Although SSIIaJ has low SSIIa activity, mutant rice lines with complete SSIIa knockout have not been isolated. Hence, the effects of SSIIa deficiency on other enzymes involved in starch biosynthesis, protein complex formation, apparent amylose content, starch structure, and starch physicochemical properties are unknown. In this study, we isolated an SSIIa-deficient mutant rice line from a chemically mutagenized japonica population and identified the SSIIa gene mutation. We evaluated the following rice lines: #1110-290, Kinmaze, and EM204. These lines expressed different SSIIa alleles (SSIIaI, SSIIaJ, and ss2a, respectively) and the same GBSSI allele derived from japonica cultivars (GBSSIJ). We discuss the effects of three SSIIa alleles (SSIIaI, SSIIaJ, and ss2a) with the GBSSIJ allele on starch biosynthesis, protein complex formation, apparent amylose content, amylopectin structure, and starch gelatinization temperature.
Materials and Methods
Plant Materials
A total of 1,500 mutant rice lines were generated by treating fertilized egg cells with 1 mM N-methyl-N-nitrosourea as described previously (Satoh and Omura, 1979). The initial screening was performed by measuring the water absorbance properties of boiled rice to isolate possible candidates for low-calorie rice lines. SSIIa-deficient EM204 (ss2a/GBSSIJ) was identified as one of 12 candidate lines by immunoblotting total proteins extracted from mature seeds using antiserum raised against rice SSIIa.
Kinmaze (the parental line, SSIIaJ/GBSSIJ), IR36 (indica rice cultivar, SSIIaI/GBSSII), and #1110-290 [SSIIa and GBSSI genes derived from indica (IR36) and japonica (Nipponbare) cultivars, respectively (SSIIaI/GBSSIJ), Crofts et al., 2017b] were used as control lines (Table 1). All rice lines were grown in a paddy field of Akita Prefectural University during the summer months under natural conditions.
Table 1.
Dehulled grain weight and genotypes of rice lines used in this study.
| Line | Genotype | Grain weight (mg) |
|---|---|---|
| IR36 | SSIIaI/GBSSII | 18.9 ± 0.2a (97)b |
| #1110-290 | SSIIaI/GBSSIJ | 16.8 ± 0.2 (87) |
| Kinmaze | SSIIaJ/GBSSIJ | 19.4 ± 0.5 (100) |
| EM204 | ss2a/GBSSIJ | 16.5 ± 0.4∗ (85) |
aValues shown are means ± SE of 20 seeds. bValues in parentheses are the percentages of the wild type (Kinmaze). ∗Significant differences between EM204 and Kinmaze (t-test, P<0.01).
Nucleotide Sequencing of SSIIa Gene From mRNA and Genomic DNA
Total RNA was extracted from developing endosperm of Kinmaze (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ) using the RNeasy Plant Mini Kit (QIAGEN). A full-length rice SSIIa cDNA was amplified using the PrimeScript II High Fidelity One-Step RT-PCR Kit (Takara) and a pair of SSIIa gene-specific primers (5′-aagcacgcgcacacactcaa-3′ and 5′-catcctattacgcatgcatgatgc-3′). Two primer pairs (5′-ccacaacgtattcagagatcgga-3′ and 5′-gaggta tcgaaggagttcatccgt-3′) were designed to cover the region of deletion in EM204 (ss2a/GBSSIJ). The genomic DNA fragment was isolated and amplified from Kinmaze (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ) leaves using the DNeasy Plant Mini Kit (QIAGEN), and then the sequence was confirmed using the above gene-specific primers.
Protein Analyses
For immunoblotting, total proteins from one mature rice seed and three developing rice seeds (with the embryo removed) were extracted with 10 volumes of denaturing extraction buffer [0.125 M Tris–HCl, pH 6.8, 8 M urea, 4% sodium dodecyl sulfate (SDS), 5% β-mercaptoethanol], and then incubated at room temperature for 2 h with rotation according to the method of Crofts et al. (2012). Soluble protein (SP), loosely bound to starch granule protein (LBP), and tightly bound to starch granule protein (TBP) were extracted from eight developing endosperm according to a previously published method (Asai et al., 2014; Crofts et al., 2015). Immunoblotting was performed as described previously (Crofts et al., 2012).
Samples used for native-PAGE/activity staining were extracted from three developing endosperm (10–15 DAF) using 1.5 volumes of buffer (for SSIIa zymogram) or 3 volumes of buffer (for the remaining zymograms) relative to seed fresh weight, and equal amounts of protein were loaded from each line (Fujita et al., 1999). SS activity staining for detecting SSI and SSIIIa was performed using glycogen as primer as described previously (Nishi et al., 2001), except that 0.5 M citrate was added to the reaction mixture. SSIIa activity staining was performed as described previously (Nishi et al., 2001) with the following modifications: 9% acrylamide native-PAGE gel containing 0.05% maize amylopectin as primer was electrophoresed at 8 mA for stacking gel and 15 mA for separation gel. The gel was run for an additional 50 min at 15 mA after the dye front reached the bottom, and then incubated with SS reaction buffer containing 0.5 M citrate adjusted to pH 10 with NaOH. BE activity was assessed using gels containing 0.0001% oyster glycogen (Yamanouchi and Nakamura, 1992). Debranching enzyme (DBE) was assessed as described previously (Fujita et al., 1999).
A 700-mg aliquot of developing seeds was extracted and fractionated by gel filtration chromatography using Superdex 200 resin according to the method of Crofts et al. (2015). Native-PAGE/SS and native-PAGE/BE activity staining was performed as described above. Native-PAGE/immunoblotting of each fraction was performed as described by Crofts et al. (2015).
Analyses of Starch Structure and Physicochemical Properties
Starch granules were purified from three grams of mature rice endosperm to determine starch structure and physicochemical properties according to the method of Fujita et al. (2003). Endosperm amylopectin chain-length distribution was determined by capillary electrophoresis (P/ACE MDQ Carbohydrate System, AB Sciex). The ratio of amylose to amylopectin was determined by gel filtration chromatography (Toyopearl HW-55S and HW-50S × 3) according to previously published methods (Fujita et al., 2007; Toyosawa et al., 2016). The thermal properties of endosperm starch were determined by differential scanning calorimetry (DSC, Seiko Instrument 6100) and analyzed as described previously (Fujita et al., 2003, 2006).
Results
SSIIa Deficiency in Mature Endosperm and Detection of the SSIIa SNP in EM204
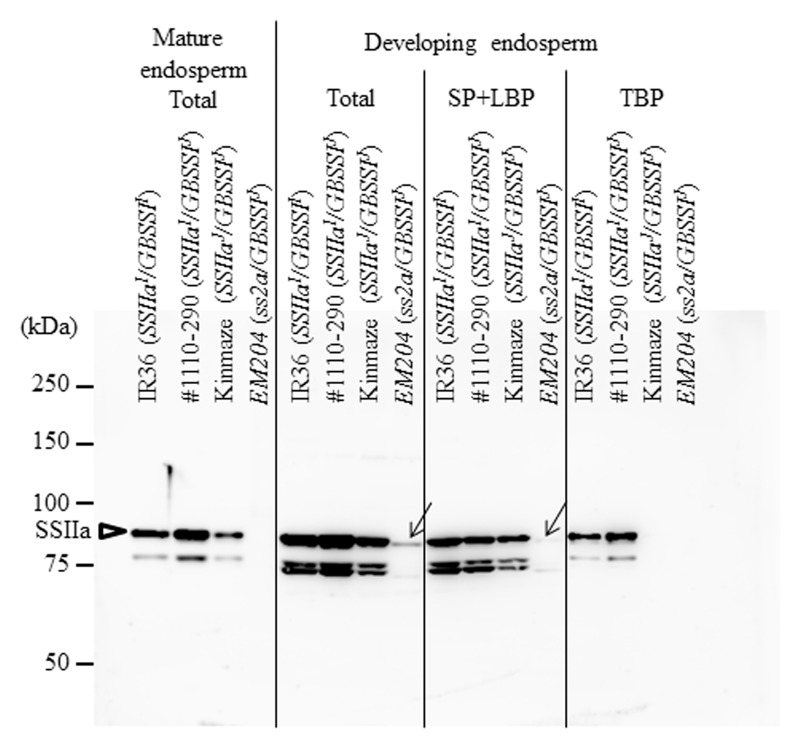
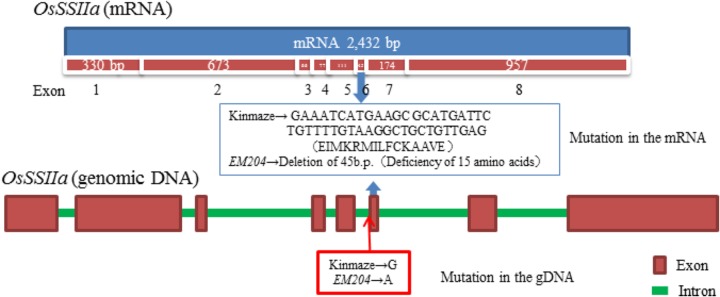
The SSIIa band (ca. 85 kDa) was not detected in the mature endosperm of EM204 (ss2a/GBSSIJ) using immunoblotting with a polyclonal antiserum of SSIIa, although it was detected in Kinmaze (SSIIaJ/GBSSIJ) (Figure 1, Mature endosperm, Total). By contrast, weak SSIIa bands with slightly smaller molecular weights were detected in the total protein fraction and in the SP+LBP fraction from EM204 (ss2a/GBSSIJ) developing endosperm (Figure 1, Developing endosperm, Total and SP+LBP). The western blot signals located around 75 kDa were likely to be truncated SSIIa since they were absent in EM204 (ss2a/GBSSIJ). To identify the mutation in the SSIIa gene in EM204 (ss2a/GBSSIJ), we compared cDNA sequences of SSIIa genes from developing endosperm in EM204 (ss2a/GBSSIJ) and Kinmaze (SSIIaJ/GBSSIJ). Exon 6 containing 45 base pairs was absent in cDNA of EM204 (ss2a/GBSSIJ) (Figure 2). DNA sequences around exon 6 of EM204 (ss2a/GBSSIJ) were determined using genomic DNA from young leaves (Figure 2). The last nucleotide in the SSIIa intron 5 in EM204 (ss2a/GBSSIJ) was mutated from guanine to adenine (Figure 2). This nucleotide substitution resulted in the splicing out of exon 6 together with introns 5 and 6, and led to the deletion of 15 amino acids in SSIIa of EM204 (ss2a/GBSSIJ; Figure 2). The slightly smaller SSIIa detected in developing EM204 endosperm (ss2a/GBSSIJ; Figure 1) might correspond to the SSIIa polypeptide with 15 amino acids deleted (from exon 6), although the amino acid sequence of truncated SSIIa in EM204 (ss2a/GBSSIJ) was not confirmed. Stereo crystallography from homology modeling of SSIIa in EM204 (ss2a/GBSSIJ) indicated that the 15 amino acids deleted from SSIIa in EM204 (ss2a/GBSSIJ) correspond to an α-helix in the N domain close to the catalytic site (Supplementary Figure S1). The mutated SSIIa in EM204 (ss2a/GBSSIJ) may not be enzymatically active or may be more rapidly degraded during endosperm development.
FIGURE 1.
Immunoblotting of mature and developing endosperm using anti-SSIIa serum. Total, total proteins; SP+LBP, soluble and loosely bound protein; TBP, tightly bound protein; IR36, indica rice cultivar, SSIIaI/GBSSII; #1110-290, SSIIaI/GBSSIJ; Kinmaze, japonica rice cultivar, SSIIaJ/GBSSIJ; EM204, ss2a/GBSSIJ. Arrows show the possible truncated SSIIa bands in EM204.
FIGURE 2.
Structure of the OsSSIIa gene [mRNA (accession No. AB115916.1 and genomic DNA (accession No. AP003509.3)] and identification of the mutation site in EM204.
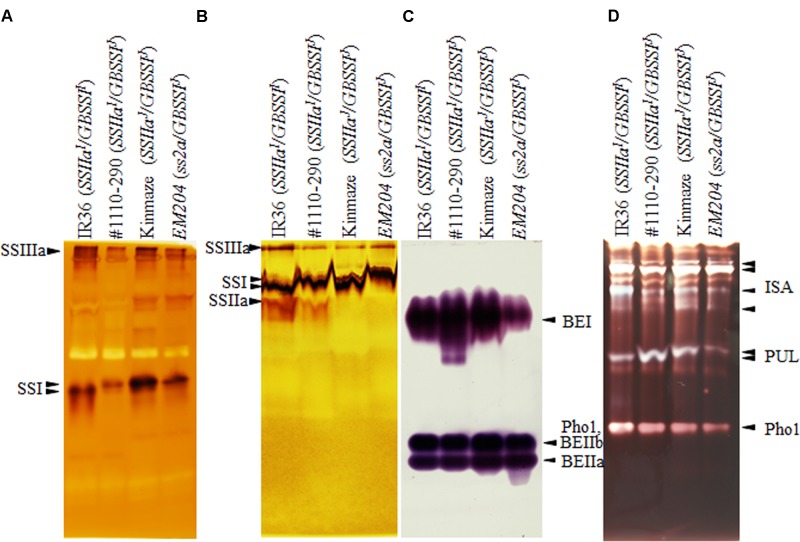
To detect SSIIa activity, we performed native-PAGE/SS activity staining using a maize amylopectin gel (Figure 3B). SSIIa activity bands were detected below the SSI activity band in IR36 (SSIIaI/GBSSII) and #1110-290 (SSIIaI/GBSSIJ), which contain the SSIIaI allele. By contrast, SSIIa activity bands were not detected in Kinmaze (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ). A previous in vitro study using recombinant SSIIa enzymes estimated that the activity of Kinmaze SSIIaJ was 10% of that of SSIIaI (Nakamura et al., 2005). The SSIIa activity of Kinmaze (SSIIaJ/GBSSIJ) was not detectable on native-PAGE/SS activity staining gels due to the detection limit of this method.
FIGURE 3.
Pleiotropic effects on starch biosynthetic enzymes revealed by native-PAGE/activity staining. (A) Starch synthase (SS) I and SSIIIa activity was detected using glycogen as primer. It is noted that SSI derived from Indica rice migrate faster than that of japonica rice (Chen and Bao, 2016; Itoh et al., 2017). (B) SSIIa activity was detected using maize amylopectin as primer; reaction was performed at pH 10 to minimize hydrolase activity (see Section “Materials and Methods”). (C) Branching enzyme (BE) activity. BEIIb and Phosphorylase 1 (Pho1) activity bands were overlapped. (D) Debranching enzyme (DBE) activity. Each isozyme labeled with arrowheads has been identified with corresponding mutant rice lines. IR36, indica rice cultivar, SSIIaI/GBSSII; #1110-290, SSIIaI/GBSSIJ; Kinmaze, japonica rice cultivar, SSIIaJ/GBSSIJ; EM204, ss2a/GBSSIJ.
Pleiotropic Effects of SSIIa Loss on Other Starch Biosynthetic Enzymes
To investigate the effects of loss of SSIIa on starch biosynthetic enzymes, SSs, BEs, DBEs, and plastidial starch phosphorylase (Pho1) were detected by native-PAGE/activity staining (Figure 3). Although the zymogram signals are not quantitative, the following pleiotropic effects were observed at least three times (replicated data were not shown). SSI and SSIIIa activity bands were slightly weaker in #1110-290 (SSIIaI/GBSSIJ) than in the other lines (Figure 3A). The BEI activities in EM204 (ss2a/GBSSIJ) and #1110-290 (SSIIaI/GBSSIJ) were weaker and stronger than those in Kinmaze (SSIIaJ/GBSSIJ) and IR36 (SSIIaI/GBSSII), respectively. BEIIa activities were comparable in the four lines (Figure 3C). Pho1 activity was weaker in EM204 (ss2a/GBSSIJ) than in the other lines (Figures 3C,D). Isoamylase (ISA) and pullulanase (PUL) activities were slightly weaker in EM204 (ss2a/GBSSIJ) than in the other lines (Figure 3D). PUL activities (Figure 3D) were weaker and stronger in IR36 (SSIIaI/GBSSII) and #1110-290 (SSIIaI/GBSSIJ), respectively, than in Kinmaze (SSIIaJ/GBSSIJ).
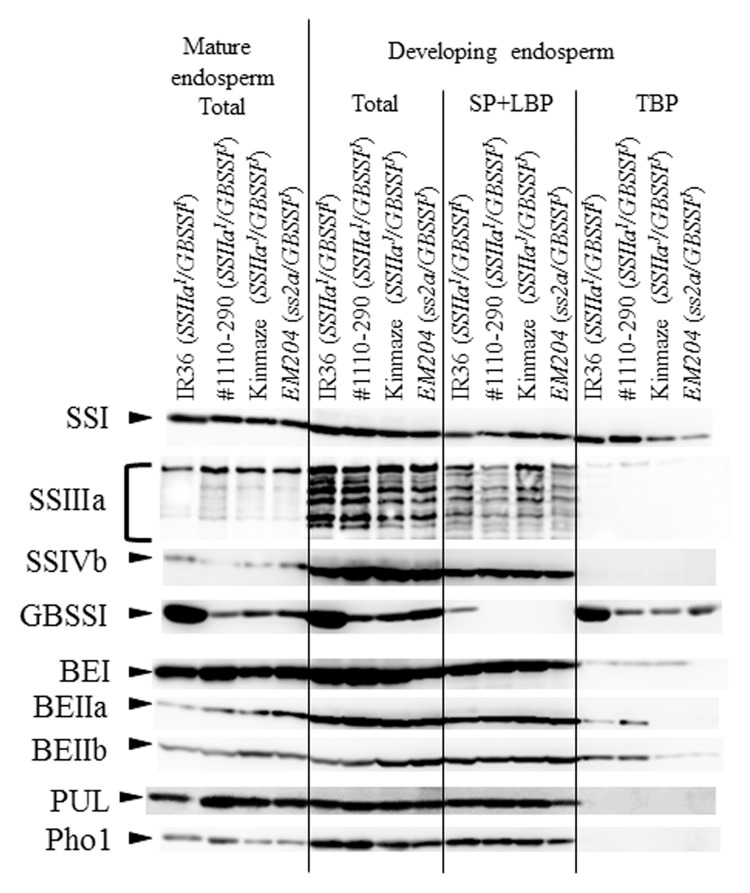
Total protein, SP+LBP, and TBP extracted from developing endosperm, and total protein extract of mature endosperm were analyzed to detect major starch biosynthetic enzymes by immunoblotting with polyclonal antisera (Figure 4). Most of the enzyme levels were similar in total protein extracts of mature endosperm from all lines, except that the GBSSI protein level was much higher in IR36 (SSIIaI/GBSSII) than in other lines. BEI, PUL, and Pho1 protein levels were higher in #1110-290 (SSIIaI/GBSSIJ) than in other lines (Figure 4, Mature endosperm, Total). In developing endosperms, SSI protein levels were lower in the TBP fractions from EM204 (ss2a/GBSSIJ) than in fractions from other lines (Figure 4, Developing endosperm, TBP). SSIIa protein bands were not detected in the TBP fractions from Kinmaze (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ) (Figure 1). SSIIIa protein band was not detected in the TBP fractions from EM204 (ss2a/GBSSIJ), whereas faint SSIIIa bands were detected in IR36 (SSIIaI/GBSSII), #1110-290 (SSIIaI/GBSSIJ), and Kinmaze (SSIIaJ/GBSSIJ) (Figure 4, Developing endosperm, TBP). Total SSIVb levels were lower in IR36 (SSIIaI/GBSSII) than in other lines. GBSSI levels were much higher in total and TBP fractions from IR36 (SSIIaI/GBSSII) than in those from other lines and were detected even in the SP+LBP fractions (Figure 4, Developing endosperm). GBSSI levels were slightly higher in TBP fractions from EM204 (ss2a/GBSSIJ) than #1110-290 (SSIIaI/GBSSIJ) and Kinmaze (SSIIaJ/GBSSIJ). BEI levels were lower in total and SP+LBP fractions from EM204 (ss2a/GBSSIJ) than in those from other lines and were not detected in the TBP fraction. BEIIa protein bands were not detected in the TBP fraction from Kinmaze (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ). BEIIb levels were lower in the TBP fractions from Kinmaze (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ) than in those from IR36 (SSIIaI/GBSSII) and #1110-290 (SSIIaI/GBSSIJ) (Figure 4, Developing endosperm, TBP). Differences between SSIIaI and SSIIaJ binding to starch granules (Figure 1, Developing endosperm, TBP) were consistent with previous reports (Umemoto et al., 2004; Nakamura et al., 2005).
FIGURE 4.
Isozyme distribution in protein fractions [Total protein (Total), soluble protein (SP), loosely bound protein (LBP), and tightly bound protein (TBP)] extracted from developing endosperm (10–15 DAF). Immunoblotting of each fraction was performed using anti-SSI, SSIIa, SSIIIa, SSIVb, GBSSI, BEI, BEIIa, and BEIIb antibodies. IR36, indica rice cultivar, SSIIaI/GBSSII; #1110-290, SSIIaI/GBSSIJ; Kinmaze, japonica rice cultivar, SSIIaJ/GBSSIJ; EM204, ss2a/GBSSIJ.
Effects of SSIIa Loss on Starch Biosynthetic Enzyme Complexes in Developing Seeds
Soluble extracts from developing seeds containing SSIIaI (#1110-290), SSIIaJ (Kinmaze), and ss2a (EM204) with GBSSIJ were fractionated by performing gel filtration chromatography with Superdex 200 resin. The fractions were analyzed by native-PAGE/SS and BE activity staining and immunoblotting using the corresponding native-PAGE gels (Figure 5). SSI and SSIIIa activity bands were found in Fractions (Fr.) 5–12 and Fr. 2–6, respectively (Figure 5A). SSIIa activity band was detected in. Fr. 8–11 of #1110-290 (SSIIaI/GBSSIJ), but not in Kinmaze (SSIIaJ/GBSSIJ) or EM204 (ss2a/GBSSIJ) (Figure 5A), confirming the native-PAGE/SS activity staining of crude extract result (Figure 3B). Immunoblotting of corresponding native-PAGE gels showed that activity bands and immunoblotting signals merged for SSI (Figure 5B) and SSIIIa (data not shown). However, immunoblotting signals of SSIIa in #1110-290 (SSIIaI/GBSSIJ) were detected in broader area than its activity bands (Figure 5C). There are two possible reasons why SSIIa activity was not detected in the faster-migrating area. The first possibility is that some SSIIa proteins happen to co-migrate with glucan hydrolase or transferase on native-PAGE; hence, the glucans generated by SSIIa may be degraded. The second possibility is that some SSIIa proteins became slightly degraded during the experimental procedure, and these inactive proteins likely migrated faster on the native-PAGE gel. The SSIIa activity band was not detected in Kinmaze (SSIIaJ/GBSSIJ), but SSIIa protein was detected by immunoblotting in Kinmaze (SSIIaJ/GBSSIJ), whereas neither the activity band nor the immunoblot signal of SSIIa was detected in EM204 (ss2a/GBSSIJ) (Figure 5C). Although some portion of SSI eluted in Fr. 6–9 (<230 kDa) in #1110-290 (SSIIaI/GBSSIJ) and Kinmaze (SSIIaJ/GBSSIJ), the majority of SSI eluted in Fr. 10–13 based on immunoblots. By contrast, more SSI immunoblotting signals in EM204 (ss2a/GBSSIJ) occurred in Fr. 6–9, with a lower content in Fr. 10–13.
FIGURE 5.
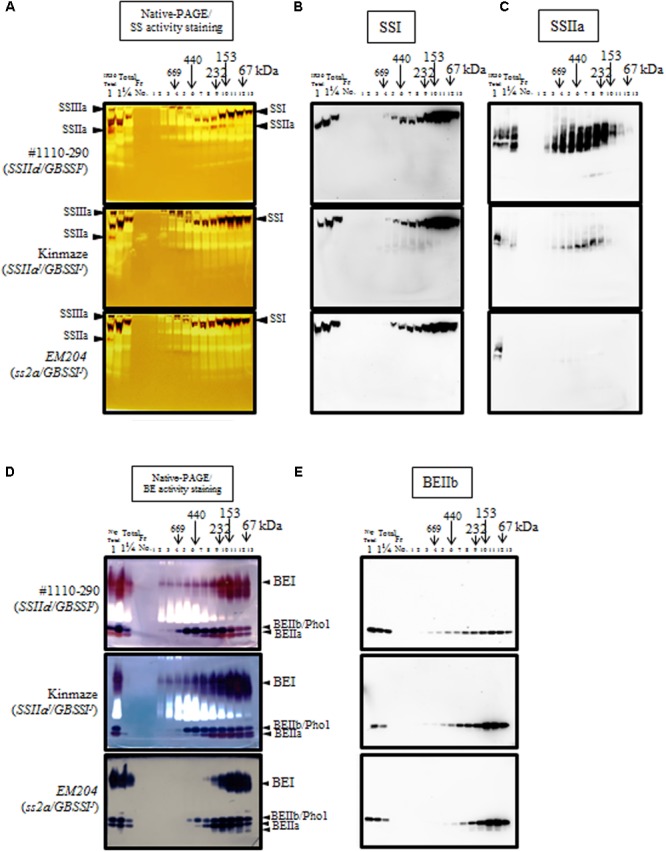
Molecular weight distribution of starch synthase (SS) (A–C) and branching enzyme (BE) (D,E) analyzed by native-PAGE/activity staining (A,D) and immunoblotting of the corresponding gel (B,C,E) loaded with the fractions obtained from gel filtration chromatography of soluble extract from rice developing endosperm. #1110-290, SSIIaI/GBSSIJ; Kinmaze and Nipponbare, japonica rice cultivar, SSIIaJ/GBSSIJ; EM204, ss2a/GBSSIJ. The left lane of each native-PAGE/activity staining gel and immunoblot is the loading control (IR36 for SS and Nipponbare for BE), except for D and E in Kinmaze, which are blanks. Total is the crude extract before gel filtration chromatography loaded with different amounts of extract (1 and ¼) for semi-quantitative comparisons. More mobile purple bands in EM204 of BE activity staining (D) are also BEIIa.
Native-PAGE/BE activity staining (Figure 5D) showed that less BEI activity eluted in high molecular weight fractions in EM204 (ss2a/GBSSIJ; Fr. 2–10) than in #1110-290 (SSIIaI/GBSSIJ) and Kinmaze (SSIIaJ/GBSSIJ). BEIIb immunoblotting showed that BEIIb protein eluted in wide molecular weight range (Fr. 3–13) in #1110-290. By contrast, the majority of BEIIb eluted in Fr. 10–13 in Kinmaze (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ), and BEIIb was not detected in Fr. >6 in EM204 (ss2a/GBSSIJ) (Figure 5D). The elution patterns of BEIIa, ISA, PUL, and Pho1 were essentially the same in #1110-290 (SSIIaI/GBSSIJ), Kinmaze (SSIIaJ/GBSSIJ), and EM204 (ss2a/GBSSIJ) (data not shown).
Seed Morphology and Grain Weight
Seeds of Kinmaze (SSIIaJ/GBSSIJ), IR36 (SSIIaI/GBSSII), and #1110-290 (SSIIaI/GBSSIJ) were translucent, whereas some of EM204 (ss2a/GBSSIJ) seeds were slightly chalky which was observed as dark shadows on a light box (Figure 6). The average seed weight (Table 1) of EM204 (ss2a/GBSSIJ; 16.5 mg) was only ca. 85% of that of Kinmaze (SSIIaJ/GBSSIJ). IR36 (SSIIaI/GBSSII) seeds had elongated shapes, and the seed weight (18.9 mg) was lower than that of Kinmaze (SSIIaJ/GBSSIJ). The #1110-290 (SSIIaI/GBSSIJ) seeds were shorter than those of IR36 (SSIIaI/GBSSII) but longer than those of Kinmaze (SSIIaJ/GBSSIJ), and the average seed weight was 16.8 mg.
FIGURE 6.

Morphology of rice grains observed using a stereo-microscope with overhead light (upper panels) and on a light box (lower panels). The parts that are chalky or opaque in the seeds appeared dark on a light box. Note that central region of some of EM204 seeds were chalky. IR36, indica rice cultivar, SSIIaI/GBSSII; #1110-290, SSIIaI/GBSSIJ; Kinmaze, japonica rice cultivar, SSIIaJ/GBSSIJ; EM204, ss2a/GBSSIJ.
Analysis of Starch Structure and Physicochemical Properties
Apparent amylose content and the ratios of amylopectin short chains and long chains (Fr. III/II) were measured by gel filtration of debranched starch from four rice lines with Toyopearl HW-55S and HW-50S × 3 (Table 2). Apparent amylose content was the highest (25.6%) in IR36 (SSIIaI/GBSSII) among the four rice lines, whereas apparent amylose content of #1110-290 (SSIIaI/GBSSIJ) was 13.7% and much lower than that of Kinmaze (20.7%, SSIIaJ/GBSSIJ). Apparent amylose content of EM204 (ss2a/GBSSIJ) was 24.1%, significantly higher than that of Kinmaze (SSIIaJ/GBSSIJ) (Table 2). Fr. III/II of IR36 (SSIIaI/GBSSII; 2.8) and #1110-290 (SSIIaI/GBSSIJ; 2.9) was lower than that of Kinmaze (SSIIaJ/GBSSIJ; 3.2), but there were no significant differences between Kinmaze (SSIIaJ/GBSSIJ; 3.2) and EM204 (ss2a/GBSSIJ; 3.3) (Table 2).
Table 2.
Carbohydrate content (weight %) in endosperm starch fractions separated by gel filtration chromatography (Toyopearl HW55S/HW50S x 3).
| Line | Genotype | Fr. Ia | Fr. II | Fr. III | III/II |
|---|---|---|---|---|---|
| IR36 | SSIIaI/GBSSII | 25.6 ± 0.5** | 19.8 ± 0.4 | 54.6 ± 0.3 | 2.8 ± 0.1* |
| #1110-290 | SSIIaI/GBSSIJ | 13.7 ± 0.3** | 21.9 ± 0.2 | 64.4 ± 0.2 | 2.9 ± 0.0* |
| Kinmazeb | SSIIaJ/GBSSIJ | 20.7 ± 0.2 | 18.9 ± 0.2 | 60.4 ± 0.2 | 3.2 ± 0.1 |
| EM 204 | ss2a/GBSSIJ | 24.1 ± 0.5** | 17.8 ± 0.2 | 58.1 ± 0.7 | 3.3 ± 0.1 |
Mean value ± SE of at least three replications. Every rice line used in this experiment was harvested in 2015. aThree fractions (Fr. I, II, III) were separated at the minima of the carbohydrate content curve detected with refractive index detectors. Fr. I, II, and III correspond to the apparent amylose content, amylopectin long chains, and amylopectin short chains, respectively. bData from Itoh et al. (2017). ∗Significant differences between Kinmaze and other lines (t-test, P<0.05). ∗∗Significant differences between Kinmaze and other lines (t-test, P<0.01).
Chain-length distribution of endosperm amylopectin was analyzed by capillary electrophoresis (Figure 7). The contents of short chains with DP = 6–12 and middle chains with DP = 13–24 from Kinmaze expressing SSIIaJ were greater and fewer, respectively, than those from IR36 expressing SSIIaI (Umemoto et al., 1999; Nakamura et al., 2005). The chain-length distributions in #1110-290 expressing SSIIaI showed similar patterns to those in IR36 (Figure 7A). By contrast, the contents of short chains with DP = 6–12 and middle chains with DP = 13–24 from EM204 (ss2a/GBSSIJ) were greater and fewer, respectively, than those from Kinmaze (SSIIaJ/GBSSIJ) (Figure 7B). These results suggest that the function of SSIIa in EM204 (ss2a/GBSSIJ) was completely impaired or at least lower than that in Kinmaze (SSIIaJ/GBSSIJ), whose recombinant SSIIa activity was estimated as only ca. 10% of that in IR36 (SSIIaI/GBSSII) in vitro (Nakamura et al., 2005).
FIGURE 7.
Capillary electrophoresis analysis of amylopectin molecular structure. (A) Amylopectin chain-length distribution patterns in mature endosperm. (B) Differential plots of Kinmaze (SSIIaJ), EM204 (ss2a), and #1110-290 (SSIIaI) expressing japonica-type GBSSI. Each panel shows one typical representative data set of at least three replications. DP, degree of polymerization. ∗Data from Crofts et al. (2017b).
The gelatinization temperature of endosperm starch from japonica rice cultivars is much lower than that of endosperm starch from indica rice cultivars (Nakamura et al., 2002). The onset, peak, and conclusion temperatures of Kinmaze (SSIIaJ/GBSSIJ), endosperm starch were 16.5°C, 13.5°C, and 10.8°C, respectively, lower than those of IR36 (SSIIaI/GBSSII) (Table 3). Those of EM204 (ss2a/GBSSIJ) were 11.5°C, 5.6°C, and 3.1°C lower than those of Kinmaze (SSIIaJ/GBSSIJ). By contrast, those of #1110-290 (SSIIaI/GBSSIJ) were 6.2°C, 5.1°C, and 5.1°C higher than those of IR36 (SSIIaI/GBSSII).
Table 3.
Thermal properties of endosperm starch as determined by differential scanning calorimetry.
| Line | Genotype | Toa (°C) | Tpb (°C) | Tcc (°C) | ΔHd (mJ/mg) |
|---|---|---|---|---|---|
| IR36 | SSIIaI/GBSSII | 61.9 ± 0.1 | 66.4 ± 0.1* | 70.4 ± 0.0* | 14.3 ± 0.5* |
| #1110-290 | SSIIaI/GBSSIJ | 68.1 ± 0.1* | 71.5 ± 0.0* | 75.5 ± 0.1* | 16.4 ± 0.8* |
| Kinmaze | SSIIaJ/GBSSIJ | 45.4 ± 0.2 | 52.8 ± 0.0 | 59.6 ± 0.2 | 9.5 ± 0.4 |
| EM204 | ss2a/GBSSIJ | 33.9 ± 0.6* | 47.2 ± 0.4* | 56.5 ± 0.2* | 7.1 ± 0.0* |
Mean value ± SE of three seeds.
aOnset temperature, bPeak temperature, cConclusion temperature, dGelatinization enthalpy of starch. ∗Significant differences between Kinmaze and other lines (t-test, P<0.01).
Discussion
Isolation of SSIIa-Deficient Mutant Rice Line
A mutant rice line lacking SSIIa protein in mature seeds was isolated in this study and designated as EM204 (ss2a/GBSSIJ). Chain-length distribution analysis of endosperm starch showed that the amount of amylopectin short chains with DP = 6–12 was larger in EM204 (ss2a/GBSSIJ) than in the parental Kinmaze (SSIIaJ/GBSSIJ), a WT japonica rice cultivar (Figure 7B). This indicates that SSIIa in japonica rice cultivars slightly elongates DP = 6–12 amylopectin chains to DP = 13–24 in vivo. SSIIa activity bands were not detected in EM204 (ss2a/GBSSIJ) (Figures 3B, 5), and SSIIa protein was not detected in any gel filtration chromatography fractions from developing endosperm (Figure 5C). Although it is possible that truncated SSIIa in EM204 (ss2a/GBSSIJ) can be translated, the truncated SSIIa in developing endosperm is likely easily degraded during seed development, resulting in lower SSIIa activity in EM204 (ss2a/GBSSIJ) than in Kinmaze (SSIIaJ/GBSSIJ) based on the enzyme stereography model (Supplementary Figure S1).
EM204 (ss2a/GBSSIJ) was initially screened as one of 12 mutant lines for low-calorie rice with high water absorption in boiled polished rice (see Materials and methods). The water content in EM204 (ss2a/GBSSIJ) boiled rice was much higher than that in Kinmaze (SSIIaJ/GBSSIJ) when polished rice grains were boiled with excess water (data not shown). We expected that the SSIIa gene would correspond to a gene involved in water absorption. However, genetic analyses of the F2 population resulting from a cross between EM204 (ss2a/GBSSIJ) and the WT (SSIIaJ/GBSSIJ) showed that loss of SSIIa and loss of water absorption did not segregate identically. It is possible that gene pathways related to water absorption of polished rice grains consist of multiple genes (data not shown). It will be necessary to use genetic analyses of EM204 (ss2a/GBSSIJ) to isolate genes related to water absorption other than the SSIIa gene.
Pleiotropic Effects of SSIIa Protein Loss on the Elution Pattern of Starch Biosynthetic Enzymes
Starch biosynthetic enzymes are thought to form protein complexes for efficient starch biosynthesis (Crofts et al., 2017a). Trimeric complexes containing SSIIa, SSI, and BEIIb have been reported in maize (Liu et al., 2012) and japonica rice (Crofts et al., 2015). This trimeric complex is likely involved in generating amylopectin clusters (Crofts et al., 2017a). Therefore, differences in the composition and function of protein complexes modulated by active SSIIa (SSIIaI), low-activity SSIIa (SSIIaJ), or loss of SSIIa (ss2a) were suspected. Native-PAGE activity staining and immunoblotting of corresponding gels loaded with the gel filtration chromatography elution fractions suggested that EM204 (ss2a) may form altered trimeric protein complexes compared with those of #1110-290 with active SSIIa (SSIIaI) or Kinmaze with low-activity SSIIa (SSIIaJ). Compared with the SSI elution pattern in #1110-290 (SSIIaI/GBSSIJ) and Kinmaze (SSIIaJ/GBSSIJ), more SSI in EM204 (ss2a/GBSSIJ) eluted in Fr. 6–9 where the trimeric protein complex elutes than in Fr. 10–13 where monomeric SSI elutes (Figure 5B). BEIIb eluted in a wide range of molecular weights in #1110-290 (SSIIaI/GBSSIJ). However, the majority of BEIIb in Kinmaze (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ) was eluted in fractions corresponding to monomeric molecular weight (Fr. 11–13); in EM204 (ss2a/GBSSIJ), BEIIb was not detected in high molecular weight fractions (<Fr. 5). Based on these results, it can be speculated that SSIIaJ in Kinmaze may form alternative protein complex with SSI and BEIIb, whereas the protein complex in EM204 (ss2a/GBSSIJ) may be generated by BEIIb associating with two SSI proteins but without SSIIa, although the alteration in actual protein complex composition is to be analyzed. The mature seed weight of EM204 (ss2a/GBSSIJ) was 15% less than its parent Kinmaze (SSIIaJ/GBSSIJ), indicating that amylopectin synthesis was reduced. Complete absence of SSIIa in EM204 (ss2a/GBSSIJ) resulted in inefficient elongation of amylopectin branches and altered the elution pattern of starch biosynthetic proteins as analyzed by gel filtration chromatography. It cannot be ruled out that the alteration of elution pattern of proteins through gel-filtration chromatography may be a result of interactions of proteins with truncated SSIIa products. However, it is hard to distinguish them due to the resolution limit of gel filtration chromatography. Further analyses are required to detect the direct evidences of protein–protein interaction such as immune-precipitation and/or the reconstitution experiments using recombinant enzymes to confirm the change in composition of trimeric protein complex.
The su2 mutants in maize lack SSIIa activity. Some su2 mutants lack only SSIIa activity and others lack both activity and protein, but all su2 mutants display increased DP = 6–12 and reduced DP = 13–24 levels in amylopectin chain-length distributions (Zhang et al., 2004), which is the same as that observed in EM204 (ss2a/GBSSIJ). Protein complex formation in su2 was analyzed using the su2 mutant with inactive SSIIa protein, and that trimeric protein complex composition remained the same as the WT (Liu et al., 2012). SSI, SSIIa, and BEIIb in this su2 mutant displayed a loss of or reduced affinity to the starch granule, which is commonly observed in japonica rice (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ) and in ss2a mutants in wheat (Yamamori et al., 2000) and barley (Morell et al., 2003). Although protein–protein interactions of starch biosynthetic enzymes are likely to be an important mechanism for efficient starch synthesis (Crofts et al., 2017a), the relationship between formation of the SSI-SSIIa-BEIIb trimeric complex, its affinity to the starch granule, and starch structure remains unknown and requires further analyses.
Effects of SSIIa and GBSSI Alleles on Starch Structure in Rice Endosperm
Amylose content is higher in the storage starches of SSIIa-deficient mutants of maize (Takeda and Preiss, 1993), wheat (Yamamori et al., 2000), barley (Morell et al., 2003), and Arabidopsis (Zhang et al., 2008) than in the corresponding WT plants (Fujita and Nakamura, 2012). Japonica rice cultivars correspond to the SSIIa mutant, whereas indica rice cultivars correspond to the WT. However, most japonica cultivars have a leaky mutation in the GBSSI gene, which complicates the determination of direct effects of loss of SSIIa on amylose content in rice.
In this study, we compared the amylose contents in three rice lines that expressed a GBSSIJ allele along with three different SSIIa alleles: SSIIaI (#1110-290), SSIIaJ (Kinmaze), and ss2a (EM204) (Table 2). The apparent amylose content of EM204 (ss2a/GBSSIJ) starch was 3.4% higher than that of the Kinmaze (SSIIaJ/GBSSIJ), indicating that loss of SSIIa in rice leads to increased amylose content as observed in other crops. This is because SSIIa deficiency reduces amylopectin synthesis and GBSSI uses the excess ADPG for amylose synthesis. Moreover, slightly higher expression of GBSSI in EM204 (ss2a/GBSSIJ) than Kinmaze (SSIIaJ/GBSSIJ) might be another explanation (Figure 4). The apparent amylose content of #1110-290 (SSIIaI/GBSSIJ), which still has an active SSIIa, was 7% lower than that of Kinmaze (SSIIaJ/GBSSIJ), which already has significantly reduced SSIIa activity (Table 2). The vigorous amylopectin synthesis in #1110-290 (SSIIaI/GBSSIJ) expressing an active SSIIa leads to a shortage of ADPG levels in the amyloplast and reduces amylose synthesis. The apparent amylose content of the near-isogenic rice lines (NILs) that have SSIIaI from indica rice (Kasalath) with Nipponbare (japonica) background (GBSSIJ) was approximately 0.9–2.1% lower than that of Nipponbare (Umemoto et al., 2004). A reduction of 7% of the apparent amylose content in #1110-290 (SSIIaI/GBSSIJ) could be due to endosperm development under high temperature because the flowering time of #1110-290 (SSIIaI/GBSSIJ) was 3 weeks earlier than that of Kinmaze (SSIIaJ/GBSSIJ). The average temperature at 20 days after flowering in #1110-290 (SSIIaI/GBSSIJ) was 25.1°C, which is 4°C higher than that in Kinmaze (SSIIaJ/GBSSIJ). The amylose content depends on the temperature during seed development (Hirano and Sano, 1998). An SNP at the splice site in the first intron of GBSSI is associated with different temperature sensitivities during seed development (Larkin and Park, 1999). Japonica rice cultivars expressing GBSSIJ accumulate fewer mature GBSSI mRNA transcripts at high temperatures than at low temperatures (Zhang et al., 2013).
The apparent amylose content of IR36 (SSIIaI/GBSSII) with high expression of GBSSII was ca. 12% higher than that of #1110-290 (SSIIaI/GBSSIJ) with low expression of GBSSIJ, as expected (Table 2). By contrast, the amylopectin chain-length distributions were not identical in both lines. The amounts of shorter chains with DP ≤ 17 and middle chains with 19 ≤ DP ≤ 30 in IR36 (SSIIaI/GBSSII) were higher and lower, respectively, than those of #1110-290 (SSIIaI/GBSSIJ) (Figure 7B). These changes in amylopectin structure result in a lower starch gelatinization temperature in IR36 (SSIIaI/GBSSII) than in #1110-290 (SSIIaI/GBSSIJ) (Table 3). Alternatively, more vigorous SSIIa activity along with low activities of the other SS isozymes [SSI (Figure 3) and GBSSIJ] in #1110-290 (SSIIaI/GBSSIJ) may lead to elongation of shorter chains with DP ≤ 17 to 19 ≤ DP ≤ 30.
Conclusion
In this study, we isolated an ss2a rice mutant designated EM204 (ss2a/GBSSIJ), which completely lacked SSIIa in mature seed and accumulated only very low levels of truncated SSIIa in developing endosperm (Figure 1). Analyses of starch biosynthetic protein complex formation in EM204 (ss2a/GBSSIJ) suggested that loss of SSIIa may be compensated by SSI, which might form an alternative trimeric protein complex (Figure 5). EM204 (ss2a/GBSSIJ) grain accumulated starch with unique properties. The gelatinization temperature (Tp°C) of EM204 (ss2a/GBSSIJ) endosperm starch was very low, 5.6°C lower than typical japonica rice (Table 3). The amylose content was significantly higher in EM204 (ss2a/GBSSIJ) than in WT (SSIIaJ/GBSSIJ) (Table 2).
The processing methods and retrogradation of starch foods during storage are greatly affected by differences in the starch gelatinization temperature. EM204 (ss2a/GBSSIJ) starches with low gelatinization temperatures (Table 3) could be used for generating foods and additives that retrograde slowly. However, the seed weight of EM204 (ss2a/GBSSIJ) was 15% lower than that of the WT (SSIIaJ/GBSSIJ) (Table 1). It will be necessary to improve the agricultural traits of EM204 (ss2a/GBSSIJ) by crossing it with elite cultivars before it can be used for practical applications.
Author Contributions
SM performed the experiments and prepared the figures and tables. NC performed the experiments and wrote the article. YS performed the experiments. YH and NO provided technical assistance to SM. TW and TK complemented the writing, and NF conceived the original research plans, supervised, and complemented the writing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer IJT and handling Editor declared their shared affiliation.
Acknowledgments
The authors thank Ms. Yuko Nakaizumi (Akita Prefectural University) for growing the rice plants, Hayashibara Co., Ltd. for providing isoamylase, and Bioedit for the English language review.
Footnotes
Funding. This research project was partially supported by The Science and Technology Research Promotion Program for Agriculture, Forestry, and Fisheries and Food Industry (25033AB and 28029C), the Grant-in-Aid for JSPS fellows from Japan Society for the Promotion of Science (No. 15J40176), and the Japan Society for the Promotion of Science (No. 16K18571).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00645/full#supplementary-material
Homology modeling comparison of the SSIIa stereo structure in Kinmaze (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ). Images were rendered in Spanner in the Sequence-To-Function Annotation Service. Proteins are represented as a ribbon diagram. N- and C-terminal domains are shown in blue and red, respectively. KTGGL motifs (green) are important for the active site. The yellow loop indicates the 15 amino acids in Kinmaze that were absent in EM204 (ss2a/GBSSIJ).
References
- Abe N., Asai H., Yago H., Oitome N. F., Itoh R., Crofts N., et al. (2014). Relationships between starch synthase I and branching enzyme isozymes determined using double mutant rice lines. 14:80. 10.1186/1471-2229-14-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Z., Tetlow I. J., Ahmed R., Morell M. K., Emes M. J. (2015). Protein-protein interactions among enzymes of starch biosynthesis in high-amylose barley genotypes reveal differential roles of heteromeric enzyme complexes in the synthesis of A and B granules. 233 95–106. 10.1016/j.plantsci.2014.12.016 [DOI] [PubMed] [Google Scholar]
- Asai H., Abe N., Matsushima R., Crofts N., Oitome N. F., Nakamura Y., et al. (2014). Deficiencies in both starch synthase IIIa and branching enzyme IIb lead to a significant increase in amylose in SSIIa-inactive japonica rice seeds. 65 5497–5507. 10.1093/jxb/eru310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Bao J. (2016). Underlying mechanisms of zymographic diversity in starch synthase I and pullulanase in rice-developing endosperm. 64 2030–2037. 10.1021/acs.jafc.5b06030 [DOI] [PubMed] [Google Scholar]
- Crofts N., Abe K., Aihara S., Itoh R., Nakamura Y., Itoh K., et al. (2012). Lack of starch synthase IIIa and high expression of granule-bound starch synthase I synergistically increase the apparent amylose content in rice endosperm. 19 62–69. 10.1016/j.plantsci.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Crofts N., Abe N., Oitome N. F., Matsushima R., Tetlow I. J., Emes M. J., et al. (2015). Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. 66 4469–4482. 10.1093/jxb/erv212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts N., Nakamura Y., Fujita N. (2017a). Critical and speculative review of the roles of multi-protein complexes in starch biosynthesis in cereals. 262 1–8. 10.1016/j.plantsci.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Crofts N., Sugimoto K., Oitome F. N., Nakamura Y., Fujita N. (2017b). Differences in specificity and compensatory functions among three major starch synthases determine the structure of amylopectin in rice endosperm. 94 399–417. 10.1007/s11103-017-0614-8 [DOI] [PubMed] [Google Scholar]
- Fujita N., Kubo A., Perigio B. F., Nakakita M., Harada K., Minaka N., et al. (1999). Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. 208 283–293. [DOI] [PubMed] [Google Scholar]
- Fujita N., Kubo A., Suh D.-S., Wong K.-S., Jane J.-L., Ozawa K., et al. (2003). Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. 44 607–618. [DOI] [PubMed] [Google Scholar]
- Fujita N., Nakamura Y. (2012). “Distinct and overlapping functions of starch synthase isoforms,” in Vol. 5 ed. Tetlow I. J. (London: The society for experimental biology; ), 115–140. [Google Scholar]
- Fujita N., Yoshida M., Asakura N., Ohdan T., Miyao A., Hirochika H., et al. (2006). Function and characterization of starch synthase I using mutants in rice. 140 1070–1084. 10.1104/pp.105.071845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Yoshida M., Kondo T., Saito K., Utsumi Y., Tokunaga T., et al. (2007). Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. 144 2009–2023. 10.1104/pp.107.102533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen-Bierwagen T. A., Liu F., Marsh R. S., Kim S., Gan Q., Tetlow I. J., et al. (2008). Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. 146 1892–1908. 10.1104/pp.108.116285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H. Y., Sano Y. (1998). Enhancement of Wx gene expression and the accumulation of amylose in response to cool temperatures during seed development in rice. 39 807–812. [Google Scholar]
- Itoh Y., Crofts N., Abe M., Hosaka Y., Fujita N. (2017). Characterization of the endosperm starch and the pleiotropic effects of biosynthetic enzymes on their properties in novel mutant rice lines with high resistant starch and amylose content. 258 52–60. 10.1016/j.plantsci.2017.02.002 [DOI] [PubMed] [Google Scholar]
- Katayama K., Komae K., Kohyama K., Kato T., Tamiya S., Komaki K. (2002). New sweet potato line having low gelatinization temperature and altered starch structure. 54 51–57. [Google Scholar]
- Kitahara K., Fukunaga S., Katayama K., Takahata Y., Nakazawa Y., Yoshinaga M., et al. (2005). Physicochemical properties of sweetpotato starches with different gelatinization temperatures. 57 473–479. 10.1002/star.200400349 [DOI] [Google Scholar]
- Larkin P. D., Park W. D. (1999). Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. 40 719–727. [DOI] [PubMed] [Google Scholar]
- Liu F., Romanova N., Lee E. A., Ahmed R., Evans M., Gilbert E. P., et al. (2012). Glucan affinity of starch synthase IIa determines binding of starch synthase I and starch-branching enzyme IIb to starch granules. 448 373–387. 10.1042/BJ20120573 [DOI] [PubMed] [Google Scholar]
- Morell M. K., Kosar-Hashemi B., Cmiel M., Samuel M. S., Chandler P., Rahman S., et al. (2003). Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. 34 173–185. 10.1046/j.1365-313X.2003.01712.x [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Aihara S., Crofts N., Sawada T., Fujita N. (2014). In vitro studies of 5 enzymatic properties of starch synthases and interactions between starch synthase I 6 and starch branching enzymes from rice. 224 1–8. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Francisco B. P., Jr., Hosaka Y., Sato A., Sawada T., Kubo A., et al. (2005). Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. 58 213–227. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Sakurai A., Inaba Y., Kimura K., Iwasawa N., Nagamine T. (2002). The fine structure of amylopectin in endosperm from Asian cultivated rice can be largely classified into two classes. 54 117–131. [Google Scholar]
- Nishi A., Nakamura Y., Tanaka N., Satoh H. (2001). Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. 127 459–472. 10.1104/pp.010127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y. (1984). Differential regulation of waxy gene expression in rice endosperm. 68 467–473. 10.1007/BF00254822 [DOI] [PubMed] [Google Scholar]
- Satoh H., Omura T. (1979). Induction of mutation by the treatment of fertilized egg cell with N-methyl-N-nitrosourea in rice. 24 165–174. [Google Scholar]
- Takeda Y., Preiss J. (1993). Structure of B90 (sugary) and W64A (normal) maize starches. 240 265–275. [Google Scholar]
- Tetlow I. J., Beisel K. G., Cameron S., Makhmoudova A., Liu F., Bresolin N. S., et al. (2008). Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. 146 1878–1891. 10.1104/pp.108.116244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow I. J., Wait R., Lu Z., Akkasaeng R., Bowsher C. G., Esposito S., et al. (2004). Protein phosphorylation in amyloplasts regulates starch-branching enzyme activity and protein–protein interactions. 16 694–708. 10.1105/tpc.017400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyosawa Y., Kawagoe Y., Matsushima R., Crofts N., Ogawa M., Fukuda M., et al. (2016). Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. 170 1255–1270. 10.1104/pp.15.01232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto T., Aoki N., Lin H., Nakamura Y., Inouch N., Sato Y., et al. (2004). Natural variation in rice starch synthase IIa affects enzyme and starch properties. 31 671–684. 10.1071/FP04009 [DOI] [PubMed] [Google Scholar]
- Umemoto T., Nakamura Y., Satoh H., Terashima K. (1999). Differences in amylopectin structure between two rice varieties in relation to the effects of temperature during grain-filling. 51 58–62. [Google Scholar]
- Umemoto T., Yano M., Satoh H., Shomura A., Nakamura Y. (2002). Mapping of a gene responsible for the difference in amylopectin structure between japonioca-type and indica-type rice varieties. 104 1–8. [DOI] [PubMed] [Google Scholar]
- Yamamori M., Fujita S., Hayakawa K., Matsuki J., Yasui T. (2000). Genetic elimination of a starch granule protein, SGP-1, of wheat generates an altered starch with apparent high amylose. 101 21–29. [Google Scholar]
- Yamanouchi H., Nakamura Y. (1992). Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. 33 985–991. 10.1093/oxfordjournals.pcp.a078351 18844776 [DOI] [Google Scholar]
- Zhang H., Duan L., Dai J. S., Zhang C. Q., Li J., Gu M. H., et al. (2013). Major QTLs reduce the deleterious effects of high temperature on rice amylose content by increasing splicing efficiency of Wx pre-mRNA. 127 273–282. 10.1007/s00122-013-2216-4 [DOI] [PubMed] [Google Scholar]
- Zhang X., Colleoni C., Ratushna V., Sirghie-Colleoni M., James M. G., Myers A. M. (2004). Molecular characterization demonstrates that the Zea mays gene sugary2 codes for the starch synthase isoform SSIIa. 54 865–879. [DOI] [PubMed] [Google Scholar]
- Zhang X., Szydlowski N., Delvallé D., D’Hulst C., James M. G., Myers A. M. (2008). Overlapping functions of the starch synthase SSII and SSIII in amylopectin biosynthesis in Arabidopsis. 8:96. 10.1186/1471-2229-8-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Homology modeling comparison of the SSIIa stereo structure in Kinmaze (SSIIaJ/GBSSIJ) and EM204 (ss2a/GBSSIJ). Images were rendered in Spanner in the Sequence-To-Function Annotation Service. Proteins are represented as a ribbon diagram. N- and C-terminal domains are shown in blue and red, respectively. KTGGL motifs (green) are important for the active site. The yellow loop indicates the 15 amino acids in Kinmaze that were absent in EM204 (ss2a/GBSSIJ).