Abstract
Colorectal cancer (CRC) is the fourth leading worldwide cause of cancer-associated mortalities. Nuclear factor-κB (NF-κB) is a transcriptional regulator of multiple genes associated with CRC. Tumor tissue were compared with normal adjacent mucosa from 30 sporadic patients with CRC were investigated. A total of 8 non-CRC patients were analyzed as a control group. In the present study, the protein expression of NF-κB/p65 was detected by immunohistochemistry, and the gene expression profiles of cyclin D1 (CCND1), prostaglandin-endoperoxide synthase 2, vascular endothelial growth factor A, matrix metallopeptidase 9, BCL2 apoptosis regulator (BCL2), BCL2 like 1, nitric oxide synthase 2, tumor necrosis factor and arachidonate lipoxygenase were detected by reverse transcription-quantitative polymerase chain reaction. NF-κB/p65 and genes expression profiles were classified according to tumor-node-metastasis (TNM) clinicopathological parameters, followed by statistical analysis. Higher protein expression of NF-κB/p65 in the cytoplasm of tumor tissues compared with adjacent normal mucosa was reported; this increment was positively associated with all clinicopathological parameters, except for tumor localization site. The selected genes demonstrated a diverse associative pattern when analyzed with clinicopathological parameters. CCND1 was positively associated with all TNM parameters and BCL2 was negatively associated with all TNM parameters, thus indicating their importance as strong molecular biomarkers for CRC. According to these results, not all selected genes regulated by NF-κB/p65 show increased expression during CRC development, whereas the transcription factor did. The present study suggests that NF-κB/p65 overexpression is necessary for CRC establishment and progression, but its transcriptional activity is not sufficient to regulate all target genes in CRC. NF-κB/p65 and the gene expression profiles reported in the present study may be therapeutically useful. Considering the heterogeneity of the disease, the particular evaluation of these molecules may allow for the selection of proper diagnosis, treatment and follow-up for patients with sporadic CRC.
Keywords: colorectal cancer, immunohistochemistry, NF-κB, cytoplasm, RT-qPCR
Introduction
Colorectal cancer (CRC) is a worldwide health problem being the fourth cause of death due to cancer (1). CRC tumorigenesis involves molecular deregulation of genes related to proliferation, tumor growth, antiapoptosis, invasiveness, metastasis and angiogenesis (2). Nuclear factor-κB (NF-κB) is a transcriptional factor that plays an important role in biological processes, comprises a family of five proteins grouped in homo or heterodimers (3). NF-κB is normally inactive, sequestered by IκBα inhibitor, but commonly has been reported active in cancer, and plays a key role in tumorigenesis by transcriptional regulation of multiple genes (4). Several reports have found higher NF-κB/p65 protein expression in CRC tissue compared to normal tissue (5–7). However, to our knowledge, the evaluation of NF-κB/p65 and genes expression profiles, in tumor tissue compared to adjacent normal mucosa from the same CRC patient, and its association with clinicopathological parameters has not been fully reported (8).
Materials and methods
Patients
Thirty patients with sporadic CRC histopathological diagnosis who underwent to colonoscopy or surgery at Hospital Civil de Guadalajara ‘Dr. Juan I. Menchaca’, Jalisco, Mexico, were enrolled in this study after informed consent request, only non-treated patients were included. Eight patients classified as non-CRC were evaluated as comparative control group. The study was performed according to the declaration of Helsinki and was approved by the local Ethics Committee.
Tissues
Both, tumor tissue and its adjacent normal mucosa were obtained from respective areas in colonic or rectal resection specimens from the same patient according to the ‘Cancer Care Quality Measures: Diagnosis and Treatment of Colorectal Cancer’ from the ‘Agency for Health Care Research and Quality’ (9). CRC tissue samples for RNA isolation, were collected in RNAlater® Stabilization Solution (AM7020; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and in 10% neutral buffered formaldehyde (11–0705; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for immunohistochemistry analysis. Non-CRC patient's tissue samples were collected during colonoscopy, before pathological analysis according to the same procedure. CRC tissues collected for RNA isolation were transported to laboratory and processed immediately. Tissues collected in 10% of neutral buffered formaldehyde were examined microscopically to confirm the diagnosis and perform subsequent immunohistochemistry analysis. Remaining tissues were stored at −80°C in case of extra analysis.
Immunohistochemistry
Tissue samples were fixed in 10% neutral buffered formaldehyde and embedded in paraffin wax. Each tissue sample was sectioned at a thickness of 4–5 µm, placed on slides, and deparaffinized by heat for 60 min at 65°C. Slides were placed in xylene and serial alcohol solutions (100, 96, 80, and 50%). All procedure was performed using EnVision™ FLEX kit (Dako; Agilent Technologies, Inc. Santa Clara, CA, USA) as follows: Slides were washed for 5 min in wash buffer, treated in epitope retrieval solution for 20 min at 90°C, and rewashed. To block endogenous peroxidase, slides were incubated in peroxidase-blocking reagent for 15 min at room temperature, washed for 5 min in wash buffer, and incubated for 1 h at room temperature with a mouse monoclonal primary antibody raised against the N-terminus of human NF-κB/p65 (dilution 1:50) (sc-8008; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Then, the slides were washed and incubated for 30 min at room temperature with secondary antibody coupled with peroxidase and rewashed. Visualization was made with 3,3′-diaminobenzidine and counterstaining with hematoxylin. Finally, slides were dehydrated with serial alcohol solutions (50, 80, 96 and 100%) and xylene. Positive and negative controls were included for each staining procedure, using a section of CRC tissue known as strongly NF-κB/p65-positive.
NF-κB/p65 staining evaluation
Evaluation of slides was made by two pathologists blinded to patient's characteristics. The slides were scored according to the method recommended by Abdullah et al (10), as follows: Intensity of staining was classified ‘in crosses’ from 0 to 3, as 0 (−) negative, 1 (+) weak, 2 (++) moderate, and 3 (+++) strong. The extent of staining referred as the percentage of positive epithelial cells in relation to the whole tumor area, was classified from 0 to 4, as 0 (0%), 1 (≤25%), 2 (26–50%), 3 (51–75%) and 4 (>75%). The final staining score was calculated by the addition of staining intensity and the extent of staining. The scores have values between 0 and 7, and scores greater than or equal to 3 were classified as positive. For negative control, primary antibody was replaced by distilled water, as an internal control from each slide, staining of macrophages was considered.
Gene expression
A group of relevant genes in CRC was selected considering that NF-κB participates in their transcriptional regulation and its particular role in tumoral processes as described below: Proliferation, CCND1 (11) and PTGS2 (12); tumor growth, TNF (13), ALOX (14), and NOS2 (15); anti-apoptosis, BCL2 and BCL2L1 (16); invasiveness and metastasis, MMP9 (17); angiogenesis, VEGFA (18).
Tissues were collected in RNAlater® RNA Stabilization Solution, transported to laboratory and processed immediately as follows: Tissue (10–20 mg) were cut into small pieces and collected in 0.5 ml of TRIzol® Reagent (cat. no. 15596; Thermo Fisher Scientific, Inc.). Then, each sample was homogenized in Tissue Lyser LT (cat. no. 85600; Qiagen GmbH, Hilden, Germany) for 3 min/25 Hz. Next steps of RNA isolation were made according to manufacturer's instructions (Life Technologies; Thermo Fisher Scientific, Inc.). Isolated RNA samples were stored at −80°C. Reverse transcription (RT-PCR) was performed using 1 µg of total RNA treated with DNase1 amplification grade (cat. no. 18068; Life Technologies; Thermo Fisher Scientific, Inc.) and the High Capacity cDNA Reverse Transcription kit (cat. no. 4368813; Thermo Fisher Scientific, Inc.) according to manufacturer's instructions. The RT-PCR conditions were: 25°C/25, 37°C/120, 85°C/5 min, and infinite hold at 4°C. RT-Quantitative-PCR (RT-qPCR) was performed using TaqMan® Gene Expression Master Mix (cat. no. 4369016; Thermo Fisher Scientific, Inc.), and TaqMan® Gene Expression Assays with FAM-MGB fluorophore-quencher system. For each gene mRNA detection, the next assays were used (PTGS2, Hs00153133_m1; BCL2, Hs00608023_m1; BCL2L1, Hs00236329_m1; CCND1, Hs00765553_m1; MMP9, Hs00234579_m1; VEGFA, Hs00900055_m1; TNF, Hs99999043_m1; NOS2, Hs01075529_m1; ALOX5, Hs01095330_m1; GUSB, Hs00939627_m1; cat. no. 4331182, Thermo Fisher Scientific, Inc.). RT-qPCR was performed in a 7900 HT Fast Real-Time PCR System linked to SDS 2.4 software (Thermo Fisher Scientific, Inc.), PCR conditions were: 50°C/2, 95°C/10 min, 95°C/15 sec and 60°C/1 min (40 cycles). Gene expression assays were validated using β-glucoronidase (GUS), β-Actin (ACTB), and Abelson (ABL) genes as constitutive control (housekeeping genes). Gene-expression profiles were calculated by relative quantification using the 2−ΔΔCq method (19).
Statistical analysis
Data was analyzed using the SPSS 20.0 software (SPSS, Inc., Chicago, IL, USA). The NF-κB/p65 staining scores differences of the 3 groups were evaluated using Kruskal-Wallis test, then differences of staining scores and positive cells percentage pairwise comparison among groups were evaluated by Mann-Whitney U test. Genes expression differences in tumor tissues vs. adjacent normal mucosa were performed by Wilcoxon signed-ranked test. To evaluate the association of NF-κB/p65 expression and gene-expression profiles with clinicopathological parameters, Pearson or Spearman rank correlation coefficient was used. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinicopathological parameters in patients
Histopathological diagnosis of CRC was confirmed by examination of hematoxylin and eosin staining before analysis. Twenty-three CRC patient's tissues corresponded to colon cancer and 7 to rectal cancer. Twenty-two men and 8 women were collected; none of them were treated with radiotherapy or chemotherapy. The average age was 60 years. Tumor staging was established by certified pathologists according to tumor-node-metastasis (TNM) classification. Tobacco and alcohol consumption was also registered; most of the patients were occasional consumers. CEA, CA 19.9 and AFP tumor markers levels were higher than normal values. Eight patients that attend ‘Colon and rectum service’, and were diagnosed CRC negative before histopathology studies, were evaluated as control group. CRC patients were classified according to TNM clinicopathological parameters for association evaluation. General features of patients are presented in Table I.
Table I.
Clinicopathological parameters of sporadic CRC patients (n=30).
| Parameters | Value (%) |
|---|---|
| Clinicala | |
| Origin of tissue | |
| Colonoscopy | 5 (16.7) |
| Surgery | 25 (83.3) |
| Age (mean, 60 years) | |
| ≤40 | 3 (10.0) |
| 41–60 | 13 (43.3) |
| >60 | 14 (46.7) |
| Sex | |
| Male | 22 (73.3) |
| Female | 8 (26.7) |
| Tobacco consumption | |
| Yes | 7 (23.3) |
| No | 11 (36.7) |
| Occasional | 12 (40.0) |
| Alcohol consumption | |
| Yes | 11 (36.7) |
| No | 2 (6.7) |
| Occasional | 17 (56.6) |
| Pathologicala | |
| TNM stage | |
| I | 5 (16.7) |
| II | 4 (13.3) |
| III | 12 (40.0) |
| IV | 9 (30.0) |
| Histopathological differentiation | |
| Well | 0 (0.0) |
| Moderate | 27 (90.0) |
| Poorly | 3 (10.0) |
| Tumor site | |
| AC | 3 (10.0) |
| TC | 4 (13.3) |
| DC | 7 (23.3) |
| SC | 9 (30.0) |
| R | 7 (23.3) |
| Tumor depth | |
| T1 | 3 (10.0) |
| T2 | 6 (20.0) |
| T3 | 11 (36.7) |
| T4 | 10 (33.3) |
| Lymph node status | |
| N0 | 9 (30.0) |
| N1 | 8 (26.7) |
| N2 | 13 (43.3) |
| Metastasis degree | |
| M0 | 21 (70.0) |
| M1 | 9 (30.0) |
| Laboratorialb | |
| Tumor markers levels | |
| CEA, ng/ml 26.52 | (1.35.59.42) |
| CA.19.9, U/ml 149.4 | (53.6.206.9) |
| AFP, U/ml 207.1 | (27.6.276.8) |
Data are presented as the number of patients (%).
Data are presented as the mean (range). AC, ascending colon; TC, transverse colon; DC, descending colon; SC, sigmoid colon; R, rectum; CEA, carcinoembryonic antigen; CA.19.9, carbohydrate antigen; AFP, ɑ-fetoprotein antigen; CRC, colorectal cancer.
NF-ĸB/p65 immunostaining scoring
In tumor tissue group, all samples expressed cytoplasmic NF-κB/p65, the intensities of staining were majority ‘moderate’ (15/30) and ‘strong’ (11/30), the group showed a mean NF-κB/p65 extent of staining of 52.3% with a standard deviation of 18.2%. NF-κB/p65 intensities of staining in normal adjacent mucosa were mostly ‘weak’ (15/30) and ‘non-staining’ (12/30), the group showed a mean NF-κB/p65 extent of staining of 27.6% with a standard deviation of 11.8%. In non-CRC tissues, NF-κB/p65 intensity of staining was majority ‘non-staining’ (5/8), a mean NF-κB/p65 extent of staining of 18.8% with a standard deviation of 7.5% was reported for this group (Fig. 1; Table II).
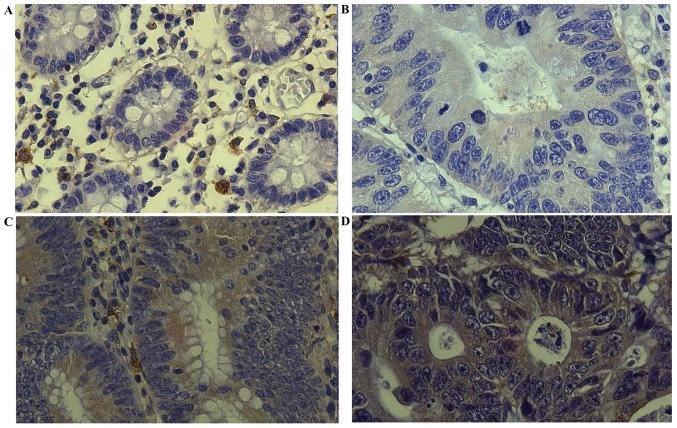
Figure 1.
Representative staining patterns. Staining intensity of NF-κB/p65 is shown. (A) Normal mucosa with negative staining (−). (B) Tumor tissue with intensity of 1 (+). (C) Tumor tissue with intensity of 2 (++). (D) Tumor tissue with intensity of 3 (+++) (Magnification, ×400). NF-κB, nuclear factor-κB.
Table II.
NF-κB/p65 immunostaining in CRC and non-CRC patient's tissues.
| Normal adjacent mucosa/Non-CRC | Tumor tissue | |||||
|---|---|---|---|---|---|---|
| Patients | Intensity of staining | Extent of staining (%) | Staining score | Intensity of staining | Extent of staining (%) | Staining score |
| CRC | ||||||
| 1 | 0 (−) | 0 (0) | 0 | 1 (+) | 2 (40) | 3a |
| 2 | 1 (+) | 2 (35) | 3a | 1 (+) | 2 (40) | 3a |
| 3 | 1 (+) | 2 (35) | 3a | 1 (+) | 2 (40) | 3a |
| 4 | 0 (−) | 0 (0) | 0 | 2 (++) | 3 (60) | 5a |
| 5 | 1 (+) | 1 (20) | 2 | 2 (++) | 2 (45) | 4a |
| 6 | 1 (+) | 2 (40) | 3a | 3 (+++) | 3 (75) | 6a |
| 7 | 0 (−) | 0 (0) | 0 | 1 (+) | 3 (60) | 4a |
| 8 | 3 (+++) | 2 (35) | 5a | 3 (+++) | 2 (50) | 5a |
| 9 | 2 (++) | 1 (20) | 3a | 3 (+++) | 4 (90) | 7a |
| 10 | 1 (+) | 1 (20) | 2 | 2 (++) | 3 (65) | 5a |
| 11 | 0 (−) | 0 (0) | 0 | 2 (++) | 3 (60) | 5a |
| 12 | 1 (+) | 1 (25) | 2 | 2 (++) | 2 (40) | 4a |
| 13 | 1 (+) | 1 (20) | 2 | 2 (++) | 2 (40) | 4a |
| 14 | 0 (−) | 0 (0) | 0 | 1 (+) | 2 (30) | 3a |
| 15 | 0 (−) | 0 (0) | 0 | 1 (+) | 2 (35) | 3a |
| 16 | 0 (−) | 0 (0) | 0 | 3 (+++) | 3 (75) | 6a |
| 17 | 0 (−) | 0 (0) | 0 | 2 (++) | 3 (65) | 5a |
| 18 | 1 (+) | 1 (25) | 2 | 2 (++) | 2 (45) | 4a |
| 19 | 0 (−) | 0 (0) | 0 | 1 (+) | 2 (30) | 3a |
| 20 | 1 (+) | 1 (20) | 2 | 2 (++) | 3 (60) | 5a |
| 21 | 0 (−) | 0 (0) | 0 | 2 (++) | 2 (40) | 4a |
| 22 | 1 (+) | 1 (20) | 2 | 3 (+++) | 2 (45) | 5a |
| 23 | 0 (−) | 0 (0) | 0 | 1 (+) | 1 (20) | 2 |
| 24 | 1 (+) | 1 (10) | 2 | 3 (+++) | 2 (50) | 5a |
| 25 | 2 (++) | 3 (60) | 5a | 2 (++) | 4 (85) | 6a |
| 26 | 1 (+) | 1 (20) | 2 | 3 (+++) | 3 (75) | 6a |
| 27 | 1 (+) | 2 (40) | 3a | 2 (++) | 3 (70) | 5a |
| 28 | 1 (+) | 2 (0) | 3a | 3 (+++) | 2 (50) | 5a |
| 29 | 1 (+) | 1 (25) | 2 | 2 (++) | 3 (70) | 5a |
| 30 | 0 (−) | 0 (0) | 0 | 1 (+) | 1 (20) | 2 |
| Non-CRC | ||||||
| 1 | 1 (+) | 1 (15) | 2 | – | – | – |
| 2 | 0 (−) | 0 (0) | 0 | – | – | – |
| 3 | 0 (−) | 0 (0) | 0 | – | – | – |
| 4 | 1 (+) | 1 (15) | 2 | – | – | – |
| 5 | 0 (−) | 0 (0) | 0 | – | – | – |
| 6 | 1 (+) | 2 (30) | 3a | – | – | – |
| 7 | 0 (−) | 0 (0) | 0 | – | – | – |
| 8 | 0 (+) | 0 (15) | 0 | – | – | – |
Samples positive to NF-κB/p65 (Staining score ≥3). Intensity of staining reported as score and num of crosses. 0 (−), no-staining; 1 (+), weak; 2 (++), moderate; and 3 (+++), strong. Extent of staining reported as score and percentage because of its variability. CRC, colorectal cancer; NF-κB, nuclear factor-κB.
An average staining score of 4.4 in tumor tissue group, 1.6 in normal adjacent mucosa and 0.8 in non-CRC tissues is reported. The analysis among 3 groups showed that they were statistically different (X2(1)=39.146, P<0.001). Pairwise comparison of staining scores among groups showed that tumor tissue was statistically higher than normal adjacent mucosa (z=−5.707, P<0.001), as well it was higher when compared to non-CRC tissue (z=−4.126, P<0.001). No difference was reported between normal adjacent mucosa and non-CRC tissue (Fig. 2).
Figure 2.
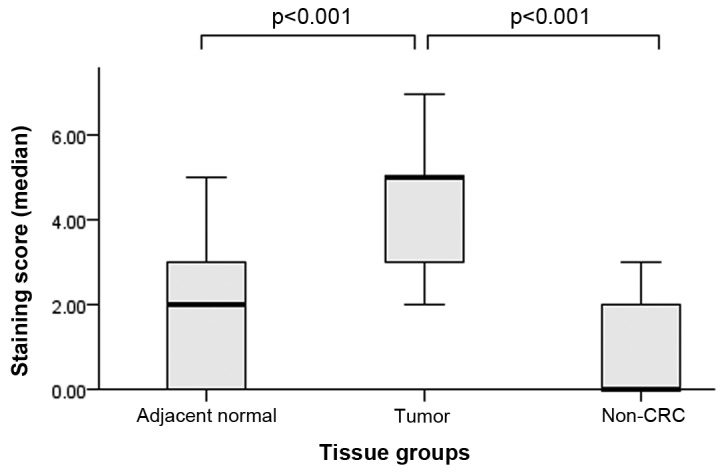
NF-ĸB/p65 staining scores analysis. Median staining score evaluation among normal adjacent mucosa, tumor tissue and non CRC tissue (X2(1)=39.146, P<0.001). Pairwise comparison among groups is also shown (tumor tissue vs. normal adjacent mucosa, z=−5.707, P<0.001), (tumor tissue vs. non-CRC tissue (z=−4.126, P<0.001). NF-κB, nuclear factor-κB.
Positive samples for NF-κB/p65 expression according to staining score (≥3), reported for each group were: 28/30 (93.3%) in tumor tissues, 8/30 (26.6%) in adjacent normal mucosa and 1/8 (12.5%) in non-CRC tissues. For statistical analysis, positive NF-κB/p65 samples were classified as ‘1’ and negative samples as ‘0’. NF-κB/p65 positive samples were statistically higher than adjacent normal tissue and non-CRC tissue groups (P<0.001). Adjacent normal tissues also showed higher NF-κB/p65 positive samples when compared to non-CRC tissue group (P<0.05) (Fig. 3).
Figure 3.
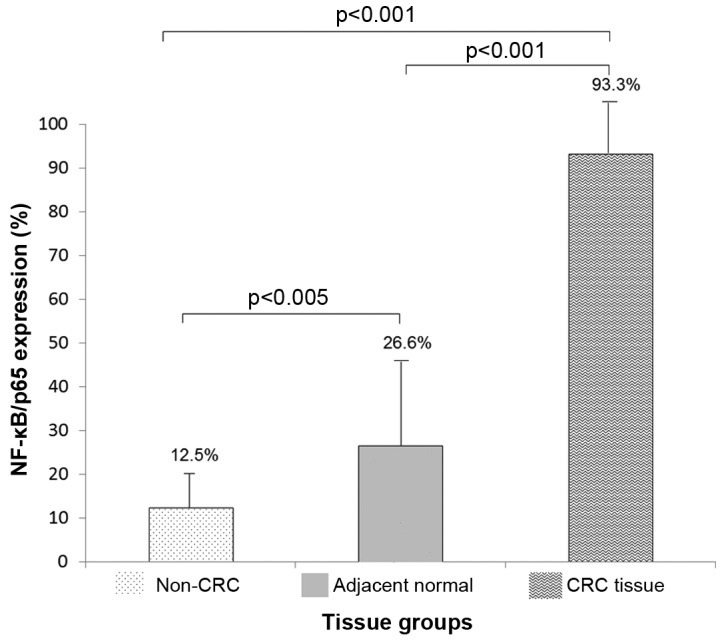
NF-ĸB/p65 positive expression. NF-ĸB/p65 (%) is significantly higher in tumor tissues vs. normal adjacent mucosa/non-CRC tissues (P<0.001).
NF-ĸB/p65 expression and clinicopathological parameters
NF-κB/p65 expression was analyzed according to clinicopathological parameters; results are described below and reported in Table III.
Table III.
NF-κB/p65 expression (%) association with clinicopathological parameters in CRC patients (n=30).
| Parameter | NF-κB/p65 expression (%) | P-value |
|---|---|---|
| CRC tissue | ||
| Tumor stage | <0.05 | |
| I | 27.00a | |
| II | 36.62b | |
| III | 63.51b | |
| IV | 70.12b | |
| Histopathology differentiation | <0.05 | |
| Well | – | |
| Moderately | 46.31b | |
| Poorly | 79.81b | |
| Tumor localization | <0.05 | |
| Ascending colon | 49.30b | |
| Transverse colon | 45.63b | |
| Descending colon | 40.74b | |
| Sigmoid colon | 52.72b | |
| Rectum | 31.33a | |
| Tumor depth | <0.05 | |
| T1 | 28.51a | |
| T2 | 34.33a | |
| T3 | 69.25b | |
| T4 | 74.84b | |
| Lymph node status | <0.05 | |
| N0 | 32.70a | |
| N1 | 49.12b | |
| N2 | 74.13b | |
| Metastasis degree | <0.001 | |
| M0 | 41.71b | |
| M1 | 73.57b | |
| Control group | ||
| Non-CRC tissue | 12.50 | |
| Normal adjacent tissue | 26.60a |
Statistically different to non-CRC tissue group (P<0.05).
Statistically different to non CRC-tissue group and normal adjacent mucosa (P<0.05). NF-κB/p65 expression (%) in tumor tissue group is compared with control groups. CRC, colorectal cancer; NF-κB, nuclear factor-κB.
NF-κB/p65 expression analyzed by tumor stages were reported as follows: I=27, II=36.6, III=63.5 and IV=70.1%. Stages II, III & IV are statistically higher than control groups (P<0.05), stage I was only statistically higher than non-CRC tissue group (P<0.05). As well, significantly increment of NF-κB/p65 expression in advanced stages compared to initial stages was reported (III+IV=66.8±3.3% vs. I+II=31.75±4.8%; P<0.05), thus NF-κB/p65 expression is positively associated to CRC progression.
In histopathological differentiation groups, NF-ĸB/p65 expression was: 79.8% in poorly differentiated group and 46.8% in moderately differentiated group, both were statistically higher than control groups (P<0.05). No patients with well differentiated tumors were collected in this study. Analysis of poorly differentiated group vs. moderately differentiated group showed a statistical increment of 33% (P<0.05), therefore NF-ĸB/p65 expression is positively associated to histopathological differentiation.
NF-κB/p65 expression analyzed by tumor localization sites is reported as follows: 49.3% in ascending colon (AC); 45.6% in transverse colon (TC); 40.7% in descending colon (DC); 52.7% in sigmoid colon (SC), and 31.3% in rectum (R). AC, TC, DC, and SC groups were statistically higher than control groups (P<0.001), while R group was only statistically higher than non-CRC tissue group. DC group vs. SC group NF-κB/p65 expression was statistically different (P<0.005). No other significative difference was reported between groups. Certainly NF-κB/p65 showed higher expression in CRC tissues, but according to previous data, there is no association with tumor localization.
NF-κB/p65 expression reported for tumor depth groups were: 28.5% in T1; 34.3% in T2; 69.2% in T3, and 74.8% in T4. T3 and T4 groups were statistically higher than control groups (P<0.05); T1 and T2 groups did not showed significative differences when compared to normal tissue group but they were statistically higher than non CRC group. T3 and T4 groups were also statistically higher than T1 and T2 groups (P<0.05). NF-κB/p65 expression is positively associated to tumor depth.
NF-κB/p65 expression analysis in lymph node status showed the next data: 32.7% in N0; 49.1% in N1, and 74.1% in N2. N1 and N2 were statistically higher than control groups (P<0.05); N0 group did not showed significative differences when compared to normal tissue group, but it was statistically higher than non CRC group. N1 and N2 groups were statistically higher than N0 (P<0.05). As well, N2 was statistically higher than N1 (P<0.05). According to these results, NF-κB/p65 expression is positively associated to lymph node status in our patients.
NF-κB/p65 expression in metastasis groups was statistically higher than control groups: M0: 41.7 and M1: 73.5%, (P<0.001). Likewise, M1 was statistically higher than M0 (P<0.001). NF-κB/p65 expression positively increments according to metastasis degree.
Housekeeping genes evaluation
Ct median of selected endogenous genes in tumor tissue compared to adjacent normal mucosa were the following: GUSB, 30.375 vs. 29.638 (P=0.489); ACTB, 29.785 vs. 28.914 (P=0.686), and ABL, 28.726 vs. 28.278 (P=0.739). There were not significant differences in any case. GUSB constitutive gene, which exhibited the minimal standard deviation, was selected as internal control for the RT-qPCR assays.
Relative quantification of gene expression
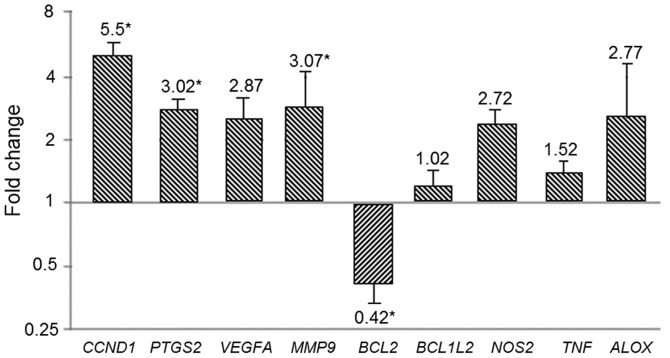
Cq data was analyzed using the 2−ΔΔCq method to obtain the relative quantification of genes. CCND1, PTGS2, and MMP9 were overexpressed in tumor tissue compared to adjacent normal mucosa (5.5; 3.02; 3.07-folds, respectively (P<0.05). While BCL2 decreased its expression (0.42-folds, P<0.05). VEGFA, BCL2L1, NOS2, TNF, and ALOX did not show significant differences (Fig. 4).
Figure 4.
Gene expression profiles. Relative expression (folds) of selected genes comparing tumor tissue relative to normal adjacent mucosa. Statistical differences are shown (*P<0.001).
Gene-expression profiles and clinicopathological parameters
Gene's expression data was analyzed according to clinicopathological parameters; results are described below and reported in Table IV.
Table IV.
Gene expression association with clinicopathological parameters in CRC patients (n=30).
| Relative quantification of gene expression (mean fold difference) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameter | CCND1 | PTGS2 | VEGFA | MMP9 | BCL2 | BCL2L1 | NOS2 | TNF | ALOX |
| Tumor stage | |||||||||
| I | 2.01±1.31 | 1.70±1.12 | 1.11±0.98 | 1.90±1.80 | 0.57±0.54 | 0.51±2.17 | 0.11±0.09 | 1.35±1.23 | 0.96±0.23 |
| II | 2.90±1.83 | 1.40±1.23 | 2.31±1.33 | 1.52±0.89 | 0.54±0.33 | 0.46±0.42 | 0.39±0.51 | 1.28±1.09 | 1.97±1.61 |
| III | 7.10±2.52 | 3.22±3.12 | 3.12±1.84 | 3.10±1.22 | 0.38±0.27 | 1.01±0.81 | 4.10±1.42 | 1.57±1.21 | 3.17±1.42 |
| IV | 10.00±2.21 | 5.81±5.03 | 5.01±1.61 | 5.83±4.21 | 0.21±0.19 | 1.21±1.13 | 6.29±1.93 | 1.91±1.73 | 4.99±1.71 |
| rs, (P-value) | 0.97 (0.02)a | 0.90 (0.94) | 0.98 (0.01)a | 0.88 (0.07) | −0.96 (0.03)a | 0.92 (0.07) | 0.95 (0.04)a | 0.89 (0.10) | 0.95 (0.01)a |
| Histological differentiation | |||||||||
| Well | – | – | – | – | – | – | – | – | – |
| Moderately | 4.71±3.73 | 2.16±2.01 | 2.66±1.91 | 1.96±1.21 | 0.55±0.72 | 0.59±0.36 | 2.46±1.12 | 1.22±1.12 | 2.13±1.22 |
| Poorly | 6.32±1.21 | 3.92±1.10 | 3.13±0.96 | 4.21±1.53 | 0.29±0.11 | 0.99±0.51 | 3.3±2.71 | 1.82±0.96 | 3.41±1.73 |
| rs (P-value) | 0.85 (0.05)a | 0.90 (0.11) | 0.78 (0.01)a | 0.79 (0.03) | −0.91 (0.01)a | −0.83 (0.12) | −0.54 (0.23) | −0.69 (0.27) | −0.89 (0.34) |
| Tumor localization | |||||||||
| Ascending colon | 6.31±2.34 | 2.71±1.71 | 5.54±2.81 | 1.79±1.11 | 0.45±0.21 | 0.80±0.31 | 2.39±1.23 | 2.47±1.31 | 1.9±0.35 |
| Transverse colon | 3.61±1.82 | 3.55±1.23 | 1.62±1.32 | 3.42±2.03 | 0.38±0.12 | 0.65±0.34 | 3.05±1.72 | 0.99±0.34 | 3.62±1.36 |
| Descending colon | 6.65±4.26 | 1.63±1.12 | 3.35±2.90 | 4.66±4.23 | 0.49±0.26 | 0.91±0.61 | 1.17±0.96 | 1.32±1.02 | 2.48±1.81 |
| Sigmoid colon | 6.98±3.81 | 5.32±4.20 | 1.83±1.12 | 4.48±3.11 | 0.49±0.24 | 0.96±0.67 | 4.41±1.23 | 1.22±0.67 | 1.05±0.43 |
| Rectum | 3.95±1.52 | 2.05±1.71 | 2.06±1.21 | 1.05±0.76 | 0.39±0.29 | 0.63±0.31 | 3.38±1.71 | 1.67±0.97 | 4.80±1.51 |
| rs, (P-value) | −0.40 (0.50) | −0.10 (0.87) | −0.21 (0.47) | 0.11 (0.37) | −0.10 (0.61) | −0.21 (0.87) | −0.28 (0.58) | −0.19 (0.59) | 0.23 (0.71) |
| Tumor depth | |||||||||
| T1 | 1.98±1.42 | 1.56±1.11 | 1.45±1.21 | 2.21±1.81 | 0.54±0.22 | 0.86±0.64 | 1.21±0.78 | 1.29±0.37 | 1.21±0.56 |
| T2 | 3.92±1.80 | 2.31±1.33 | 2.88±1.80 | 1.94±1.22 | 0.49±0.18 | 0.62±0.47 | 2.24±1.35 | 1.87±0.46 | 1.69±0.79 |
| T3 | 6.38±2.01 | 2.99±1.94 | 3.34±1.23 | 3.62±1.73 | 0.34±0.28 | 0.73±0.38 | 3.62±1.92 | 1.51±0.28 | 3.87±1.23 |
| T4 | 9.72±4.33 | 5.26±1.72 | 3.85±1.71 | 4.55±2.01 | 0.31±0.171 | 0.95±0.71 | 4.45±2.41 | 1.41±0.41 | 4.31±1.54 |
| rs, (P-value) | 0.94 (0.01)a | 0.78 (0.03)a | 0.64 (0.07) | 0.81 (0.06) | −0.89 (0.01)a | 0.54 (0.08) | 0.91 (0.01)a | 0.24 (0.35) | 0.82 (0.01)a |
| Lymph node status | |||||||||
| N0 | 1.91±1.22 | 1.22±0.95 | 1.23±0.67 | 1.49±1.35 | 0.74±2.73 | 0.83±0.28 | 1.29±1.32 | 1.09±0.97 | 1.96±1.13 |
| N1 | 4.62±1.85 | 3.21±1.41 | 3.01±1.44 | 3.73±1.82 | 0.32±1.73 | 0.76±0.64 | 2.37±1.93 | 1.11±0.92 | 2.59±1.51 |
| N2 | 9.97±4.21 | 4.66±2.74 | 4.43±2.91 | 4.02±2.11 | 0.20±2.12 | 0.78±0.10 | 4.98±2.61 | 2.36±1.21 | 3.76±1.44 |
| rs, (P-value) | 0.84 (0.03)a | 0.79 (0.04)a | 0.90 (0.03)a | 0.71 (0.08) | −0.92 (0.01)a | 0.41 (0.26) | 0.78 (0.02)a | 0.62 (0.26) | 0.61 (0.14) |
| Metastasis degree | |||||||||
| M0 | 2.94±1.13 | 2.33±1.33 | 1.24±1.13 | 1.70±0.87 | 0.56±0.48 | 0.78±0.19 | 1.38±1.22 | 1.17±1.02 | 1.36±1.23 |
| M1 | 8.06±3.71 | 3.73±1.72 | 4.52±1.71 | 4.46±2.82 | 0.28±0.15 | 0.80±0.45 | 4.38±2.93 | 1.87±1.13 | 4.18±3.11 |
| rs, (P-value) | 0.91 (0.04)a | 0.51 (0.12) | 0.79 (0.03)a | 0.31 (0.31) | −0.95 (0.03)a | 0.11 (0.58) | 0.81 (0.03)a | 0.21 (0.48) | 0.89 (0.06) |
Fold change normalized with GUSB endogenous gene.
Statistically significant correlation (P<0.05). CRC, colorectal cancer; CCND1, cyclin D1; PTGS2, prostaglandin-endoperoxide synthase 2; VEGFA, vascular endothelial growth factor A; MMP9, matrix metallopeptidase 9; BCL2, BCL2, apoptosis regulator; BCL2L1, BCL2 like 1; NOS2, nitric oxide synthase 2; TNF, tumor necrosis factor; ALOX, arachidonate lipoxygenase; T, tumor; N, node; M, metastasis.
Positive association of CCND1, VEGFA, NOS2, and ALOX as well as a negative association of BCL2 with tumor stage progression was reported (P<0.05). Significant overexpression in the advanced stages group compared to initial stages in CCND1 (III+IV=8.55±1.45 vs. I+II=2.45±0.45; P<0.05), VEGFA (I+II=1.2±0.3 vs. III+IV=4.55±1.3; P<0.05), NOS2 (I+II=0.25±0.14 vs. III+IV=5.2±2.1; P<0.05) and ALOX (I+II=1.46±0.5 vs. III+IV=4.08±0.19; P<0.05) corroborate the positive association. No significant difference was found in gene expression of PTGS2, MMP9, BCL2L1, and TNF during tumor progression.
Gene expression association with histopathological differentiation groups, was statistically positive in the case of CCND1, MMP9 and VEGFA, while BCL2 expression was negatively associated (P<0.05). No association of histopathological differentiation with PTGS2, MMP9, BCL2L1, NOS2, ALOX and TNF gene expression was reported.
No association between tumor localization site and expression of any evaluated gene was reported.
In the case of tumor depth, positive association was observed with CCND1, PTGS2, NOS2 and ALOX expression, negative association with BCL2 expression was also reported (P<0.05). VEGFA, MMP9, BCL2L1, and TNF expression did not showed any association.
Lymph node status and gene expression was positively associated in the case of CCND1, PTGS2, VEGFA, and NOS2 (P<0.05). Negative association with BCL2 was also reported (P<0.05). No association with MMP9, BCL2L1, TNF, and ALOX was found.
Metastasis degree and gene expression was positively associated in the case of CCND1, VEGFA and NOS2, and negative association was reported with BCL2 (P<0.05). No association was observed in PTGS2, MMP9, BCL2L1, TNF, and ALOX.
Discussion
In the present study, NF-κB/p65 and genes expression association with clinicopathological parameters of CRC patients was investigated. We reported higher NF-κB/p65 cytoplasmic expression in tumor tissue compared to normal adjacent mucosa; our findings are consistent with previous studies (20). NF-κB/p65 expression in CRC showed discrepancy in stained protein localization, nevertheless nuclear staining has been mainly reported (20,21). NF-κB/p65 detected in previous reports, similar as our study, indicates released IκBα, but we were not able to confirm the transcriptional activity. We hypothesize that NF-κB/IκBα binding alterations could masked the nuclear localization signal in p65 as other studies suggest (22).
NF-κB/p65 expression by CRC stages has been commonly evaluated. Higher levels in advanced stages compared to initial stages, evaluated by immunohistochemistry are reported in this study. According to our results, NF-κB/p65 increment was positively associated with tumor stage progression. We suggest that NF-κB/p65 cytoplasmic expression may play a key role in CRC progression probably by molecular changes in its downstream pathway. In addition, positive association between NF-κB/p65 expression with histopathology differentiation, tumor depth, lymph node status and metastasis degree was reported, but no association with tumor localization was observed in any case. NF-κB/p65 association with clinicopathological parameters has not been entirely described. A meta-analysis in solid tumors, reported a positive association with lymph node status and metastasis degree (23), but the conclusions are still in contradiction. Our results suggest that NF-κB/p65 is involved in CRC establishment and progression by promoting clinicopathological parameters development.
Gene expression analyzed in tumor tissue vs. normal adjacent mucosa, showed overexpression in CCND1, PTGS2 and MMP9, decreased expression of BCL2, while no significant differences in expression of BCL2L1, VEGFA, TNF, ALOX and NOS2 was reported. Overexpression of CCND1 in tumor tissue was observed in this study as in others (24), considering that CCND1 promotes the transition of G1- to S-phase of cell cycle it may play a key role in tumor cells proliferation in the evaluated tissues. PTGS2 was similarly reported overexpressed in tumor tissue as others studies did using different methodologies (25–27), but no alteration of PTGS2 expression has been also observed, these contradictory reports suggest that PTGS2 activity in CRC is still unclear. According to our results, we hypothesize that proliferation and survival processes due to PTGS2 overexpression plays an important role in CRC. MMP9 overexpression observed in this study agreed with previous reports that relate its expression with invasiveness and metastasis (28–30). In this study we just report decreased expression of BCL2 in CRC, higher expression in tumor tissue than in normal mucosa had been reported (31). BCL2 promotes tumorigenesis by inhibition of apoptosis, according to our results; we suggest that once CRC is established it began to decrease its activity and consecutively its gene expression. Previous studies report higher expression of BCL2L1, VEGFA, TNF, ALOX and NOS2 in tumor tissue than in normal mucosa, in this study we did not observed difference in expression of these genes. BCL2L1 is associated with apoptosis and opposite of our results its overexpression has been previously reported (32). VEGFA overexpression has been observed and associated with advanced tumor stages and poor clinical outcome (33), but one study reported no difference at protein level similar as our results (34). TNF is involved in tumor promotion and progression; previous reports have found expression of TNF in 94% of tumors (35) in disagreement to our results. ALOX activity is related with tumor growth and invasiveness of solid tumors, opposite to our results ALOX overexpression in CRC has been reported (36). Similarly, NOS2 overexpression in CRC has been observed and related to angiogenesis (37), while according to our results, decreased expression of NOS2 in CRC tumor was previously reported (38).
Even though we did not found significant differences in expression of all selected genes between tumor and normal adjacent mucosa, we analyze the association of these genes with clinicopathological parameters due to their importance in tumorigenesis. The results of these analyses are discussed below: The strong positive association of CCND1 with all TNM parameters, excluding tumor localization, confirmed its role during CRC tumorigenesis; as well its association with tumor progression suggests its potential as tumor marker in early stages. The activity of MMP9 during invasiveness processes in CRC was confirmed in this study due to its positive association with tumor stages, but we can found any association with metastasis as other groups did (28–30). Antiapoptosis activity of BCL2 has been associated to different cancer types, as well as CRC, in this study BCL2 was negatively associated with all TNM parameters, supporting the highest expression level in normal adjacent mucosa of CRC patients we found; this data is completely disagreeing with previous reports (31), we suggest that others antiapoptotic factors play more important role than BCL2 does in our CRC patients, even though we considered that is necessary to evaluate BCL2 protein expression to reinforce our results. The role of VEGFA in solid tumors is confirmed with the positive association in lymph node status and metastasis degree CRC groups, according to others reports the poor clinical outcome is related to VEGFA expression (33), we confirmed this data in our CRC advanced group when VEGFA is overexpressed, so we also suggest its role as a possible tumor marker in advanced disease. Surprisingly, TNF and BCL1L2 genes that has been several times reported associated with different types of solid tumors (32,35), did not showed association with any clinicopathological parameter, differences in their expression between tumor vs. normal adjacent mucosa was reported neither. Further studies should analyze the TNF and BCL1L2 proteins vs. NF-κB/p65 activity in human CRC samples. ALOX overexpression influences in CRC development, and it is observed according to the positive association with tumor stages and tumor depth reported in this study. Tumor growth mediated by inflammation, supposed to be one of the most important processes related to ALOX overexpression (36), this data is supported by our results. NOS2 showed a positive association with TNM parameters, but it was not significantly associated to histopathological differentiation and tumor localization. NOS2 expression was not statistical different in tumor vs. normal adjacent mucosa, nevertheless we suggest that NOS2 activity is related to tumorigenesis, absolute quantification analyses could help in future studies to confirm the NOS2 role in CRC establishment and development (38).
According to results discussed above, not all selected genes regulated by NF-κB/p65 increment their expression as the transcription factor did; probably these genes are partially regulated by others transcription factors in CRC. These results suggest that NF-κB activity is necessary but not sufficient in CRC establishment. To our knowledge the present study reports for the first time, the selected genes expression and NF-κB/p65 association with clinicopathological parameters in sporadic CRC. Due to NF-κB/p65 nucleus translocation is required for its transcriptional activity; we suggest that others quantitative techniques are necessary to statistically associate NF-κB activity with gene expression profiles in CRC; conditions of our experiments did not permit us to work more deeply on it because some of our patients are already under treatment (non-inclusion criterion) or because we are not able to take more tissue samples (the patient did not continue attending hospital or because they passed away). Meta-analysis association studies are indispensable to completely understand the behavior of these molecules in CRC.
Higher NF-κB/p65 expression in CRC tissue compared to normal adjacent mucosa from the same patient is reported, and this increment was positively associated with clinicopathological parameters, except for tumor localization. The monitoring and regulation of this transcription factor may be therapeutically useful in CRC patients.
NF-κB regulated genes showed an irregular expression pattern when compared CRC tissue vs. normal adjacent mucosa; CCND1, PTGS2 and MMP9 were overexpressed. VEGFA, BCL2L1, NOS2, TNF and ALOX did not changed, while BCL2 decreases its expression level. Results of genes expression association with clinicopathological parameters are summarized below; CCND1 was positively associated with all TNM parameters. PTGS2 was associated with tumor depth and lymph node status. VEGFA showed a positively association with lymph node status and metastasis degree. MMP9 was positively associated with tumor stages. NOS2 showed a positive association with tumor stages, tumor depth, lymph node status and metastasis degree. ALOX was positively associated with tumor stages and tumor depth. BCL1L2 and TNF did not showed association with any clinicopathological parameter, while BCL2 was negatively associated with all TNM parameters. Tumor localization site was not related with any of the evaluated genes. According to the association of these genes with different clinicopathological parameters, they may be considered for the selection of proper diagnosis, treatment and follow-up for CRC patients.
Acknowledgements
This study was partially supported by COECYTJAL 5-2010-1-1083 grant. The authors would like to thank Dr. Bustos-Rodríguez F and Dr. Valenzuela-Pérez JA for their technical support in this study. The authors would like to thank the Ph.D. Program of Molecular Biology in Medicine, University of Guadalajara.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Peddareddigari V, Wang D, Dubois RN. The tumor microenvironment in colorectal carcinogenesis. Cancer Microenviron. 2010;3:149–166. doi: 10.1007/s12307-010-0038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto K, Maeda S, Hikiba Y, Nakagawa H, Hayakawa Y, Shibata W, Yanai A, Ogura K, Omata M. Constitutive NF-kappaB activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth. Clin Cancer Res. 2009;15:2248–2258. doi: 10.1158/1078-0432.CCR-08-1383. [DOI] [PubMed] [Google Scholar]
- 5.Puvvada SD, Funkhouser WK, Greene K, Deal A, Chu H, Baldwin AS, Tepper JE, O'Neil BH. NF-kB and Bcl-3 activation are prognostic in metastatic colorectal cancer. Oncology. 2010;78:181–188. doi: 10.1159/000313697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewander A, Gao J, Adell G, Zhang H, Sun XF. Expression of NF-κB p65 phosphorylated at serine-536 in rectal cancer with or without preoperative radiotherapy. Radiol Oncol. 2011;45:279–284. doi: 10.2478/v10019-011-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdullah M, Rani AA, Sudoyo AW, Makmun D, Handjari DR, Hernowo BS. Expression of NF-kB and COX2 in colorectal cancer among native Indonesians: The role of inflammation in colorectal carcinogenesis. Acta Med Indones. 2013;45:187–192. [PubMed] [Google Scholar]
- 8.Yu LL, Yu HG, Yu JP, Luo HS, Xu XM, Li JH. Nuclear factor-kappaB p65 (RelA) transcription factor is constitutively activated in human colorectal carcinoma tissue. World J Gastroenterol. 2004;10:3255–3260. doi: 10.3748/wjg.v10.i22.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patwardhan MB, Samsa GP, McCrory DC, Fisher DA, Mantyh CR, Morse MA, Prosnitz RG, Cline KE, Gray RN. Cancer care quality measures: Diagnosis and treatment of colorectal cancer. Evid Rep Technol Assess (Full Rep) 2006:1–116. [PMC free article] [PubMed] [Google Scholar]
- 10.Abdullah M, Sudoyo AW, Pranowo BS, Rini D, Sutrisna B, Rani AA. Expression of NF-kappaB and COX-2 in young versus older patients with sporadic colorectal cancer. Acta Med Indones. 2009;41:70–74. [PubMed] [Google Scholar]
- 11.Balcerczak E, Pasz-Walczak G, Kumor P, Panczyk M, Kordek R, Wierzbicki R, Mirowski M. Cyclin D1 protein and CCND1 gene expression in colorectal cancer. Eur J Surg Oncol. 2005;31:721–726. doi: 10.1016/j.ejso.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Roelofs HM, Morsche Te RH, van Heumen BW, Nagengast FM, Peters WH. Over-expression of COX-2 mRNA in colorectal cancer. BMC Gastroenterol. 2014;14:1. doi: 10.1186/1471-230X-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanilov S, Drodeva Z, Stanilova S. Higher TNF-alpha production detected in colorectal cancer patients monocytes. Med Biotechnol. 2011;26:107–110. [Google Scholar]
- 14.Rao CV, Janakiram NB, Mohammed A. Lipoxygenase and cyclooxygenase pathways and colorectal cancer prevention. Curr Colorectal Cancer Rep. 2012;8:316–324. doi: 10.1007/s11888-012-0146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yagihashi N, Kasajima H, Sugai S, Matsumoto K, Ebina Y, Morita T, Murakami T, Yagihashi S. Increased in situ expresión of nitric oxide synthase in human colorectal cancer. Virchows Arch. 2000;436:109–114. doi: 10.1007/PL00008208. [DOI] [PubMed] [Google Scholar]
- 16.Zeestraten EC, Benard A, Reimers MS, Schouten PC, Liefers GJ, van de Velde CJ, Kuppen PJ. The prognostic value of the apoptosis pathway in colorectal cancer: A review of the literature on biomarkers identified by immunohistochemistry. Biomark Cancer. 2013;5:13–29. doi: 10.4137/BIC.S11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koskensalo S, Hagström J, Linder N, Lundin M, Sorsa T, Louhimo J, Haglund C. Lack of MMP-9 expression is a marker for poor prognosis in Dukes' B colorectal cancer. BMC Clin Pathol. 2012;12:24. doi: 10.1186/1472-6890-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guba M, Seeliger H, Kleespies A, Jauch KW, Bruns C. Vascular endothelial growth factor in colorectal cancer. Int J Colorectal Dis. 2004;19:510–517. doi: 10.1007/s00384-003-0576-y. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Kojima M, Morisaki T, Sasaki N, Nakano K, Mibu R, Tanaka M, Katano M. Increased nuclear factor-kB activation in human colorectal carcinoma and its correlation with tumor progression. Anticancer Res. 2004;24:675–681. [PubMed] [Google Scholar]
- 21.Yu HG, Yu LL, Yang Y, Luo HS, Yu JP, Meier JJ, Schrader H, Bastian A, Schmidt WE, Schmitz F. Increased expression of RelA/nuclear factor-kappa B protein correlates with colorectal tumorigenesis. Oncology. 2003;65:37–45. doi: 10.1159/000071203. [DOI] [PubMed] [Google Scholar]
- 22.Sun SC, Ganchi PA, Béraud C, Ballar DW, Greene WC. Autoregulation of the NF-kappa B transactivator RelA (p65) by multiple cytoplasmic inhibitors containing ankyrin motifs. Proc Natl Acad Sci USA. 1994;91:1346–1350. doi: 10.1073/pnas.91.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu D, Wu P, Zhao L, Huang L, Zhang Z, Zhao S, Huang J. NF-kB expression and outcomes in solid tumors: A systematic review and meta-analysis. Medicine (Baltimore) 2015;94:e1687. doi: 10.1097/MD.0000000000001687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutter T, Doi S, Carnevale KA, Arber N, Weinstein IB. Expression of cyclins D1 and E in human colon adenocarcinomas. J Med. 1997;28:285–309. [PubMed] [Google Scholar]
- 25.Antonacopoulou AG, Tsamandas AC, Petsas T, Liava A, Scopa CD, Papavassiliou AG, Kalofonos HP. EGFR, HER-2 and COX-2 levels in colorectal cancer. Histopathology. 2008;53:698–706. doi: 10.1111/j.1365-2559.2008.03165.x. [DOI] [PubMed] [Google Scholar]
- 26.Cressey R, Pimpa S, Tontrong W, Watananupong O, Leartprasertsuke N. Expression of cyclooxygenase-2 in colorectal adenocarcinoma is associated with p53 accumulation and hdm2 overexpression. Cancer Lett. 2006;233:232–239. doi: 10.1016/j.canlet.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 27.Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994;107:1183–1188. doi: 10.1016/0016-5085(94)90246-1. [DOI] [PubMed] [Google Scholar]
- 28.Chu D, Zhao Z, Zhou Y, Li Y, Li J, Zheng J, Zhao Q, Wang W. Matrix metalloproteinase-9 is associated with relapse and prognosis of patients with colorectal cancer. Ann Surg Oncol. 2012;19:318–325. doi: 10.1245/s10434-011-1686-3. [DOI] [PubMed] [Google Scholar]
- 29.Langers AM, Verspaget HW, Hawinkels LJ, Kubben FJ, van Duijn W, van der Reijden JJ, Hardwick JC, Hommes DW, Sier CF. MMP-2 and MMP-9 in normal mucosa are independently associated with outcome of colorectal cancer patients. Br J Cancer. 2012;106:1495–1498. doi: 10.1038/bjc.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng ZS, Huang Y, Cohen AM, Guillem JG. Prediction of colorectal cancer relapse and survival via tissue RNA levels of matrix metalloproteinase-9. J Clin Oncol. 1996;14:3133–3140. doi: 10.1200/JCO.1996.14.12.3133. [DOI] [PubMed] [Google Scholar]
- 31.Sun N, Meng Q, Tian A. Expressions of the anti-apoptotic genes Bag-1 and Bcl-2 in colon cancer and their relationship. Am J Surg. 2010;200:341–345. doi: 10.1016/j.amjsurg.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 32.Krajewska M, Moss SF, Krajewski S, Song K, Holt PR, Reed JC. Elevated expression of Bcl-X and reduced Bak in primary colorectal adenocarcinomas. Cancer Res. 1996;56:2422–2427. [PubMed] [Google Scholar]
- 33.Altomare DF, Rotelli MT, Pentimone A, Rossiello MR, Martinelli E, Guglielmi A, De Fazio M, Marino F, Memeo V, Colucci M, Semeraro N. Tissue factor and vascular endothelial growth factor expression in colorectal cancer: Relation with cancer recurrence. Colorectal Dis. 2007;9:133–138. doi: 10.1111/j.1463-1318.2006.01158.x. [DOI] [PubMed] [Google Scholar]
- 34.Cao D, Hou M, Guan YS, Jiang M, Yang Y, Gou HF. Expression of HIF-1alpha and VEGF in colorectal cancer: Association with clinical outcomes and prognostic implications. BMC Cancer. 2009;9:432. doi: 10.1186/1471-2407-9-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimm M, Lazariotou M, Kircher S, Höfelmayr A, Germer CT, von Rahden BH, Waaga-Gasser AM, Gasser M. Tumor necrosis factor-α is associated with positive lymph node status in patients with recurrence of colorectal cancer indications for anti-TNF-α agents in cancer treatment. Anal Cell Pathol (Amst) 2010;33:151–163. doi: 10.1155/2010/891869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soumaoro LT, Lida S, Uetake H, Ishiguro M, Takagi Y, Higuchi T, Yasuno M, Enomoto M, Sugihara K. Expression of 5-lipoxygenase in human colorectal cancer. World J Gastroenterol. 2006;12:6355–6360. doi: 10.3748/wjg.v12.i39.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moochhala S, Chhatwal VJ, Chan ST, Ngoi SS, Chia YW, Rauff A. Nitric oxide synthase activity and expression in human colorectal cancer. Carcinogenesis. 1996;17:1171–1174. doi: 10.1093/carcin/17.5.1171. [DOI] [PubMed] [Google Scholar]
- 38.Kojima M, Morisaki T, Tsukahara Y, Uchiyama A, Matsunari Y, Mibu R, Tanaka M. Nitric oxide synthase expression and nitric oxide production in human colon carcinoma tissue. J Surg Oncol. 1999;70:222–229. doi: 10.1002/(SICI)1096-9098(199904)70:4<222::AID-JSO5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]