Abstract
Overdue treatment and prognostic evaluation lead to low survival rates in patients with lung adenocarcinoma (LUAD). To date, effective biomarkers for prognosis are still required. The aim of the present study was to screen differentially expressed genes (DEGs) as biomarkers for prognostic evaluation of LUAD. DEGs in tumor and normal samples were identified and analyzed for Kyoto Encyclopedia of Genes and Genomes/Gene Ontology functional enrichments. The common genes that are up and downregulated were selected for prognostic analysis using RNAseq data in The Cancer Genome Atlas. Differential expression analysis was performed with 164 samples in GSE10072 and GSE7670 datasets. A total of 484 DEGs that were present in GSE10072 and GSE7670 datasets were screened, including secreted phosphoprotein 1 (SPP1) that was highly expressed and DEGs ficolin 3, advanced glycosylation end-product specific receptor (AGER), transmembrane protein 100 that were lowly expressed in tumor tissues. These four key genes were subsequently verified using an independent dataset, GSE19804. The gene expression model was consistent with GSE10072 and GSE7670 datasets. The dysregulation of highly expressed SPP1 and lowly expressed AGER significantly reduced the median survival time of patients with LUAD. These findings suggest that SPP1 and AGER are risk factors for LUAD, and these two genes may be utilized in the prognostic evaluation of patients with LUAD. Additionally, the key genes and functional enrichments may provide a reference for investigating the molecular expression mechanisms underlying LUAD.
Keywords: lung adenocarcinoma, differentially expressed genes, functional analysis, prognostic biomarker
Introduction
Lung adenocarcinoma (LUAD) is one of the three major histopathological subtypes along with squamous cell carcinoma (SqCLC) and large cell carcinoma (1). Currently, LUAD has become the most common lung cancer with increasing morbidity. This uptrend of incidence may be due to increasing smoking rate and air pollution (2). Furthermore, LUAD is diagnosed at late stages (stage III and IV), when the cancer has spread to nearby tissues and metastasis has occurred (3). As well as overdue diagnosis that leads to delayed treatment, the failure of prognostic evaluation also contributes to the low survival rate of LUAD (4,5). Therefore, there remains to be an urgent requirement for effective biomarkers for prognosis, where the outcome of patients with LUAD can be evaluated in time to provide adjuvant therapy.
Due to cancer largely being a genetic disease, the latest approaches to identify genes as biomarkers are based on microarray technology. Oncogenomic analysis has the ability to generate a wealth of data, which identifies the complex gene expression patterns in cancer (6,7). These gene expression patterns, including point mutations, structural variants at the DNA level and changes at the epigenetic level, are frequently studied to identify the association with the occurrence, development or the survival time of patients with cancer (8,9). A number of studies have identified specific genes that function as diagnostic or prognostic biomarkers for several types of cancer, including in LUAD (10–12). Additionally, gene expression profiling by microarray technology has been used to investigate the molecular mechanisms underlying a number of other diseases as well (13).
The reliability of biomarker identification relies on not only the analysis methods but also the database (14). The Cancer Genome Atlas (TCGA) provides comprehensive data on the molecular basis of various types of cancer (15). Significant gene patterns observed in one or several independent databases may also act in a similar genetic subtype of cancer (16). This may also be the case for different types of cancer. However, researchers must have the ability to evaluate and determine if this common pattern affects clinical phenotypes, including survival by using survival or clustering analysis with another independent dataset.
In the present study, the common differentially expressed genes (DEGs) were screened, including secreted phosphoprotein 1 (SPP1) with high expression and ficolin 3 (FCN3), advanced glycosylation end-product specific receptor (AGER; RAGE), transmembrane protein 100 (TMEM100) demonstrating low expression in tumor tissues. These four key genes (SPP1, FCN3, AGER and TMEM100) were verified using an additional dataset. The gene expression model was consistent with the two previous datasets that were analyzed. The dysregulation of highly expressed SPP1 and lowly expressed AGER significantly reduced the median survival time of patients with LUAD. These two genes were verified and indicated stable significance as risk factors for LUAD. This discovery enables these prognostic marker genes to be utilized in the evaluation of patients with LUAD. Additionally, the key genes and functional enrichments may provide reference to investigate the molecular expression mechanism underlying LUAD.
Materials and methods
Data source
Gene expression profiling of three individual datasets for microarray analysis [GSE19804 (17), GSE10072 (18) and GSE7670 (19)] were downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). There were 120 samples in GSE19804, including 60 LUAD tissues and 60 normal tissues. GSE10072 consisted of 107 samples (49 tumor samples and 58 normal tissues). GSE7670 contained 54 samples in total (27 tumor tissues and 27 normal lung tissues). Datasets GSE10072 and GSE7670 were employed for the identification of DEGs on the Affymetrix Gene Chip™ Human Genome U133A arrayplatform (Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA). GSE19804 was used to verify the significance of selected DEGs. This verification was based on Affymetrix Gene Chip Human Genome U133 Plus 2.0 (Affymetrix; Thermo Fisher Scientific, Inc.). Finally, the prognostic analysis of the selected genes was performed with the gene expression data and follow-ups of patients with LUAD by using RNAseq (Illumina HiSeq 2500; Illumina, Inc., San Diego CA, USA) in TCGA (https://cancergenome.nih.gov/).
Quality control
The quality assessment of the Affymetrix Gene Chip datasets was performed using the affyPLM package in R software (version 3.6; https://www.r-project.org) (20). In the present analysis, the linear modeling procedures were applied at a probe-level. The Relative Log Expression (RLE) and Normalized Unscaled Standard Errors (NUSE) were generated to test the consistency of the data trends. RLE and NUSE were displayed in box plots. Then AffyRNAdeg package in R software (version 3.6; https://www.r-project.org) was used to test the degradation of RNA. Finally, datasets with consistent trends as well as improved RNA quality were selected for the following analysis.
Data preprocessing
To ensure the integrity and comparability of data sets for analysis, the gcrma package (version 3.6) (https://www.bioconductor.org/packages/release/bioc/html/gcrma.html) (21) was used to standardize and adjust for background intensities in the Aymetrix array data, eliminating system errors between gene chips. The correlation between gene expression levels of the samples is an important indicator to evaluate the reliability of experiments and sample selection. Therefore, the overall correlation of the selected samples was analyzed. A correlation chart of all the samples was obtained using Pearson's correlation coefficient (22).
Screening of DEGs
The DEGs of the samples between tumor and normal samples were identified using the limma package in R (http://bioconductor.org/packages/release/bioc/html/limma.html) (23). The threshold used is as follows: (fold change, logFC) log2>1 and P<0.05. Then, the differential expression of these genes was visualized in a Venn diagram, ‘Volcano plots’ or heatmaps were generated using ggplot2 in R (24).
Functional analysis of DEGs
To further understand the potential pathways that these DEGs may participate in or regulate, functional analysis was performed in The Database for Annotation, Visualization and Integrated Discovery (DAVID) (25). All DEGs were firstly annotated using the functional annotation tool and then analyzed for enriched pathways in Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms. For every functional item, DAVID calculated the P-value of the enrichment and false discovery rate using Benjamini correction. The threshold for significantly enriched pathways was P<0.05.
Verification analysis
The common up- and down-regulated genes were selected from the top DEGs in two individual datasets (GSE7670 and GSE10072). To evaluate the expression of these candidate DEGs for the prognosis of patients with LUAD, an independent dataset GSE19804 containing 120 samples (60 tumor tissues and 60 normal tissues) was analyzed using Affymetrix Human Genome U133 Plus 2.0.
Prognostic and statistical analysis
Survival analysis was performed by using SPSS version 24 (26,27) to investigate the prognostic impact of selected DEGs on patients with LUAD. The data are presented as the mean ± standard deviation. Significance analysis was performed using Kaplan-Meier analysis with the log-rank test. Log-rank (Mantel-Cox) test was used with a significance level of P<0.05. All statistical analysis was performed with GraphPad Prism (GraphPad Software, Inc., La Jolla, CA, USA). The P-value of the enrichment and false discovery rate was obtained using Benjamini correlation. P<0.05 was considered to indicate a statistically significant difference.
Results
Data source and quality control
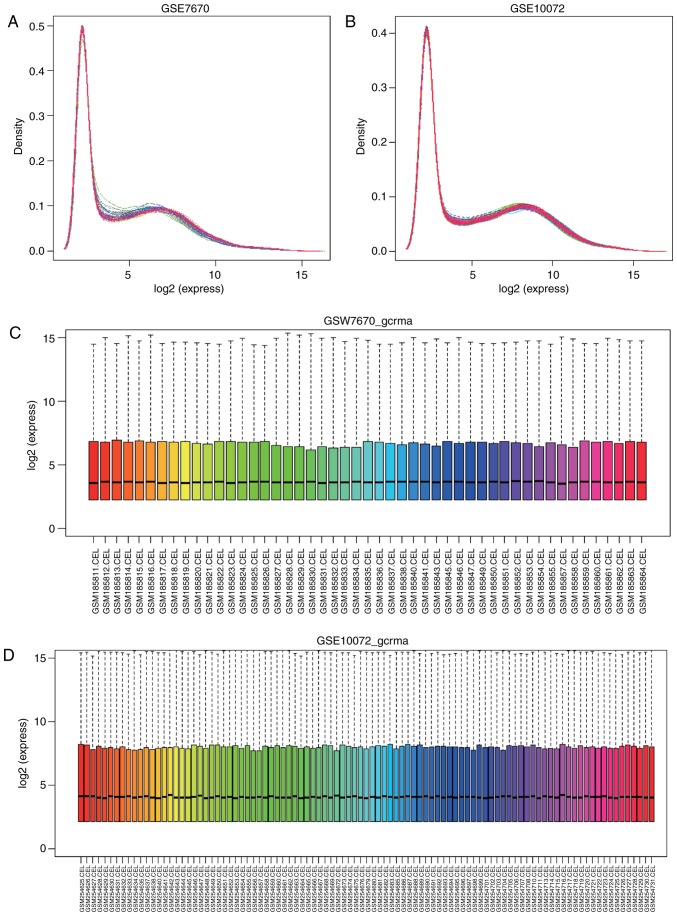
Combining correlation analysis of the raw data was performed with linear regression in affyPLM. The RLE box plot indicated that the majority of the gene expression data from samples in GSE7670 and GSE10072 were consistent (Fig. 1). More sensitive quality analysis of the data was displayed in the NUSE box plot. It was not able that threes ample chips (GSM185844, GSM185848, GSM185856, GSM185858 and GSM185862) in the dataset GSE7670 had deviations >1.05, and therefore these were removed. Furthermore, two heavily deviated chips (GSM254636, GSM254639, GSM254670, GSM254671, GSM254677, GSM254678, GSM254684, GSM254700, GSM254709 and GSM254712) were excluded.
Figure 1.
Expression density curve of the standardized samples. (A) Expression density curve of gene data in the GSE7670 dataset following standardization. (B) Expression density curve of gene data in GSE10072 following standardization. (C) Box plot of gene data in GSE7670 following standardization. (D) Box plot of gene data in GSE10072 following standardization.
Data preprocessing
Following quality assessment, all data were standardized with gcrma (Fig. 1). Expression density curves and box charts illustrated that the expression ranges of the 2 datasets were between 0 and 16. The expression value of gene data in GSE7670 and GSE10072 datasets were concentrated around 4following standardization, indicating a similar expression trend.
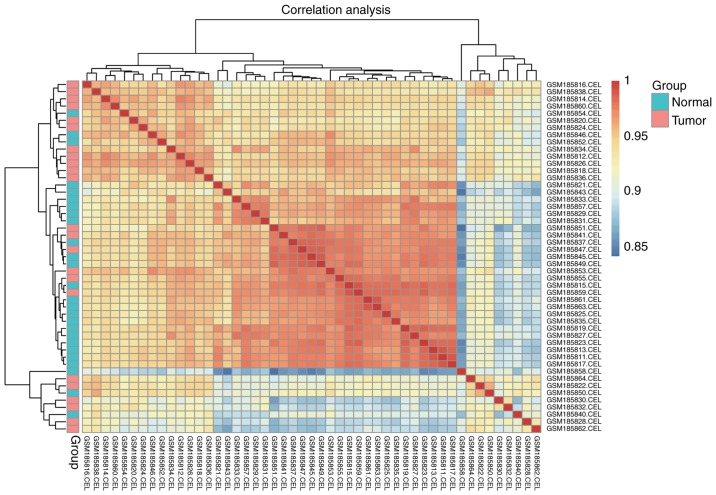
The standardized and the logarithmically converted data were conveyed into cor function in R for Pearson's correlation coefficient. Hierarchial diagram of Pearson's correlation coefficient was obtained (Fig. 2). In Fig. 2, the minimum value of the correlation coefficient between samples inGSE7670 was 0.84. The majority of the normal samples were clustered together. The tumor samples could not be clustered together due to marked differences in expression. The minimum value in GSE10072 (0.84) was a little higher compared with the value in GSE7670 (0.87).
Figure 2.
Hierarchical diagram of the correlations of gene expression between standardized samples. Hierarchical diagram of standardized gene samples in the GSE7670 dataset. Samples were analyzed with Pearson's correlation coefficient. Groups are on the left of the diagram, where red indicates tumor samples and blue indicates normal samples.
Screening of DEGs
DEGs were identified using the limma package. In the GSE7670dataset, a total of 626 DEGs were selected following comparison of 23 LUAD samples and 25 normal tissues. Following removing the duplicates 510 DEGs with annotation remained, including 264 upregulated and 362 downregulated genes (Fig. 3). Following the comparison of 51 lung tumor samples with the 46 normal samples in the GSE10072 dataset, 2,361 DEGs (2,340 were annotated) were obtained, including 1,030 DEGs that were significantly upregulated and downregulated. Following the removal of duplicates, 1,855 DEGs with annotation remained. When the DEGs in the GSE7670 and GSE10072 datasets were matched, there were 484 common genes. Of these, 196 genes were upregulated, and 288 genes were downregulated.
Figure 3.

‘Volcano plots’ for identification of differentially expressed genes in the GSE7670 dataset. Each point represents a gene from the individual geneset. Data points highlighted in blue represent upregulated genes and red points indicate downregulated genes. The black dots represent genes without significant difference in expression. The log10 ratio of expression (normal/tumor) is presented on the y-axis, and logFC is depicted on the x-axis. logFC, fold change.
Functional analysis of DEGs
In GO terms, DEGs were significantly enriched in 250 items, including 167 items in biological process (BP) and 40 in molecular function (MF). For functions in BP, more DEGs were enriched in nitric oxide transport, negative regulation of hepatocyte proliferation, caveola assembly and elastic fiber assembly. In MF, the top three functions were transforming growth factor β-activated receptor activity, small molecule binding and retinoid binding. For functions in cellular component (CC), collagen type I trimer, lamellar body and clathrin-coated endocytic vesicle were the top three popular enrichments. The pathway analysis of KEGG indicated that ECM-receptor interaction was highly enriched among the eight signaling pathways.
Verification analysis
The common up and downregulated genes were selected from the top five DEGs in two individual datasets (GSE7670 and GSE10072), respectively. There were one upregulated DEG (SPP1) and three downregulated DEGs (FCN3, AGER and TMEM100), which were obtained as candidate biomarkers. To evaluate the expression of these candidate DEGs for the prognosis of patients with LUAD, an independent dataset GSE19804, which contains 120 samples (60 tumor tissues and 60 normal tissues), was analyzed on Affymetrix Human Genome U133 Plus 2.0.
In the independent validation analysis, the expression of genes FCN3, AGER and TMEM100all significantly decreased in tumor tissues from patients with LUAD compared with normal tissues. By contrast, the expression of SPP1was increased in tumor tissues compared with normal tissues (Fig. 4). These upand downregulated genes were indicated to participate in the development and/or prognosis of LUAD.
Figure 4.
Expression of four candidate genes in the GSE19804 dataset. Data for LUAD tissue samples are indicated as ‘tumor’, and ‘normal’ represents normal tissues. ***P<0.001. SPP1, secreted phosphoprotein 1; FCN3, ficolin 3; AGER, advanced glycosylation end-product specific receptor; TMEM100, transmembrane protein 100.
Prognostic analysis and statistical analysis
To further analyze the prognostic impact of the DEGs, survival analysis on the expression data of patients with LUAD was performed using the TCGA database. There were 543 samples available, including 249 males and 294 females (>60 years, n=369; ≤60, n=174) as listed in Table I. In statistical analysis, the lowly expressed genes FCN3 and TMEM100 did not affect the survival of the patients in the samples tested (Fig. 5). However, the upregulated SPP1 (high expression) and downregulated AGER (low expression) significantly affected the survival of patients with LUAD. The median survival time of patients with downregulated SPP1 was 1,454 days, whilst that of patients with upregulated SPP1 expression was 1,258 days [log-rank (Mantel-Cox) test P=0.0463; hazard ratio (HR), 1.366; 95% confidence interval (CI), 1.006–1.865]. The median survival time for patients with downregulated AGER (1,229 days) was 150 days less compared with patients with upregulated AGER (1,379 days). A low expression of AGER indicated poor effects on the prognosis of patients with LUAD [log-rank (Mantel-Cox) test P=0.0238; HR, 1.426; 95% CI, 1.050–1.945]. This data indicated that the genes SPP1 and AGER were risk factors that affect the survival time of patients with LUAD.
Table I.
Follow-ups and candidate common differentially expressed genes of patients with lung adenocarcinoma.
| spp1 | FCN3 | AGER | TMEM100 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Factor | Total | Low | High | Low | High | Low | High | Low | High |
| Sex | |||||||||
| Male | 249 | 124 | 125 | 125 | 124 | 125 | 124 | 123 | 126 |
| Female | 294 | 147 | 147 | 145 | 149 | 146 | 148 | 148 | 146 |
| Age, years | |||||||||
| >60 | 369 | 184 | 185 | 174 | 195 | 180 | 189 | 179 | 190 |
| ≤60 | 174 | 87 | 87 | 96 | 78 | 91 | 83 | 92 | 82 |
SPP1, secreted phosphoprotein 1; FCN3, ficolin 3; AGER, advanced glycosylation end-product specific receptor; TMEM100, transmembrane protein 100.
Figure 5.
Survival analysis of differentially expressed genes. Survival analysis of (A) SPP1, (B) FCN3, (C) AGER and (D) TMEM100. Blue, genes with low expression; red, genes with high expression. P<0.05; log-rank (Mantel-Cox) test. SPP1, secreted phosphoprotein 1; FCN3, ficolin 3; AGER, advanced glycosylation end-product specific receptor; TMEM100, transmembrane protein 100.
Discussion
Lung cancer is the most common malignant tumor globally with high occurrence and mortality (28–31). It is indicated that lung cancer is not only a genetic disease; it is also affected by environmental factors. For example, it was determinedthat~85% of smokers were ata higher risk of lung cancer compared with non-smokers (32). Additionally, the occurrence and mortality of lung cancer were also indicated to be associated with air pollution (33,34).
As one of the main subtypes of lung cancer, LUAD presents similar characteristics in occurrence and mortality. Although LUAD and SqCLC are types of non-small cell lung cancer, LUAD is more likely to occur in non-smoking females, particularly young females (age 20–35) (35,36). The rate of morbidity of LUAD is lower compared with squamous and undifferentiated carcinoma (37–39). However, the outcomes and survival time of LUAD are poor with an overall five-year survival rate of only 15% (40).
Current studies focusing on the identification of biomarkers are becoming more frequent. However, present research remains insufficient for clinical application. Key genes that are associated with the outcomes and prognosis of patients with LUAD may have a considerable effect in preventing the delay of treatments by predicting survival time (3,36).
In the present study, two independent databases GSE10072 (containing 58 normal tissues and 49 tumor samples) andGSE7670 (containing 27 normal and tumor samples) were employed. With these comprehensive 164 samples, the DEGs were screened and the expression patterns and functional enrichments of the DEGs were identified. A total of 484 common DEGs were identified, including SPP1 with a high expression and FCN3, AGER and TMEM100 with a low expression in tumor tissues. To confirm this identification, these four DEGs were verified in an additional database, GSE19804. It was indicated that the expression patterns of these four genes in the verification database are in accordance with those in GSE10072 and GSE7670. The differences in expression patterns were all significant. Furthermore, the dysregulation of these candidate genes was associated with the survival rate of the patients. The upregulation of SPP1 and downregulation of AGER suggested that these were risk factors for LUAD.
SPP1 was reported to promote the survival of cancer cells and regulate tumor-associated angiogenesis and inflammation (41). Additionally, SPP1 also served an important role in the development and metastasis of lung cancer (42) by activating the nuclear factor-κB signaling pathway in LUAD (43). In the study by Wesselkamper et al (44), it was discovered that the expression of SPP1was notably increased in mice with acute lung injury.
AGER is a protein that is associated with diabetic angiopathy and thymic hyperplasia by functioning in toll-like receptor 4and advanced glycation end product (AGE)-RAGE signaling pathways (45). Under normal conditions, AGER mediates macrophages (46). However, under pathological conditions, the cross-linking reaction between the AGE and extracellular matrix is promoted, resulting in increased thickness and permeability of the endovascular membrane (47). It was also identified that the RAGE gene was suppressed in tumor tissues (48). The expression and mutation of RAGE were discovered to be highly increased in esophageal cancer but reduced in lung cancer (49). Additionally, a high expression of RAGE was indicated as a signal for lung inflammation and aggravation of other lung diseases (50). In combination with the results in the present study, SPP1 and AGER a close association with LUAD.
FCN3 is a member of the FCN/opsonin p35 lectin family. It was identified that activating the complement pathway in association with mannan-binding lectin serine peptidases and small MBL-associated proteins (51). Additionally, in ovarian cancer (OC), the differential expression of FCN3 has been discovered predominantly in serum molecules (52). In addition, FCN3 was also identified in a previous study inSqCLC (53). Although the differentially expressed TMEM100 has not been identified in lung cancer, the overexpression of TMEM100 inhibited the invasion, migration and proliferation of lung cancer cells (54). As the expression of TMEM100 is notably decreased in tumor tissues compared with normal tissues, it could be indicated that it was reliable as a prognostic biomarker for patients with LUAD. In survival analysis, SPP1, FCN3, AGER and TMEM100 worked well to cluster the patient samples in an independent database. These up and downregulated genes were indicated to participate in the development and or prognosis of LUAD.
In conclusion, among the 484 common DEGs, a high expression of SPP1 and a low expression of FCN3, AGER and TMEM100 were identified in tumor tissues from patients with LUAD. The four key genes (SPP1, FCN3, AGER and TMEM100) were verified using an independent dataset GSE19804. The present study indicated that these two genes (SPP1 and AGER) were risk factors for LUAD. Additionally, these four genes were able to cluster the patient samples in survival analysis, thereby indicating that these gene candidates may serve as prognostic markers to evaluate the prognosis of patients with LUAD. However, in order to investigate the exact functions and the mechanisms that underly LUAD, further research on functional analysis is required. Furthermore, a validation of the associations of the identified DEGs with LUAD is required in future studies. The application of these four candidate genes in clinical treatment and survival evaluation may be promising.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and analyzed in the present study are included in this published article.
Authors' contributions
WZ was responsible for the majority of the analysis and manuscript writing. JF QC, CL, BQ, and QL contributed to data collection and selection.
Ethics and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Li QD, Wang MS, Li FJ, Li QH, Ma XJ, Wang da N. Smoking and air pollution exposure and lung cancer mortality in Zhaoyuan County. Int J Hyg Environ Health. 2013;216:63–70. doi: 10.1016/j.ijheh.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Chen M, Liu X, Du J, Wang XJ, Xia L. Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers. Oncotarget. 2017;8:133–144. doi: 10.18632/oncotarget.13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corner J. Is late diagnosis in lung cancer inevitable? Rt Nursing. 2004 [Google Scholar]
- 5.Mirror: Deadly delay; most lung cancer victims diagnosed too late to save them. Mirror. 2009 [Google Scholar]
- 6.Merino-Zamorano C, Delgado P, de Retana Fernández S, Fernández-Cadenas I, Rodríguez-Luna D, Montaner J, Hernández-Guillamon M. Identification of plasma biomarkers of human intracerebral hemorrhage subtypes through microarray technology. J Stroke Cerebrovasc Dis. 2016;25:665–671. doi: 10.1016/j.jstrokecerebrovasdis.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YH, Shen L, Shen Y, Chen XD, Jiang LS. Study on key genes and regulatory networks associated with osteoporosis by microarray technology. Genetic Test Mol Biomarkers. 2013;17:625–630. doi: 10.1089/gtmb.2013.0055. [DOI] [PubMed] [Google Scholar]
- 8.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 9.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 10.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 12.Gordon GJ, Jensen RV, Hsiao LL, Gullans SR, Blumenstock JE, Ramaswamy S, Richards WG, Sugarbaker DJ, Bueno R. Translation of microarray data into clinically relevant cancer diagnostic tests using gene expression ratios in lung cancer and mesothelioma. Cancer Res. 2002;62:4963–4967. [PubMed] [Google Scholar]
- 13.Wheelan SJ, Murillo Martínez F, Boeke JD. The incredible shrinking world of DNA microarrays. Mol Biosyst. 2008;4:726–732. doi: 10.1039/b706237k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Li Q, Geng L, Chen X, Bi K. Use of the local false discovery rate for identification of metabolic biomarkers in rat urine following genkwa Flos-induced hepatotoxicity. PLoS One. 2013;8:e67451. doi: 10.1371/journal.pone.0067451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cline MS, Craft B, Swatloski T, Goldman M, Ma S, Haussler D, Zhu J. Exploring TCGA pan-cancer data at the UCSC cancer genomics browser. Scientific Rep. 2013;3:2652. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nam DH, Jin JY, Joo KM. Method for preparing patient-specific glioblastoma animal model, and use thereof. US Patent 9750826B2. 2015 Filed June 6, 2013; issued April 16. [Google Scholar]
- 17.Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC, Chuang EY. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010;19:2590–2597. doi: 10.1158/1055-9965.EPI-10-0332. [DOI] [PubMed] [Google Scholar]
- 18.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH, Huang CY. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heber S, Sick B. Quality assessment of Affymetrix GeneChip data. OMICS. 2006;10:358–368. doi: 10.1089/omi.2006.10.358. [DOI] [PubMed] [Google Scholar]
- 21.Wu C, Irizarry R, Gentry J. gcrma: Background adjustment using sequence information. 2005 [Google Scholar]
- 22.Huang HC, Zheng S, Zhao Z. Application of Pearson correlation coefficient (PCC) and Kolmogorov-Smirnov distance (KSD) metrics to identify disease-specific biomarker genes. BMC Bioinformatics. 2010;11(Suppl 4):S23. doi: 10.1186/1471-2105-11-S4-P23. [DOI] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickham H. Ggplot2: Elegant Graphics for Data Analysis. Springer; New York, NY: 2016. [DOI] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Pallant J. SPSS Survival Manual. McGraw-Hill Education; Buckingham, UK: 2013. [DOI] [Google Scholar]
- 27.Norusis MJ. SPSS: SPSS for Windows, base system user's guide release 6.0. SPSS, Inc; 1993. [Google Scholar]
- 28.Batura-Gabryel H, Foremska-Iciek J. Lung cancer in the elderly-increasing epidemiological problem of 21st century. Rocz Akad Med Bialymst. 2005;50(Suppl 1):S152–S155. [PubMed] [Google Scholar]
- 29.Goeckenjan G. Lung cancer-historical development, current status, future prospects. Pneumologie. 2010;64:555–559. doi: 10.1055/s-0030-1255636. (In German) [DOI] [PubMed] [Google Scholar]
- 30.Medenica M, Medenica M, Bojović O, Soldatović I, Durutović I. Changing trends in incidence of lung cancer by histological type in Montenegro. Srp Arh Celok Lek. 2014;142:23–28. doi: 10.2298/SARH1402023M. [DOI] [PubMed] [Google Scholar]
- 31.Rothschild SI. Advanced and metastatic lung cancer-what is new in the diagnosis and therapy? Praxis (Bern 1994) 2015;104:745–750. doi: 10.1024/1661-8157/a002058. (In German) [DOI] [PubMed] [Google Scholar]
- 32.Moyer VA U.S. Preventive Services Task Force: Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement, corp-author. Ann Intern Med. 2014;160:330–338. doi: 10.7326/M14-0334. [DOI] [PubMed] [Google Scholar]
- 33.Raaschou-Nielsen O, Andersen ZJ, Beelen R, Samoli E, Stafoggia M, Weinmayr G, Hoffmann B, Fischer P, Nieuwenhuijsen MJ, Brunekreef B, et al. Air pollution and lung cancer incidence in 17 European cohorts: Prospective analyses from the european study of cohorts for air pollution effects (ESCAPE) Lancet Oncol. 2013;14:813–822. doi: 10.1016/S1470-2045(13)70279-1. [DOI] [PubMed] [Google Scholar]
- 34.Fajersztajn L, Veras M, Barrozo LV, Saldiva P. Air pollution: A potentially modifiable risk factor for lung cancer. Nat Rev Cancer. 2013;13:674–678. doi: 10.1038/nrc3572. [DOI] [PubMed] [Google Scholar]
- 35.Levi F, Bosetti C, Fernandez E, Hill C, Lucchini F, Negri E, La Vecchia C. Trends in lung cancer among young European women: The rising epidemic in France and Spain. Int J Cancer. 2007;121:462–465. doi: 10.1002/ijc.22694. [DOI] [PubMed] [Google Scholar]
- 36.Kang HG, Lee SY, Jeon HS, Choi YY, Kim S, Lee WK, Lee HC, Choi JE, Bae EY, Yoo SS, et al. A functional polymorphism in CSF1R gene is a novel susceptibility marker for lung cancer among never-smoking females. J Thorac Oncol. 2014;9:1647–1655. doi: 10.1097/JTO.0000000000000310. [DOI] [PubMed] [Google Scholar]
- 37.Chen M, Wang X, Xia L. Comparison of alternative splicing in lung adenocarcinoma and lung squamous carcinoma. Shenzhen Daxue Xuebao. 2017;34:33. [Google Scholar]
- 38.Dzhorbenadze RA, Gagua RK, Gerzmava OKh. Lung cancer morbidity rate among the Georgian population according to sex and age and expected morbidity prognosis. Georgian Med News. 2005:65–68. (In Russian) [PubMed] [Google Scholar]
- 39.Suciu B, Bud V, Copotoiu C, Brânzaniuc K, Copotoiu R, Fodor D, Butiurca V. Factors affecting early morbidity and mortality in non-small cell lung cancer surgery, the experience of Surgical Clinic No. 1, Tg. Mureş. Rev Med Chir Soc Med Nat Iasi. 2011;115:116–126. [PubMed] [Google Scholar]
- 40.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubbard NE, Chen QJ, Sickafoose LK, Wood MB, Gregg JP, Abrahamsson NM, Engelberg JA, Walls JE, Borowsky AD. Transgenic mammary epithelial osteopontin (spp1) expression induces proliferation and alveologenesis. Genes Cancer. 2013;4:201–212. doi: 10.1177/1947601913496813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang XM, Li J, Yan MX, Liu L, Jia DS, Geng Q, Lin HC, He XH, Li JJ, Yao M. Integrative analyses identify osteopontin, LAMB3 and ITGB1 as critical pro-metastatic genes for lung cancer. PLoS One. 2013;8:e55714. doi: 10.1371/journal.pone.0055714. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Psallidas I, Stathopoulos GT, Maniatis NA, Magkouta S, Moschos C, Karabela SP, Kollintza A, Simoes DC, Kardara M, Vassiliou S, et al. Secreted phosphoprotein-1 directly provokes vascular leakage to foster malignant pleural effusion. Oncogene. 2013;32:528–535. doi: 10.1038/onc.2012.57. [DOI] [PubMed] [Google Scholar]
- 44.Wesselkamper SC, Case LM, Henning LN, Borchers MT, Tichelaar JW, Mason JM, Dragin N, Medvedovic M, Sartor MA, Tomlinson CR, Leikauf GD. Gene expression changes during the development of acute lung injury: Role of transforming growth factor beta. Am J Respir Crit Care Med. 2005;172:1399–1411. doi: 10.1164/rccm.200502-286OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.AGER: Advanced glycosylation end product-specific receptor. 2018 Feb 11; [Google Scholar]
- 46.Xu Y, Toure F, Qu W, Lin L, Song F, Shen X, Rosario R, Garcia J, Schmidt AM, Yan SF. Advanced glycation end product (AGE)-receptor for AGE (RAGE) signaling and up-regulation of Egr-1 in hypoxic macrophages. J Biol Chem. 2010;285:23233–23240. doi: 10.1074/jbc.M110.117457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chong SF, Lee JH, Zelikin AN, Caruso F. Tuning the permeability of polymer hydrogel capsules: An investigation of cross-linking density, membrane thickness, and cross-linkers. Langmuir. 2011;27:1724–1730. doi: 10.1021/la104510e. [DOI] [PubMed] [Google Scholar]
- 48.Stav D, Bar I, Sandbank J. Usefulness of CDK5RAP3, CCNB2, and RAGE genes for the diagnosis of lung adenocarcinoma. Int J Biol Markers. 2007;22:108–113. doi: 10.1177/172460080702200204. [DOI] [PubMed] [Google Scholar]
- 49.Jing RR, Cui M, Sun BL, Yu J, Wang HM. Tissue-specific expression profiling of receptor for advanced glycation end products and its soluble forms in esophageal and lung cancer. Genet Test Mol Biomarkers. 2010;14:355–361. doi: 10.1089/gtmb.2009.0064. [DOI] [PubMed] [Google Scholar]
- 50.Beucher J, Boëlle PY, Busson PF, Muselet-Charlier C, Clement A, Corvol H. French C F Modifier Gene Study Investigators: AGER-429T/C is associated with an increased lung disease severity in cystic fibrosis. PLoS One. 2012;7:e41913. doi: 10.1371/journal.pone.0041913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Entrez gene: FCN3 ficolin (collagen/fibrinogen domain containing) 3 [Homo sapiens (human)] 2018 Jun 20; [Google Scholar]
- 52.Szala A, Sawicki S, Swierzko AS, Szemraj J, Sniadecki M, Michalski M, Kaluzynski A, Lukasiewicz J, Maciejewska A, Wydra D, et al. Ficolin-2 and ficolin-3 in women with malignant and benign ovarian tumours. Cancer Immunol Immunother. 2013;62:1411–1419. doi: 10.1007/s00262-013-1445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi I, Sadraei Hashemi N, Duan ZH, Shi T. Aberrant signaling pathways in squamous cell lung carcinoma. Cancer Inform. 2011;10:273–285. doi: 10.4137/CIN.S8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ou D, Yang H, Hua D, Xiao S, Yang L. Novel roles of TMEM100: Inhibition metastasis and proliferation of hepatocellular carcinoma. Oncotarget. 2015;6:17379–17390. doi: 10.18632/oncotarget.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed in the present study are included in this published article.