Abstract
Oncolytic herpes simplex virus-1 (oHSV-1) vectors are promising therapeutic agents for cancer. The deletion of the γ34.5 gene eliminates the neurovirulence but attenuates virus replication at the same time. The carboxyl-terminus of protein phosphatase 1 regulatory subunit 15A (also known as MyD116/GADD34) is homologous to that of γ34.5; hence, it may substitute for γ34.5 to enhance the replication and cytotoxicity of the virus. To investigate whether the C-terminus of MyD116 can enhance the anti-tumour efficacy of G47Δ on human breast cancer cells, a GD116 mutant was constructed by inserting a γ34.5-MyD116 chimaera into the G47Δ genome using a bacterial artificial chromosome and two recombinase systems (Cre/loxP and FLPE/FRT). A GD-empty mutant containing only the cytomegalovirus sequence was also created as a control using the same method. Next, the replication and cytotoxicity of these two virus vectors were evaluated in breast cancer cells. Compared with the GD-empty vector, GD116 possessed an enhanced replication capability and oncolytic activity in MCF-7 and MDA-MB-231 cells. On the fifth day after infection with GD116 at MOIs of 0.01 and 0.1, 49.2 and 82.8% of MCF-7 cells, respectively, were killed, with 35.0 and 50.2% of MDA-MB-231 cells, respectively, killed by GD116 at MOIs of 0.1 and 0.3. Additionally, the insertion of the γ34.5-MyD116 chimaera promoted virus replication in MDA-MB-468 at 48 h after infection, although no increased cytotoxic effect was observed. The findings of the present study indicate that the C terminus of the MyD116 gene can be substituted for the corresponding domain of the γ34.5 gene of oHSV-1 to promote the replication of the virus in infected cells. Furthermore, the novel virus mutant GD116 armed with a γ34.5-MyD116 chimaera has enhanced anti-tumour efficacy against human breast cancer cells in vitro.
Keywords: oncolytic herpes simplex virus-1, breast cancer, G47Δ, γ34.5 gene, ICP34.5, MyD116 gene
Introduction
Breast cancer is the most common malignancy and the leading cause of cancer-associated mortality in women worldwide (1,2). Each year, >1.5 million new cases of breast cancer are reported around the world (3), and it has been estimated that 10–12% of women will develop breast cancer over the course of their lives (4). Although relapse-free survival of patients with breast cancer has been improved by modern chemo-, endocrine and targeted therapies, improved therapeutic options for advanced disease are urgently required (5).
Oncolytic herpes simplex virus-1 (oHSV-1) is a potential therapeutic vector for human cancer (6,7). Several oncolytic HSV mutants have already been used in clinical trials for various solid tumours (7–9). Talimogene laherparepvec (T-VEC), an oHSV expressing granulocyte-macrophage colony-stimulating factor, was approved by the U.S. Food and Drug Administration for the treatment of melanoma in 2015 (10). G47Δ is a third-generation oHSV-1 that lacks the γ34.5 neurovirulence gene and possesses an additional deletion of the α47 gene (11,12). Previous studies have demonstrated that G47Δ can effectively target breast cancer cells in vitro and in vivo (13–16). However, certain breast cancer cell lines remain insensitive to G47Δ (data not shown). Infected cell protein 34.5 (ICP34.5), encoded by the γ34.5 gene, has diverse activities: It is critical for the replication and neurovirulence of oHSV-1 in humans and in animal models (17). oHSV-1 mutants without γ34.5 were highly attenuated for neurovirulence, although deletion of γ34.5 also compromises the replication of the virus (18). An early report found that a stretch of 64 amino acids at the C-terminus of the γ34.5 gene was homologous to the C termini of murine protein phosphatase 1 regulatory subunit 15A (also known as MyD116/GADD34) (19). In addition, the C terminus of MyD116 can substitute for the corresponding domain of the γ34.5 gene of oHSV-1 to preclude the premature shutoff of total protein synthesis in infected human cells (19). Therefore, we hypothesised that the insertion of the MyD116 C-terminus might enhance the replication and virulence of G47Δ.
To test this hypothesis, a bacterial artificial chromosome (BAC) and two recombinase systems (Cre/loxP and FLPE/FRT) were used to reconstruct a novel oHSV-1 mutant, GD116, in which a chimeric gene (γ34.5-MyD116) consisting of the N-terminus of the γ34.5 gene and the C-terminus of the MyD116 gene was inserted (19). In this study, the novel vector, GD116, exhibited enhanced anti-tumour effects on breast cancer in vitro.
Materials and methods
Cells
The human breast cancer cell lines MCF-7, SK-BR-3 and MDA-MB-231 were provided by Dr Musheng Zeng (State Key Laboratory of Oncology in Southern China, Sun Yat-sen University Cancer Center, Guangzhou, China), and the MDA-MB-468 cell line was obtained from Dr Xiaoming Xie (Sun Yat-sen University Cancer Center, Guangzhou, China). The Vero cell line (Cercopithecus aethiops kidney cells) was purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cell types were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), which was supplemented with 4.5 g/l glucose and 10% foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere containing 5% CO2.
Plasmids
The plasmid pRB4871, containing the γ34.5-MyD116 chimaera was kindly provided by Professor Bernard Roizman (Marjorie B. Kovler Viral Oncology Laboratories, University of Chicago, Chicago, IL, USA) (19). The pG47Δ-BAC backbone plasmid and the pVec91 shuttle plasmid were obtained from Professor Samuel D. Rabkin (Molecular Neurosurgery Laboratory, Massachusetts General Hospital, Boston, MA, USA), and the pG47Δ-BAC backbone plasmid was created by a two-step replacement procedure, in which G47Δ DNA was cloned into a BAC vector for propagation in Escherichia coli, as described previously (20). The γ34.5-MyD116 chimaera fragment from pRB4871 was ligated into the BamHI-StuI site of pVec91 [containing LacZ, loxP, and FRT sites; multiple cloning sites; lambda stuffer sequences; and a cytomegalovirus (CMV) promoter] to generate pVec91-116.
Virus construction
The recombinant oHSV-1 vectors were constructed using a BAC and Cre/loxP and FLP/FRT recombinase systems, as reported previously (20,21). Mixtures of the pG47Δ-BAC plasmid (2 µg) and pVec91-116 or pVec91 (200 ng) were incubated with Cre recombinase (New England BioLabs, Ipswich, MA, USA) at 37°C for 30 min and then electroporated into E. coli DH10B using a Gene Pulser Xcell™ (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The bacteria were streaked onto LB plates containing chloramphenicol (15 µg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and kanamycin (30 µg/ml; Sigma-Aldrich; Merck KGaA), and positive clones containing the pG47Δ-BAC-116 recombinant and pG47Δ-BAC-empty plasmids were selected. Next, the recombinant plasmids were digested with HindIII and separated by electrophoresis on 0.75% agarose gels in TBE buffer (Tris-base, boric acid and EDTA) at 2.5 cm/V for 12 h with a 1 kb DNA extension ladder (Invitrogen; Thermo Fisher Scientific, Inc.). Co-transfection of pG47Δ-BAC-116 or pG47Δ-BAC-empty and a FLP recombinase expression plasmid, pOG44 (Invitrogen; Thermo Fisher Scientific, Inc.), into Vero cells was performed with Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following this, the removal of the FRT-flanked BAC sequences resulted in the generation of recombinant viruses. The progeny viruses were further selected by limiting dilution and were finally designated ‘GD116’ and ‘GD-empty’.
Analyses of viral DNA
Viral DNA was isolated using the Hirt Supernatant DNA Purification kit (GenMed Scientifics Inc., Shanghai, China), and polymerase chain reaction (PCR) was performed using recombinant Taq DNA polymerase (Takara Bio, Inc., Otsu, Japan). Primers (CMV forward 5′-ACTGCTTACTGGCTTATCG-3′ and reverse, 5′-GTCTAACTCGCTCGTCTC-3′) were used to amplify a specific 113 bp fragment for detection of the CMV sequence. Primers (γ34.5-MyD116 forward, 5′-GCGGCTCAGATTGTTCAA-3′ and γ34.5-MyD116 reverse, 5′-CGGACTGTGGAAGAGATG-3′) were used to amplify a 284 bp product that was specific for the γ34.5-MyD116 chimaera. The samples were initially denatured for 5 min at 94°C; followed by 35 cycles of denaturation at 94°C for 30 sec, re-naturation at 52°C for 30 sec and extension at 72°C for 20 sec; with a final extension at 72°C for 10 min. pVec91 or pRB4871 plasmids and PBS were used as the positive and blank controls, respectively. The PCR products were separated by size in 1% agarose gels.
Western blotting
A total of 5×106 Vero cells were mock treated or treated with viruses at an MOI (multiplicity of infection) of 2 for 24 h, and then cells were lysed using a Whole Cell Lysis kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). A total of 30 µg protein (quantified using the BCA protein quantification kit; Nanjing KeyGen Biotech Co., Ltd.) was separated by 15% SDS-PAGE and transferred to a 0.22-µm thick nitrocellulose membrane (EMD Millipore, Billerica, MA, USA). Following blocking with 5% non-fat dry milk in TBS-Tween-20 (TBS-T, 0.1% Tween) for 1 h at room temperature, the membrane was incubated overnight at 4°C with primary antibodies against ICP34.5 (amino terminus) (diluted 1:500, rabbit polyclonal; cat no. HX5191; TSZ Biosciences, San Fransisco, CA, USA; http://www.tsz-biological.com/) or β-actin (diluted 1:1,000; rabbit monoclonal; cat no. 4970; Cell Signaling Technology, Inc., Danvers, MA, USA). The membrane was then washed and blotted with a horseradish peroxidase (HRP)-linked anti-rabbit secondary antibody (diluted 1:6,000; cat no. 7074; Cell Signaling Technology, USA) for 1 h at room temperature. The Protein antibody complexes were visualized with the chemiluminescent HRP substrate (EMD Millipore) using a chemiluminescence imaging system (Tanon-5200; Tanon, Guangzhou, China).
Virus replication assay
Cells were seeded in 12-well plates at 1×105 cells per well and infected at 70–80% confluence with GD116 or GD-empty at a MOI of 0.1 (Vero, MCF-7, SK-BR-3 and MDA-MB-468) or 0.3 (MDA-MB-231). The inoculum was removed after 2 h and replaced with DMEM supplemented with 1% heat-inactivated FBS (iFBS). The cells and the supernatant were harvested at the indicated times (24 and 48 h) post-infection, processed with three freeze/thaw cycles, and virus titres were determined by plaque assays on Vero cells. In brief, Vero cells were seeded in 12-well plates at 1×105 cells/well and infected with 500 µl virus dilute (10−4/ml, 10−5/ml, 10−6/ml or 10−7/ml) at 100% confluence. Then, the infected Vero cells were cultured at 37°C in 5% CO2 for an additional 48 h prior to subsequent X-gal staining for plaque counting. The X-gal staining was performed using the in situ β-galactosidase staining kit (Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer's protocol. The virus titres were calculated using the following equation: Virus titres=plaque numbers/0.5 × dilution ratio. Experiments were repeated at least three times.
Cell susceptibility assay
Cells were seeded in 48-well plates at 8,000 cells per well. After 16 h, the cells were infected with either GD116 or GD-empty at MOIs of 0.01 and 0.1 (MCF-7, SK-BR-3 and MDA-MB-468) or at MOIs of 0.1 and 0.3 (MDA-MB-231). Cells were incubated for up to 5 days in DMEM medium supplemented with 1% iFBS at 37°C. The cell viability was assessed daily with an MTT assay and expressed as a percentage of the mock-infected control. Formazan crystals were dissolved using 400 µl DMSO and the visualization wavelength was 490 nm. Meanwhile, infection was monitored using β-galactosidase activity via X-gal staining at the indicated times (1, 2, 3, 4 and 5 days post-infection). Experiments were repeated at least three times for each condition in quadruplicate.
Statistical analysis
Student's t-test was used for statistical analyses. P<0.05 were considered to be statistically significant. Data are presented as the mean ± standard deviation. SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA) was used for all of the statistical analyses.
Results
Cloning of the γ34.5-MyD116 chimaera into the shuttle plasmid
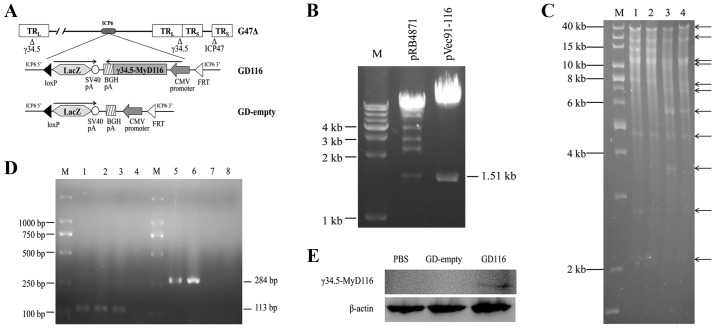
The genomic structure of the mutants is depicted in Fig. 1A. The pRB4871 plasmid was cleaved using the BamHI and StuI restriction endonucleases, and then a 1.51 kb fragment containing the γ34.5-MyD116 chimaera was purified from an agarose gel and directionally cloned into the pVec91 vector to generate pVec91-116. To identify the recombinant plasmid pVec91-116, pRB4871 and pVec91-116 were digested with BamHI and StuI, and the products were separated on a 1% agarose gel. As shown in the map of double restriction endonuclease digestions (Fig. 1B), the desired 1.51 kb bands on an agarose gel confirmed the successful construction of the recombinant pVec91-116 vector.
Figure 1.
Construction and characterization of the GD116 and GD-empty vectors. (A) A diagram depicting the genomic structure of GD116 and GD-empty. (B) Restriction digestion with BamHI and StuI revealed the presence of a 1.51-kb insert from the plasmids pRB4871 and pVec91-116. Lane M, 1 kb DNA ladder. (C) HindIII restriction digestion confirmed the structures of the integrated plasmids obtained following the first step of Cre recombination. Lane M, 1 kb DNA ladder; lanes 1 and 2, pG47Δ-BAC; lane 3, pG47Δ-BAC-Vec91-empty; lane 4, pG47Δ-BAC-Vec91-116. (D) Polymerase chain reaction amplification of the CMV and γ34.5-MyD116 sequences using recombinant virus genomic DNAs as a template. Lane M, DL2000 ladder; lane 1, pVec91; lanes 2 and 6, GD116; lanes 3 and 7, GD-empty; lanes 4 and 8, PBS; lane 5, pRB4871. (E) Expression of the γ34.5-MyD116 protein was confirmed by western blotting. Vero cells were treated with PBS (left), GD-empty (middle) or GD116 (right). CMV, cytomegalovirus.
Construction of recombinant oHSVs with or without the γ34.5-MyD116 chimaera
The generation of two novel oHSV-1 mutants was performed by a previously described two-step replacement procedure (22). In the first step, the shuttle plasmid pVec91-116 or pVec91-empty were integrated into pG47Δ-BAC using Cre recombinase in a tube and an integrated BAC clone (pG47Δ-BAC-Vec91-116 or pG47Δ-BAC-empty) was isolated in E. coli by selection with chloramphenicol and kanamycin. The integrated plasmids were collected and the DNA structures of the plasmids were confirmed by gel analyses following Hind III restriction endonuclease digestion (Fig. 1C).
In the second step, the integrated plasmid pG47Δ-BAC-Vec91-116 or pG47Δ-BAC-empty and the FLP expression plasmid were co-transfected into Vero cells, and the BAC backbone and lambda stuffer sequences were excised by FLP recombinase. As a result, the novel oHSV-1 mutants were generated and were designated GD116 and GD-empty.
To verify the insertion of the target sequences into the novel mutants, specific PCR analyses were conducted using pVec91 and pRB4871 plasmids as the positive controls. The PCR products were checked by electrophoresis on 1% agarose gels. GD116 and GD-empty produced the desired 113 bp band on an agarose gel when used as templates to amplify the CMV sequence (Fig. 1D). However, only GD116 generated the expected size band of 284 bp when γ34.5-MyD116-specific PCR products were detected (Fig. 1D). To determine the expression of the γ34.5-MyD116 protein, Vero cells were treated with PBS, GD-empty or GD116. Western blot analysis revealed that the expression of γ34.5-MyD116 was shown only in the GD116-treated group (Fig. 1E). In addition, the two mutants could form blue plaques on Vero cells by X-gal staining 48 h after infection, which confirmed the insertion and expression of the LacZ gene (data not shown).
Replication of GD116 and GD-empty in vitro
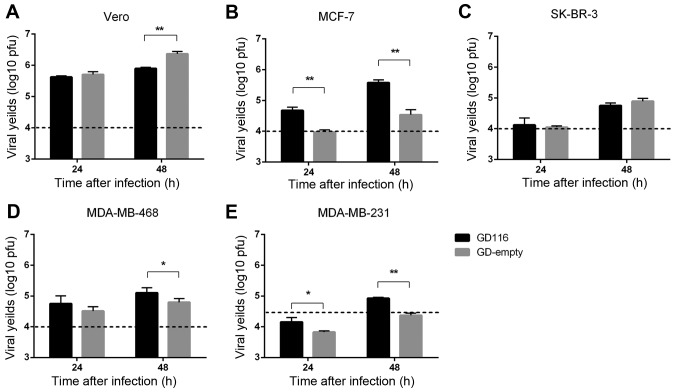
Viral replication is one of the key determinants of oHSV-1 efficacy against tumour cells. The replication of the two mutants was compared in Vero cells and a panel of human breast cancer cell lines at 24 and 48 h after infection (Figs. 2A-E). Compared with GD-empty, GD116 had a significantly increased viral yield in breast cancer MCF-7 and MDA-MB-231 cell lines at 24 and 48 h (Fig. 2B and E). Additionally, the viral yields of GD116 in MDA-MB-468 cells were greater than that of GD-empty at 48 h (Fig. 2D). However, the insertion of γ34.5-MyD116 did not significantly increase the replication rate of oHSV-1 in SK-BR-3 cells (Fig. 2C), and even reduced the viral replication in Vero cells (Fig. 2A).
Figure 2.
Replication of GD116 and GD-empty in Vero cells and a panel of human breast cancer cell lines. (A-D) Cells (1×105 cells per well) as indicated were infected with GD116 or GD-empty at a MOI of 0.1, and the virus yields were determined at 24 and 48 h post-infection. (E) MDA-MB-231 cells (1×105 cells per well) were infected at a MOI of 0.3. *P<0.05 and **P<0.01, respectively. MOI, multiplicity of infection; pfu, plaque-forming unit.
In vitro cytotoxicity of GD116 and GD-empty in breast cancer cell lines
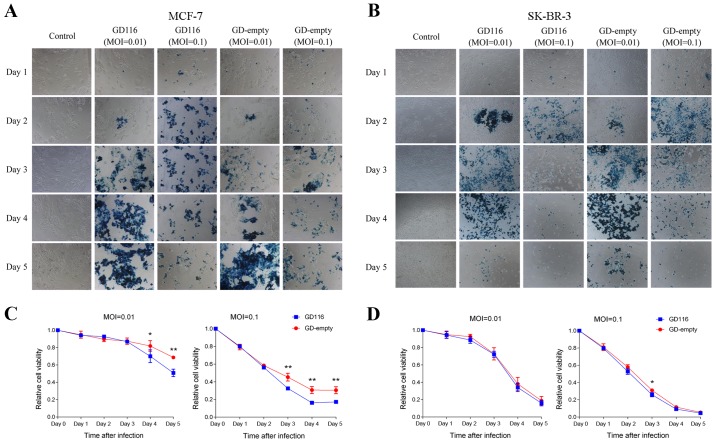
Next, the oncolytic activity of these two mutants was examined in human breast cancer cell lines. The breast cancer cells MCF-7, SK-BR-3 and MDA-MB-468 were infected with GD116 or GD-empty at MOIs of 0.01 and 0.1, whereas MDA-MB-231 cells were infected with these two mutants at MOIs of 0.1 and 0.3. In addition, cells infected with the virus could express LacZ and thus were stained blue by X-gal; staining of MCF-7 and SK-BR-3 cells with X-gal was presented in Fig. 3A and B. Similar to what was observed with virus replication, GD116 was more effective at inhibiting the growth of MCF-7 cells at MOIs of 0.01 and 0.1 (Fig. 3C). On the fifth day after infection with GD116 at MOIs of 0.01 and 0.1, 49.2 and 82.8% of MCF-7 cells were killed, respectively, which was higher than 31.3 and 69.6%, respectively, with GD-empty infection (Fig. 3C). However, only a slightly increased efficacy was observed with GD116 than with GD-empty in SK-BR-3 cells (74.2 vs. 69.0%, respectively) on day 3 at an MOI of 0.1 (Fig. 3D). Aside from that effect, no significant difference in cytotoxicity was observed between the two mutants at the other indicated times (Fig. 3D).
Figure 3.
Cytotoxicity of GD116 and GD-empty in MCF-7 and SK-BR-3 cells in vitro. Monolayers of cells were mock-infected or infected with viruses at multiplicities of infection of 0.01 and 0.1 and further incubated at 37°C for 5 days. (A and B) X-gal staining of MCF-7 and SK-BR-3 cells. Infected cells expressing LacZ were stained positive (blue) with X-gal. Original magnification, ×100. (C and D) Cell viability was assessed using an MTT assay from days 1 to 5 following infection. The relative cell viability was normalised to that of the mock-infected control. Statistical comparisons (independent t-test) were between two viruses at the indicated times. *P<0.05 and **P<0.01.
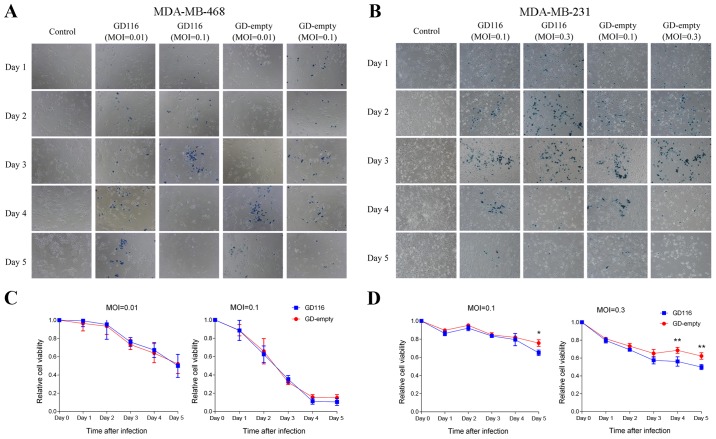
X-gal staining of MDA-MB-468 and MDA-MB-231 cells was presented in Fig. 4A and B. As shown in Fig. 4C, GD116 had a similar cytotoxic effect to GD-empty in MDA-MB-468 cells. However, the insertion of the γ34.5-MyD116 chimaera could significantly increase the oncolytic activity of GD116 on MDA-MB-231 cells (Fig. 4D). On day 5 after infection, 35.0 and 50.2% of MDA-MB-231 cells were killed by GD116 at MOIs of 0.1 and 0.3, respectively; however, the killing activity of GD-empty at the different MOIs was only 24.3 and 37.8%, respectively (Fig. 4D). In vitro, the spread of oHSV-1 was associated with cytotoxicity; therefore, X-gal staining visually showed the difference in oncolytic activity between GD116 and GD-empty in different breast cancer cell lines (Figs. 3A and B, and 4A and B).
Figure 4.
Cytotoxicity of GD116 and GD-empty against MDA-MB-468 and MDA-MB-231 cells in vitro. Monolayers of cells were mock-infected or infected with viruses at multiplicities of infection of 0.01 and 0.1 (MDA-MB-468) or 0.1 and 0.3 (MDA-MB-231) and further incubated at 37°C for 5 days. (A and B) X-gal staining of MDA-MB-468 and MDA-MB-231 cells. Infected cells expressing LacZ were stained positive (blue) with X-gal. Original magnification, ×100. (C and D) Cell viability was assessed by an MTT assay from days 1–5 following infection. The relative cell viability was normalised to that of the mock-infected control. Statistical comparisons (independent t-test) were made between two viruses at the indicated times. *P<0.05 and **P<0.01.
Discussion
oHSV-1 vectors that specifically replicate in and kill tumour cells, sparing normal cells, are promising cancer therapeutic agents. In the oHSV-1 genome, γ34.5 is transcribed as a leaky-late gene that encodes ICP34.5, whose expression can be detected as early as 2–3 h post-infection (23). ICP34.5 is involved in multiple aspects of viral pathogenesis, and one of its key roles is to promote neurovirulence. oHSV-1 mutants with deletions in ICP34 are unable to replicate in neuronal cells and are not neurovirulent (24). In this regard, ICP34.5 has currently been deleted in all oHSV vectors in clinical trials for treating malignant gliomas, such as 1716 and G207 (18,25). However, at the C-terminus of ICP34.5, a region homologous to murine MyD116 and GADD34 can bridge phosphorylated eukaryotic initiation factor 2α (eIF2α) and protein phosphatase-1α (PP1α) to prevent the accumulation of the former and dephosphorylate it (26,27). As a result, ICP34.5 can counteract PKR-mediated innate immune responses and allow translation to proceed (26,28). Therefore, mutants with γ34.5 gene deletions reveal markedly reduced lethality in the peripheral or intracerebral routes of infection but replicate poorly in certain confluent cell types in vitro (23). Thus, it is necessary to reconstruct novel oHSV-1 mutants that maintain low neurovirulence yet replicate efficiently in cancer cells.
The present study used a BAC-based recombinase system to reconstruct novel oHSV-1 mutants. This recombinase system enabled the rapid generation of recombinant oHSV-1 vectors with the desired transgene inserted in the place of the deleted ICP6 locus. The C-terminus of MyD116/GADD34 was used to substitute that of γ34.5 and to create a novel oHSV-1 mutant, GD116. Compared with GD-empty, the mutation enhanced the replication and increased the cytotoxicity of GD116 in breast cancer MCF-7 and MDA-MB-231 cell lines. Although the insertion of the γ34.5-MyD116 chimaera promoted the replication of the virus in MDA-MB-468 cells, no increased cytotoxic effect was obtained. The insertion of γ34.5-MyD116 slightly inhibited viral replication in Vero cells, but the viral cytotoxicity was not attenuated in cancer cells. Therefore, the murine MyD116/GADD34 gene could be used to substitute the γ34.5 gene to promote the cytotoxicity of oHSV-1 without increasing the neurovirulence. However, substitution of the carboxyl-terminus of the γ34.5 gene with the corresponding domain of murine MyD116 did not markedly enhance the virulence of G47Δ in breast cancer cell lines, and even SK-BR-3 cells were not sensitive to the mutation of G47Δ.
Recently, a region in the N-terminus of γ34.5 [amino acids (aa) 68 to 87] that interacts with Beclin 1 and inhibits autophagy, which contributes to neurovirulence in a PKR-dependent fashion, has been identified (29,30). Deletion of the Beclin 1-binding domain (BBD) (aa 68–87) resulted in mutants that could induce autophagy in neurons and was attenuated for neurovirulence following intracerebral inoculation (18). In addition, BBD deletion mutants remain able to dephosphorylate eIF2α and inhibit host protein shutoff, and thus the replication and cytotoxicity are not attenuated. Therefore, promoting the anti-tumour efficacy by expressing BBD-deleted ICP34.5 for oHSV-1 mutants with the deletion of the two copies of the γ34.5 gene, such as G47Δ may represent a promising strategy for generation of a therapeutic vector for the treatment of breast cancer.
Acknowledgements
The authors would like to thank Professor Bernard Roizman for providing the pRB4871 plasmid and Professor Samuel D. Rabkin for providing the pG47Δ-BAC plasmid and the shuttle plasmid pVec91.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81172523 and 81372815; principle investigator, Renbin Liu).
Availability of data and materials
The datasets generated and analyzed in the present study are included in this published article.
Authors' contributions
LC, HJ and RL conceived and designed the experiment. LC, HJ and JF performed the experiments. JW, PH and YR collected the data and performed the statistical analyses. LC wrote the manuscript. JW and RL reviewed and revised the manuscript. All authors read and approved the final manuscript.
Ethics and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Huang J, Li H, Ren G. Epithelial-mesenchymal transition and drug resistance in breast cancer (Review) Int J Oncol. 2015;47:840–848. doi: 10.3892/ijo.2015.3084. [DOI] [PubMed] [Google Scholar]
- 3.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, Ettl J, Patel R, Pinter T9, Schmidt M, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study. Lancet Oncol. 2015;16:25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 4.Kleibl Z, Kristensen VN. Women at high risk of breast cancer: Molecular characteristics, clinical presentation and management. Breast. 2016;28:136–144. doi: 10.1016/j.breast.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Hartkopf AD, Fehm T, Wallwiener D, Lauer UM. Oncolytic virotherapy of breast cancer. Gynecol Oncol. 2011;123:164–171. doi: 10.1016/j.ygyno.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Kuruppu D, Tanabe KK. HSV-1 as a novel therapy for breast cancer meningeal metastases. Cancer Gene Ther. 2015;22:506–508. doi: 10.1038/cgt.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andtbacka RH, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, Delman KA, Spitler LE, Puzanov I, Agarwala SS, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 8.Campadelli-Fiume G, Petrovic B, Leoni V, Gianni T, Avitabile E, Casiraghi C, Gatta V. retargeting strategies for oncolytic herpes simplex viruses. Viruses. 2016;8:63. doi: 10.3390/v8030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Fulci G, Wakimoto H, Cheema TA, Buhrman JS, Jeyaretna DS, Rachamimov Stemmer AO, Rabkin SD, Martuza RL. Combination of oncolytic herpes simplex viruses armed with angiostatin and IL-12 enhances antitumor efficacy in human glioblastoma models. Neoplasia. 2013;15:591–599. doi: 10.1593/neo.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.http://www.webcitation.org/6drvCltG7. [Oct 27;2015 ];U.S. Food and Drug Administration: FDA approves first-of-its-kind product for the treatment of melanoma. [Google Scholar]
- 11.Todo T. Active immunotherapy: Oncolytic virus therapy using HSV-1. Adv Exp Med Biol. 2012;746:178–186. doi: 10.1007/978-1-4614-3146-6_14. [DOI] [PubMed] [Google Scholar]
- 12.Nigim F, Esaki SI, Hood M, Lelic N, James MF, Ramesh V, Stemmer-Rachamimov A, Cahill DP, Brastianos PK, Rabkin SD, et al. A new patient-derived orthotopic malignant meningioma model treated with oncolytic herpes simplex virus. Neuro Oncoly. 2016;18:1278–1287. doi: 10.1093/neuonc/now031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan J, Jiang H, Cheng L, Liu R. The oncolytic herpes simplex virus vector, G47Δ, effectively targets tamoxifen-resistant breast cancer cells. Oncol Rep. 2016;35:1741–1749. doi: 10.3892/or.2015.4539. [DOI] [PubMed] [Google Scholar]
- 14.Zeng WG, Li JJ, Hu P, Lei L, Wang JN, Liu RB. An oncolytic herpes simplex virus vector, G47Δ, synergizes with paclitaxel in the treatment of breast cancer. Oncol Rep. 2013;29:2355–2361. doi: 10.3892/or.2013.2359. [DOI] [PubMed] [Google Scholar]
- 15.Liu R, Varghese S, Rabkin SD. Oncolytic herpes simplex virus vector therapy of breast cancer in C3(1)/SV40 T-antigen transgenic mice. Cancer Res. 2005;65:1532–1540. doi: 10.1158/0008-5472.CAN-04-3353. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Hu P, Zeng M, Rabkin SD, Liu R. Oncolytic herpes simplex virus treatment of metastatic breast cancer. Int J Oncol. 2012;40:757–763. doi: 10.3892/ijo.2011.1266. [DOI] [PubMed] [Google Scholar]
- 17.Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol. 2007;81:12128–12134. doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanai R, Zaupa C, Sgubin D, Antoszczyk SJ, Martuza RL, Wakimoto H, Rabkin SD. Effect of γ34.5 deletions on oncolytic herpes simplex virus activity in brain tumors. J Virol. 2012;86:4420–4431. doi: 10.1128/JVI.00017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He B, Chou J, Liebermann DA, Hoffman B, Roizman B. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the gamma(1)34.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J Virol. 1996;70:84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuhara H, Ino Y, Kuroda T, Martuza RL, Todo T. Triple gene-deleted oncolytic herpes simplex virus vector double-armed with interleukin 18 and soluble B7-1 constructed by bacterial artificial chromosome-mediated system. Cancer Res. 2005;65:10663–10668. doi: 10.1158/0008-5472.CAN-05-2534. [DOI] [PubMed] [Google Scholar]
- 21.Tsuji T, Nakamori M, Iwahashi M, Nakamura M, Ojima T, Iida T, Katsuda M, Hayata K, Ino Y, Todo T, Yamaue H. An armed oncolytic herpes simplex virus expressing thrombospondin-1 has an enhanced in vivo antitumor effect against human gastric cancer. Int J Cancer. 2013;132:485–494. doi: 10.1002/ijc.27681. [DOI] [PubMed] [Google Scholar]
- 22.Kuroda T, Martuza RL, Todo T, Rabkin SD. Flip-Flop HSV-BAC: Bacterial artificial chromosome based system for rapid generation of recombinant herpes simplex virus vectors using two independent site-specific recombinases. BMC Biotechnol. 2006;6:40. doi: 10.1186/1472-6750-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korom M, Davis KL, Morrison LA. Up to four distinct polypeptides are produced from the γ34.5 open reading frame of herpes simplex virus 2. J Virol. 2014;88:11284–11296. doi: 10.1128/JVI.01284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown SM, Harland J, MacLean AR, Podlech J, Clements JB. Cell type and cell state determine differential in vitro growth of non-neurovirulent ICP34.5-negative herpes simplex virus types 1 and 2. J Gen Virol. 1994;75:2367–2377. doi: 10.1099/0022-1317-75-9-2367. [DOI] [PubMed] [Google Scholar]
- 25.Zemp FJ, Corredor JC, Lun X, Muruve DA, Forsyth PA. Oncolytic viruses as experimental treatments for malignant gliomas: Using a scourge to treat a devil. Cytokine Growth Factor Rev. 2010;21:103–117. doi: 10.1016/j.cytogfr.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 26.He B, Gross M, Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Zhang C, Chen X, Yu J, Wang Y, Yang Y, Du M, Jin H, Ma Y, He B, Cao Y. ICP34.5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging eukaryotic initiation factor 2alpha (eIF2alpha) and protein phosphatase 1. J Biol Chem. 2011;286:24785–24792. doi: 10.1074/jbc.M111.232439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis KL, Korom M, Morrison LA. Herpes simplex virus 2 ICP34.5 confers neurovirulence by regulating the type I interferon response. Virology. 2014;468–470:330–339. doi: 10.1016/j.virol.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Tang S, Guo N, Patel A, Krause PR. Herpes simplex virus 2 expresses a novel form of ICP34.5, a major viral neurovirulence factor, through regulated alternative splicing. J Virol. 2013;87:5820–5830. doi: 10.1128/JVI.03500-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed in the present study are included in this published article.