Abstract
Type I interferons (IFNs) are critical in animal antiviral regulation. IFN-mediated signalling regulates hundreds of genes that are directly associated with antiviral, immune and other physiological responses. The signalling pathway mediated by mechanistic target of rapamycin (mTOR), a serine/threonine kinase regulated by IFNs, is key in regulation of cellular metabolism and was recently implicated in host antiviral responses. However, little is known about how animal type I IFN signalling coordinates immunometabolic reactions during antiviral defence. Here, using porcine reproductive and respiratory syndrome virus (PRRSV), we found that the genes in the mTOR signalling pathway were differently regulated in PRRSV-infected porcine alveolar macrophages at different activation statuses. Moreover, mTOR signalling regulated PRRSV infection in MARC-145 and primary porcine cells, in part, through modulating the production and signalling of type I IFNs. Taken together, we determined that the mTOR signalling pathway involves PRRSV infection and regulates expression and signalling of type I IFNs against viral infection. These findings suggest that the mTOR signalling pathway has a bi-directional loop with the type I IFN system and imply that some components in the mTOR signalling pathway can be utilized as targets for studying antiviral immunity and for designing therapeutic reagents.
Keywords: type I IFNs, mTOR signalling pathway, PRRSV, antiviral immunity
Introduction
Animal innate immunity is the frontline defence in restricting viral invasion or replication and launching early antiviral responses [1]. Through a cascade of signalling transduction, virus-infected cells or surrounding activated cells are elicited to increase the production of immune effectors. Prominent among these are interferons (IFNs), an essential family of animal cytokines with pivotal influences on biological action, especially those related to antiviral defence [2, 3]. The IFN family is divided into three classes, type I, II and III IFNs, which are distinguished by their distinct receptor complexes [4]. Type I IFNs are perceived by a heterodimeric receptor complex (IFNAR1/2) and form the largest group which comprises more than 10 subtypes including IFN-α, IFN-β, IFN-ε, IFN-κ and IFN-ω, generally found in most mammalian species. However, type I IFNs also include species-specific subtypes such as IFN-δ (pigs and horses), IFN-τ (cattle), IFN-ζ (mice) and IFN-αω (pigs, horses and cattle). Furthermore, IFN-α, IFN-δ and IFN-ω are subtypes that contain multiple genes. For example, the swine genome contains 25, 11 and 8 functional genes of IFN-α, IFN-δ, and IFN-ω, respectively, which contributes to the diverse features of type I IFNs in immune regulation [5, 6].
Type I IFNs are primarily thought of as antiviral mediators and play an indispensable role in regulation of antiviral immunity [7]. Upon viral infection, various cellular pattern recognition receptors (PRRs) are involved in regulation of type I IFN production through association with diverse adaptor proteins resulting in activation and nuclear translocation of transcription factors including IFN regulatory factor (IRF) 3, IRF7 and NF-κB to specifically bind to various promoters of type I IFN genes [1, 3, 7]. Once type I IFNs are induced, they are secreted by infected cells and stimulate target cells to induce type I IFN action signalling [8]. The binding of type I IFNs to their specific receptors (IFNAR1/IFNAR2) on the surface of target cells leads to an intracellular signalling cascade and culminates in the stimulation of hundreds of IFN-stimulated genes (ISGs), which exert various antiviral or virostatic activities [9]. Beyond this classic IFNAR-mediated IFN signalling, other important signalling cascades, including the mitogen-activated protein kinase (MAPK) pathway, the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) pathway, and the pathway mediated by the mechanistic target of rapamycin (mTOR), also potentially interact with type I IFNs for regulatory transcription of diverse ISGs [10–12].
The protein of mTOR is an evolutionarily conserved serine/threonine kinase, which acts as an overriding node for maintenance of homeostasis in animal cells [13]. In mammalian cells, mTOR kinase is a central component that forms two functionally distinct multi-subunit complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). The mTORC1 consists of five components: mTOR, the catalytic subunit of the complex; regulatory-associated protein of mTOR (Raptor); mammalian lethal with Sec13 protein (mLST8); proline-rich AKT substrate 40 kDa (PRAS40); and DEP-domain-containing mTOR-interacting protein (Deptor). In addition to the mTOR and the two other shared subunits of mLST8 and Deptor, mTORC2 also contains a rapamycin-insensitive companion of mTOR (Rictor), mammalian stress-activated protein kinase interacting protein (mSIN1), and protein observed with Rictor 1 and 2 (Protor 1/2). Upon stimulation of external signals such as nutrients, cytokines or growth factors, the mTOR signalling pathway is initiated via the upstream PI3K-AKT cascade [13]. It has been shown that both mTORC1- and mTORC2-signalling cascades are critical for the production and signalling of type I IFNs [12, 14, 15]. For example, Cao et al. reported that the PI3K-mTOR-p70 S6 kinase pathway is required for Toll-like receptor (TLR)-induced type I IFN production in plasmacytoid dendritic cells (pDCs) [16]. The type I IFN system also coordinates mTOR signalling to selectively regulate transcription and translation of various ISGs [15, 17, 18]. A recent study further demonstrated that an Unc51-like kinase (ULK1) acts as a link between the type I IFN response and the mTOR signalling pathway, and that the mTOR-ULK1 pathway is critical for gene transcription mediated by the cis-elements (such as ISREs and GAS) in type I IFN genes [19]. In addition, the mTOR signalling pathway can be activated/regulated by type I IFN responses [15, 20]. However, functions of the mTOR signalling pathway in antiviral immunity, particularly via the regulation of the type I IFN response, remain poorly defined in viral diseases in pigs.
In this study, we focus on the function of the mTOR signalling pathway in the production and signalling of type I IFNs during an antiviral response. Cells infected by porcine reproductive and respiratory syndrome virus (PRRSV) were used to decipher the interaction of type I IFNs and mTOR signalling. In PRRSV pathogenesis, the virus directly infects porcine tissue macrophages (primarily those in the lung and reproductive tract) and monocyte-derived dendritic cells (mDCs), which leads to immune compromise partly resulting from the viral suppression of IFN-α responses. Our previous studies showed that PRRSV infection of macrophages and mDCs is significantly associated with the activation status of these porcine monocytic cells. For example, the alternatively activated (M2) macrophages by IL-4 or IL-10 are significantly more susceptible than the macrophages that are activated by either type I or type II IFNs [21]. Interestingly, through interactions with these cytokines and other cell mediators, mTOR signalling plays a key role in regulating the activation statuses of macrophages [22]. Here, we demonstrate that the mTOR signalling pathway is critical in regulation of PRRSV infection, and that the effect of mTOR in PRRSV infection is mechanistically through regulation of the expression and signalling of type I IFNs.
Results and discussion
Genes in the mTOR signalling pathway were differentially expressed in PRRSV-infected macrophages at different activation statuses
Macrophages are one of the most important immune effector and regulator cells and display multiple roles in both innate and adaptive immune responses [23]. To maintain their various functions, macrophages undergo phenotypical polarization in response to diverse environmental stimulants [24, 25]. Typical activation statuses characterized in macrophage polarization include classical (M1) and alternative (M2) statuses [21, 24]. M1 status is induced in response to IFN-γ and bacterial products, such as lipopolysaccharides (LPS) [21]. M2 status is further categorized into three subclasses: M2a, induced by Th2-type cytokines, IL-4 or IL-13; M2b, obtained by triggering Fcγ receptors plus a TLR stimulus; and M2c, activated by glucocorticoid (GC), IL-10 and/or TGF-β [21].
Previously, we showed that many signalling pathways are significantly modulated in PRRSV-infected macrophages at different activation statuses [21] and it has been shown that mTOR signalling involves regulating macrophage polarization [26]. Genome-wide analysis of gene regulation was conducted in PRRSV-infected porcine alveolar macrophages at different activation states as described [21, 27]. The differentially expressed genes (DEGs) involved in the mTOR signalling pathway, including mTOR complexes, their upstream regulators and downstream effectors, were extracted for further analysis (Fig. S1a, available in the online Supplementary Material). Some of these displayed significantly differential expression and were selected as candidate targets to analyse antiviral regulation of macrophages by mTOR signalling (Fig. S1b, c). RNA-Seq analysis revealed that a large number of genes in the mTOR signalling pathway were significantly and differentially regulated, including mTOR, Rictor, AKT3, IKBKB, EIF4E, RPS6KB2, EIFEBP2, ULK1 and ULK2 in PRRSV-infected macrophages at different activation statuses. mTOR kinase, the pivotal component for both mTORC1 and mTORC2, was up-regulated in IFN-γ (M1) and IL-4 (M2a)-stimulated cells and down-regulated in LPS (M1) and IL-10 (M2c) treatments, but less regulated by antiviral IFN-α1, indicating that mTOR is more related to macrophage activation status, or linking to antiviral regulation through cell polarization. Rictor, the key subunit of mTORC2, was greatly up-regulated by IFN-α1. Two downstream effectors of mTORC1 and mTORC2, RPS6KB2 (also called p70 S6 kinase) and AKT3 respectively, were differentially regulated by macrophage polarization. The results imply that mTORC2 may play a crucial role in macrophage polarization and antiviral regulation, considering Rictor, AKT and p70 S6 kinase are closely relevant to mTORC2 activity [13]. Furthermore, ULK1, acting as a crosslink for type I IFNs-mTOR, was down-regulated by IFN-γ, LPS and IFN-α1, and up-regulated by IL-4 and IL-10, which implies that type I IFN signalling correlates with the mTOR signalling pathway to regulate anti-PRRSV infection [12]. Thus, genes related to the mTOR signalling pathway presented a large group of DEGs in PRRSV-infected macrophages at different activation statuses, suggesting that the mTOR signalling pathway closely involves antiviral regulation in macrophages. Previous reports have indicated that mTOR signalling is critical for inter-regulation with macrophage polarization in autoimmune diseases and parasitic infections; however, the involvement of mTOR signalling in viral infection in polarized macrophages has not been previously studied [11]. We therefore focus on examining the significantly differential expression of mTOR-mediated gene responsive pathway based on comparative transcriptomes revealed in macrophages at different activation statuses upon viral infection, rather than a transcriptomic comparison between infected and non-infected tissues/cells, which has been well-documented in previous studies [21]. The significant regulation of mTOR gene responsive pathway in PRRSV-infected macrophages at different activation statuses suggests a potential target to regulate the dynamic interaction between macrophage activation status and PRRSV infection.
Pharmaceutical regulation of mTOR signalling affects PRRSV infection
Two mTOR inhibitors, rapamycin and PP242, and one mTOR activator, MHY1485, were used for pharmaceutical regulation of mTOR signalling. Rapamycin and its analogues are first-generation mTOR inhibitors that associate with 12 kDa FK506-binding protein (FKBP12) to form a complex interacting with the FKBP12-rapamycin binding (FRB) domain in mTOR kinase and in turn disrupt the formation of mTORC1 [28]. Only mTORC1 activity is inhibited by rapamycin in a short time period, and prolonged rapamycin treatment also affects mTORC2 activity [29]. PP242 is a non-selective inhibitor that targets the adenosine triphosphate (ATP)-binding site of mTOR kinase and suppresses both mTORC1 and mTORC2 activities [30]. In contrast, MHY1485 is a novel, potent and selective cell-permeable mTOR activator [31]. Thus, rapamycin, PP242 and MHY1485 were selected to comparatively modulate the mTOR signalling pathway and to examine the effects on IFN production and antiviral response.
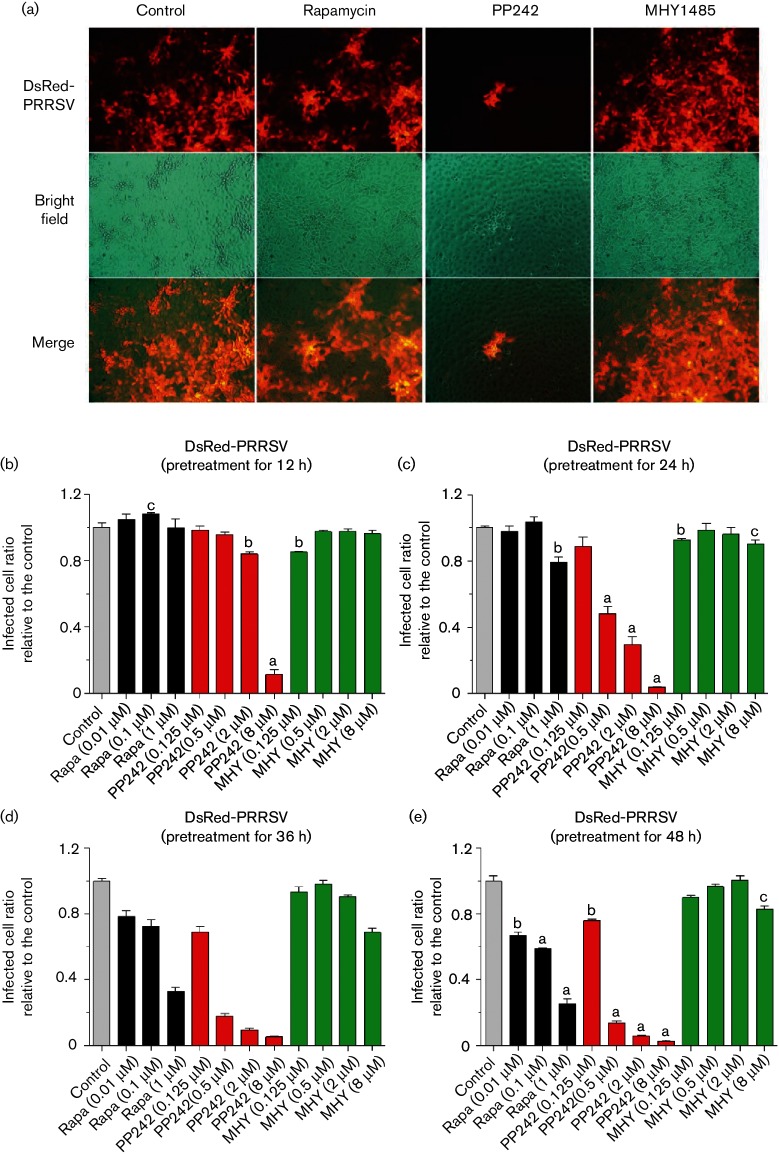
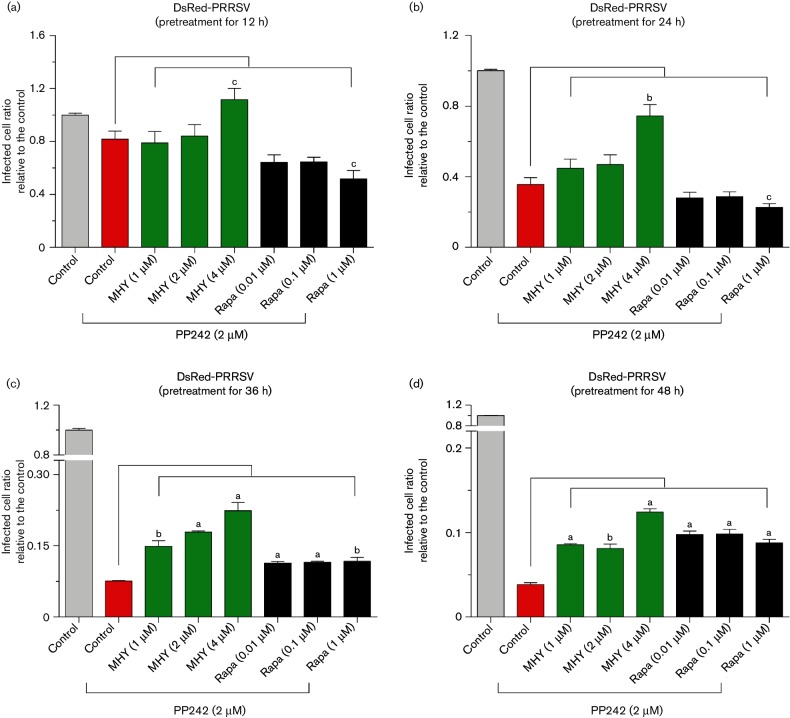
The pharmaceutic effect of mTOR mediators was first evaluated in MARC-145 cells, a cell line derived from monkey kidney and well-known for its permissiveness to PRRSV infection. Similar to other studies, rapamycin, PP242 and MHY1485 at doses lower than 1, 8 and 8 µM respectively, caused little cytotoxicity (monitored with an [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] MTT assay, data not shown) or changes in cell morphology during 12–48 h (Figs 1 and 2) [28, 31]. In control MARC-145 cells, nearly all cells were infected by PRRSV, shown by the virus replication-competent expression of the red fluorescent protein (DsRed). Treatment of cells with both mTOR inhibitors substantially repressed PRRSV infection, especially, the non-selective inhibitor, PP242, which exerted significantly higher effects than rapamycin to suppress nearly 60–90 % of the viral infection at 0.5–8.0 µM (Figs 1a–e and S2). In contrast, the mTOR activator, MHY1845, had a negligible effect in promotion of PRRSV infection in MARC-145 cells, likely because the basal mTOR activity in the cells was sufficient to support the virus replication and further activation added very little. However, MHY1485 at a high concentration of 8 µM slightly inhibited PRRSV infection (Fig. 1c–e), which could be relevant to its inhibitory effect on cell autophagy—a process that might help PRRSV enter cells [31, 32]. Cells treated with both rapamycin and PP242 showed little additive effect on viral repression at a short time treatment, with most virostatic effect from PP242, which non-selectively inhibited both mTORC1 and mTORC2 activity (Figs 2a–d and S2). The combination of PP242 and rapamycin slightly reversed the suppressive effect of PP242 at a long time treatment, probably due to rapamycin relieving the suppression of mTORC2 activity by PP242 as rapamycin only suppresses mTORC1 at a short time period. The addition of the mTOR activator, MHY1485, at 1–4 µM, partly reversed the effect of PP242 in suppression of PRRSV replication (Figs 2a–d and S2), which verified the involvement of the mTOR pathway in the antiviral response from both pharmaceutical loss-of-function and gain-of-function.
Fig. 1.
Regulation of PRRSV infection by mTOR inhibitors and activator in MARC-145 cells. (a) Cells were treated with 1 µM rapamycin, 2 µM PP242 or 2 µM MHY1485 for 24 h, then infected with DsRed-labelled PRRSV (m.o.i. of 1), visualized and imaged with fluorescence microscopy at 36 h post-infection (h p.i.). (b–e) mTOR inhibitors and activator regulate PRRSV infection in a dose- and time-dependent manner in MARC-145 cells. Cells were pretreated with serial dilutions of mTOR inhibitors, rapamycin and PP242, or activator MHY1485 for 12, 24, 36 or 48 h, then infected with DsRed-labelled PRRSV (m.o.i. of 1), and quantified with a SpectraMax i3 (Molecular Devices) at 36 h p.i.; n=3; a, b and c indicate p<0.001, 0.01 and 0.05, respectively, relative to the control. Rapa, Rapamycin; MHY, MHY1485.
Fig. 2.
mTOR activator MHY1485 reverses the repression of PRRSV replication by PP242. (a–d) Cells were pretreated with 2 µM PP242 for 12, 24, 36 or 48 h, then infected with DsRed-labelled PRRSV along with MHY1485 or rapamycin at different concentrations, and quantified with a SpectraMax i3 at 36 h p.i.; n=3; a, b and c indicate p<0.001, 0.01, and 0.05, respectively, relative to the control. Rapa, Rapamycin; MHY, MHY1485.
The time-course evaluations of rapamycin, PP242 and MHY1485 relative to their efficacy in modulation of PRRSV infection mirrored what was observed in the dose-dependence experiments; PP242 at 0.5–8 µM was consistently effective through 12–48 h (Fig. 1b–e). Rapamycin exerted suppression only after the long time treatment (Fig. 1b–e). In addition, the reverse effect of the activator, MHY1485, against PP242 was clear at the long time treatment of PP242 during the co-treatment (Fig. 2a–d). As a non-selective inhibitor, PP242 has suppressive effects on both mTORC1 and mTORC2 complexes; interestingly, a recent study showed that the prolonged treatment actually strengthened the suppressive effect of rapamycin on mTORC2 [29]. In summary, antiviral regulation could be achieved via pharmaceutical regulation of mTOR activity, and the non-selective inhibitor, PP242, significantly inhibited viral propagation in both dose- and time-dependent manners in MARC-145 cells. Our observations indicate that mTORC2 plays more of a role than mTORC1 in regulation of virus replication in cells. To our knowledge, this is one of the first studies that have associated mTORC2 with antiviral regulation in PRRSV infection.
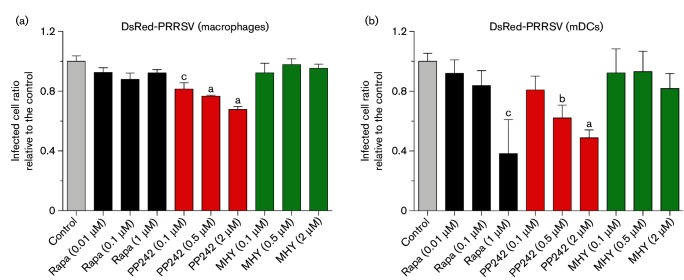
Studies of PRRSV pathogenesis in pigs have concluded that PRRSV is monocytotropic. The primary cell targets, which are highly permissive to PRRSV, consist of tissue macrophages in lungs and the reproductive tract and monocyte-derived DCs (mDCs) [33]. Thus, we used porcine primary alveolar macrophages and mDCs derived from blood monocytes to validate the results obtained in MARC-145 cells. Compared to MARC-145 cells, primary porcine macrophages and mDCs are more sensitive to the mTOR mediators. The physiological concentrations of rapamycin, PP242 and MHY1485, which caused little observed/measurable cytotoxic effect in the tested periods, are lower than 1–2 µM. Similar to what we observed in MARC-145 cells, PP242 but not rapamycin showed significant suppression of PRRSV infection in the porcine macrophages. However, because primary macrophages comprise very diverse subsets of cells compared with the established cell line with respect to cell susceptibility to the virus and cell responsiveness to mTOR inhibitors, the suppression of PP242 on the PRRSV infection only attained 20–40 % of the control cells. The most interesting observation was in mDCs, where rapamycin and PP242 acted similarly in suppression of PRRSV propagation, indicating that the expression levels and interaction between mTORC1 and mTORC2 in mDCs were probably different from those in porcine macrophages and especially MARC-145 cells. PP242 displayed an inhibitory effect on PRRSV infection in both macrophages and mDCs with comparable dose-dependence as shown in MARC-145 cells (Fig. 3a, b). Unexpectedly, rapamycin showed inhibition on PRRSV infection only in mDCs. The activator, MHY1485, had no effect on PRRSV infection in both macrophages and mDCs as shown in MARC-145 cells. Clearly, the effect of pharmaceutical regulation of antiviral response via the mTOR signalling pathway is also dependent on cell types. In general, MARC-145 cells, an established cell line, are more uniform and nearly 100 % permissive to PRRSV; in contrast, porcine primary cells including macrophages and mDCs comprise diverse cell subsets, and are only partially permissive to PRRSV in vitro [33]. Therefore, we observed a more significant and reproducible effect in MARC-145 cells than in porcine primary cells; both mTOR inhibitors, rapamycin and PP242, had a better suppressive effect against PRRSV infection in MARC-145 cells. In addition, the prolonged effect of rapamycin observed in MARC-145 cells, was unable to be demonstrated in porcine cells due to the shorter duration of treatment. However, the suppressive effect was clearly demonstrated in porcine primary cells even with reasonably different kinetics (Figs 1b–e and 3a, b). These results suggest that inhibition of the mTOR signalling pathway provides a potential route to regulate anti-PRRSV responses and that host antiviral immunity is significantly inter-regulated by the mTOR signalling pathway.
Fig. 3.
Repression of PRRSV infection by mTOR inhibitors in porcine alveolar macrophages and mDCs. Cells were incubated with DsRed-labelled PRRSV (m.o.i. of 0.5) for 1 h, and then treated with mTOR inhibitors (rapamycin or PP242) or activator (MHY1485) at indicated concentrations for another 20 h. Infected macrophages (a) and mDCs (b) were quantified using a SpectraMax i3 at 20 h p.i. n=4; a, b and c indicate p<0.001, 0.01 and 0.05, respectively, relative to the control. Rapa, Rapamycin; MHY, MHY1485.
Regulation of type I IFN response by mTOR signalling
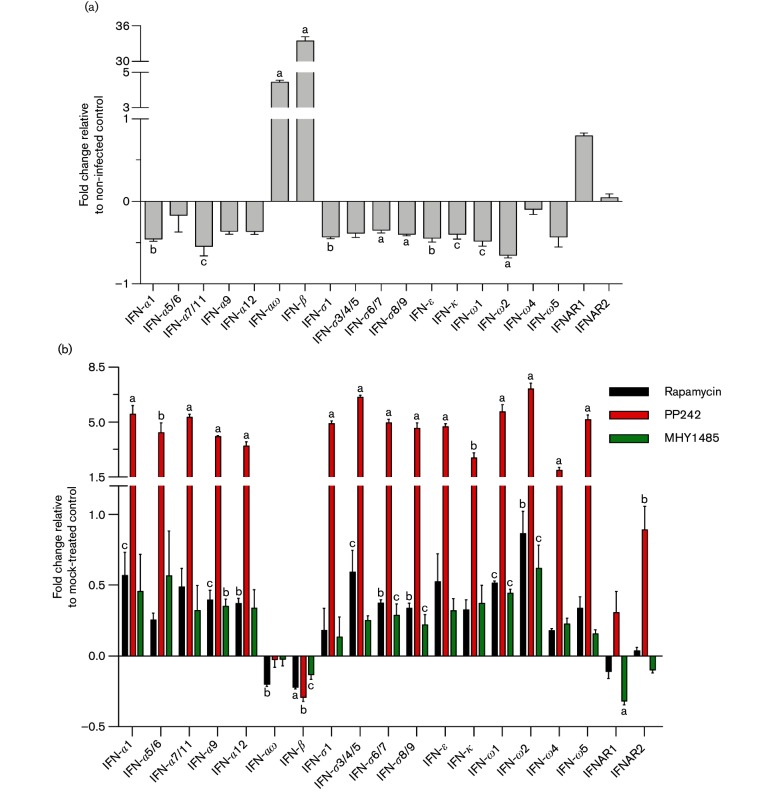
IFNs are a family of cytokines critically important in regulation of antiviral responses. However, most viruses, including PRRSV, have evolved diverse mechanisms to evade host IFN-mediated antiviral responses [3, 34]. For example, several non-structural or structural proteins encoded by PRRSV are capable of functionally inhibiting the components in IFN signalling including IRF3, NF-κB, NEMO and ISGs [35, 36]. This virus-mediated early suppression on both the production and action of type I IFNs (in particular IFN-α subtypes) in primary infection sites (i.e. tissue macrophages and mDCs) is involved in the pathogenic success of PRRSV in pigs. The signalling pathway mediated by mTOR has been well-studied for its key function in regulation of cell growth and cell metabolisms relevant to lipids, protein and RNA [13]. Although mTOR activity has been implied to correlate with IFN signalling, the involvement of mTOR signalling in PRRSV infection and relevant antiviral responses have not been studied [12, 14, 15]. To determine if mTOR signalling is involved in type I IFN production upon PRRSV infection, we analysed the expression of type I IFN genes in virus-infected porcine cells with pharmaceutical modulation of mTOR signalling. Porcine alveolar macrophages were infected with PRRSV along with mTOR inhibitors or an activator, and the expression of type I IFN and their specific receptor genes was examined using validated real time pcr (RT-PCR) assays [37]. We found that PRRSV infection repressed the expression of most type I IFN genes including IFN-α1/5/6/9/12, IFN-δ1/3/4/5 and IFN-ω1/2/5. Among these IFNs, IFN-α6, IFN-α9 and IFN-ω5 exert the highest anti-PRRSV activity as shown in our previous studies (Fig. 4a) [37, 38]. Meanwhile, the porcine IFN-αω and IFN-β subtypes, which generally had less virostatic activity than porcine IFN-α subtypes, were stimulated by PRRSV infection (Fig. 4a). Corresponding to this observation, we and others have postulated that family-wide evaluations and subtype-specific regulation of porcine type I IFNs are critical for studying PRRSV pathogenesis and antiviral responses, rather than only evaluating a few IFN subtypes (primarily IFN-α1 and IFN-β) [5, 37].
Fig. 4.
mTOR inhibitors or mTOR activator modulate gene expression of type I IFNs and their receptors in PRRSV-infected porcine alveolar macrophages. Cells were incubated with DsRed-labelled PRRSV (m.o.i. of 0.5) for 1 h, and then treated with mTOR inhibitors (rapamycin 1 µM or PP242 2 µM) or activator (MHY1485 2 µM) for 12 h. Total RNAs were extracted to analyse expression of type I IFNs and their receptors in cells infected with PRRSV or not (a) and PRRSV-infected cells with/without inhibitor or activator treatments (b) by specific primers using two-step RT-PCR. Control was normalized to 0; n=3; a, b and c indicate p<0.001, 0.01 and 0.05, respectively to the control.
Upon treatment with mediators of the mTOR pathway, we observed that PP242, a non-selective mTORC inhibitor, significantly reversed the suppression of all analysed type I IFN subtypes except for IFN-αω and IFN-β (Fig. 4b). In comparison to PP242, the mTORC1 inhibitor rapamycin and activator MHY1485 showed much less effect on PRRSV-induced inhibition of type I IFN gene expression. In addition, we also examined the expression of cellular receptors for type I IFNs, IFNAR1 and IFNAR2. PP242 treatment stimulated expression levels of IFNAR1 and IFNAR2 genes; in contrast, the mTOR activator MHY1485 suppressed expression of both IFN receptor genes. This indicates that suppression of the mTOR signalling pathway by the inhibitor PP242 induces a cellular stress response, which strengthens IFN responses via synergistic stimulation of both IFN production and signalling perceiving by IFN receptors. In addition, comparison between the effects of rapamycin and PP242 also revealed that the mTOR signalling pathway, especially mTORC2 signalling, has a predominant role in regulating the transcription of genes relevant to type I IFN production and signalling (Fig. 4b). Furthermore, we measured IFN-α proteins secreted in culture supernatants collected from MARC-145 cells treated with mTOR inhibitors or activator. We showed that the mTOR inhibitor, PP242, increased IFN-α production, the most effective IFN subtype that acts against PRRSV, potentially providing a way to counteract viral suppression on IFN production (Fig. 5a) [5, 37]. In summary, our data show an inter-systemic regulation of antiviral responses at the cellular level, suppressing viral infection via modulation of mTOR signalling associated with cell metabolic status. Thus, the crosstalk between the mTOR and type I IFN signalling pathways implies multifunctional properties of mTOR and type I IFNs in regulation of both metabolic and immune responses, which reveals more targets for antiviral regulation.
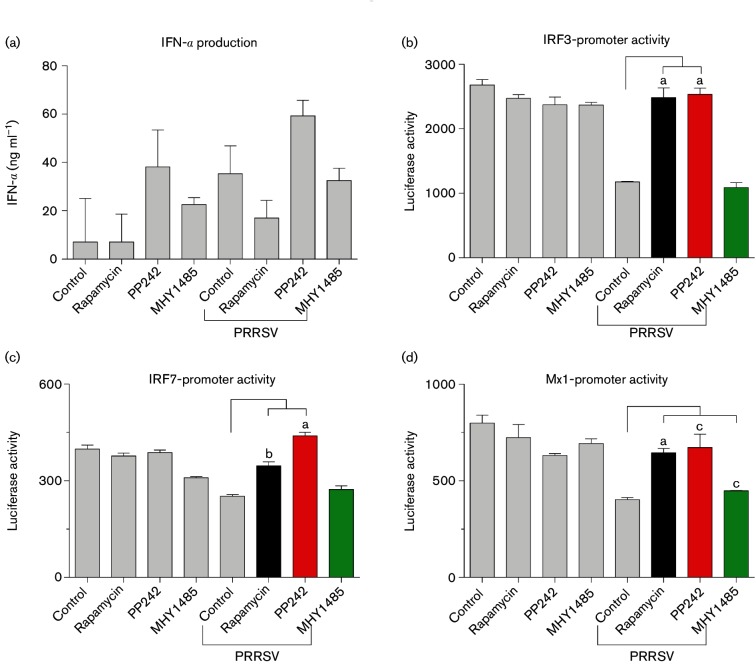
Fig. 5.
Inhibition of the mTOR signalling pathway promotes type I IFN production and signalling. MARC-145 cells were treated with mTOR inhibitors (rapamycin and PP242) and activator (MHY1485) for 24 h, and then infected with DsRed-labelled PRRSV (m.o.i. of 1) for another 24 h. Cell culture supernatants were collected and PRRSV was inactivated with UV illumination. Supernatants were used to measure IFNs with an ELISA for detection of IFN-α subtypes (a), or a bioassay in MARC-145 cells stably transformed with an IRF3-, IRF7- or Mx1-promoter driven luciferase reporter system (b–d); n=3; a, b and c indicate p<0.001, 0.01 and 0.05, respectively, relative to the control.
Through interaction with their specific receptors, type I IFNs induce expression of a myriad of IFN-stimulated genes (ISGs) to exert antiviral and other biological functions [1, 3, 9]. ISGs are a collection of hundreds of genes up-regulated in response to IFN production that have diverse roles in antiviral regulation. Typical ISGs, including PRRs, IRFs and other signal transducing proteins, function in magnifying type I IFN signalling or directly inactivating viruses [9]. IRFs, particularly IRF3 and IRF7, are critical transcription factors in mediation of signalling of both type I IFN production and action. Not only functioning in stimulating type I IFN production, IRF3 and IRF7 are also up-regulated by type I IFNs produced upon viral infection to form a positive regulatory loop in type I IFN signalling. Therefore, IRF3 and IRF7 are maker genes of ISGs to indicate activation of type I IFN signalling. Using the promoter-reporter system constructed with central promoter elements of human IRF3, IRF7 and Mx1 genes [37], we showed that PRRSV infection suppressed promoter activity of IRF3, IRF7 and Mx1, and the suppression was successfully reversed by mTOR inhibitors rapamycin and PP242 at physiological concentrations (Fig. 5b–d). In contrast, the mTOR activator, MHY1485, had little effect. In eliciting IFN gene expression in different cell types, IRF3 acts to activate the IFN-β gene, and IRF7 primarily activates the IFN-α gene [39]. Our data showed that PP242 increased IRF7 promoter activity more than rapamycin, which correlates with PP242’s higher effect in stimulating IFN-α expression (Fig. 4b), and viral repression. In general, most porcine IFN-α subtypes exert much higher anti-PRRSV activity than IFN-β. In this context, the interferon-induced dynamin-like GTPase gene, Mx1, encodes a typical effector ISG to directly restrict viral infection, and serves as an indicator for type I IFN action [39]. Similarly, we showed that inhibition of mTOR signalling by rapamycin, and in particular PP242, enhanced the promoter activity of the Mx1 gene, indicating higher antiviral IFN responses elicited by inhibition of mTOR signalling (Fig. 5d).
Collectively, our findings indicate that PRRSV replication and antiviral response in cells are significantly regulated through modulating mTOR signalling, which in turn affects both cell metabolic and immune statuses. Furthermore, through detection of type I IFN production at mRNA and protein levels as well as determination of ISG-stimulating activity, we found that the regulation of mTOR signalling in antiviral responses is, at least in part, mediated by changing type I IFN production and action signalling. Although mTOR signalling is involved in multiple physiological and metabolic processes, our results clearly revealed that the interaction between the type I IFN and mTOR pathways likely plays a major role in anti-PRRSV regulation. Together with direct analyses of type I IFN gene expression and protein secretion, the data imply that modulation of the mTOR signalling pathway, in particular those aspects mediated by mTORC2, could significantly potentiate type I IFN signalling, and reverse PRRSV-suppression on type I IFN signalling. Thus, polarization of cell metabolic statuses through modulation of the mTOR signalling pathway may provide an alternative to potentiate IFN responses for antiviral regulation.
Genetic silencing of mTOR signalling regulates PRRSV infection
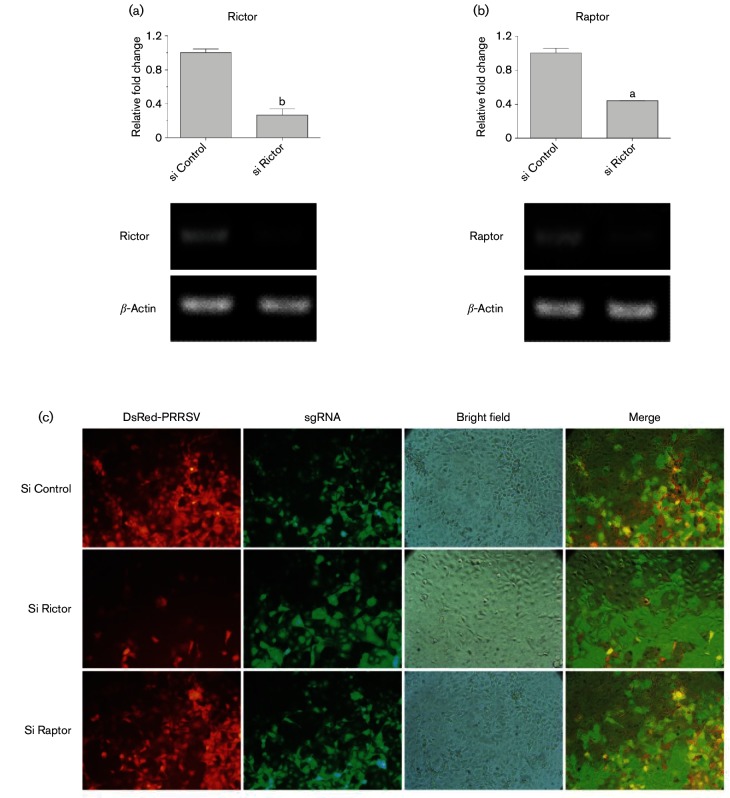
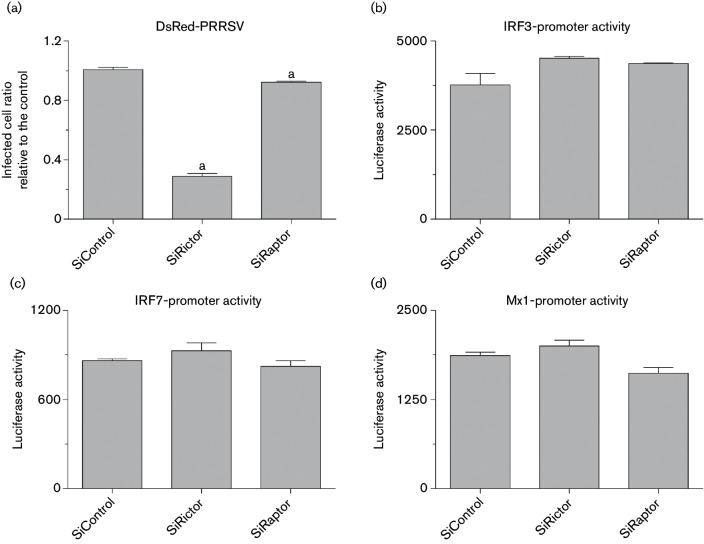
After pharmaceutical demonstration of the involvement of mTOR signalling in regulation of IFN-mediated cell antiviral responses, we genetically manipulated two key subunits of mTORC1 and mTORC2 to confirm this observation. Studies showed that knockout of Raptor or Rictor correspondingly blocked mTORC1 or mTORC2 pathway activity, respectively [40, 41]. Using the newly developed genome editing system, CRISRP/Cas9, we suppressed transcription of either Rictor or Raptor in nearly 60 % of MARC-145 cells (Fig. 6a, b). Consistent with our observation using mTOR inhibitors, genetic suppression of Rictor, the essential factor for mTORC2, significantly suppressed PRRSV infection in MARC-145 cells (Figs 6c and 7a). In contrast, PRRSV infection was only slightly inhibited by silencing Raptor, the component of mTORC1. Therefore, our tests reproducibly showed that inhibition of mTORC2 activity either pharmaceutically by PP242 or through gene silencing of Rictor induced protection against PRRSV infection in MARC-145 cells. Analyses of culture supernatants using bioassays of the ISG promoter-reporter system verified that gene silencing of Rictor enhanced the up-regulation of promoter activity of ISG genes including IRF3, IRF7 and Mx1, plausibly through increasing type I IFN production in Rictor-silent cells (Fig. 7b–d). The data obtained from both pharmaceutical treatments and genetic manipulation indicate that signalling mediated by mTORC2, but not mTORC1, is associated more with cell antiviral response, and that suppression of mTORC2 activity may provide a selective target to regulate anti-PRRSV immune responses. Compared with studies on mTORC1, little is known about biological functions mediated by mTORC2. Our results imply a potential immune regulation role of mTORC2 rather than the primary metabolic regulation mediated by mTORC1 [42]. Further studies are required to understand signalling cascades and molecular mechanisms of mTORC2 in the regulation of antiviral immunity, in particular, its potential to be targeted for antiviral regulation in vivo.
Fig. 6.
Suppression of Rictor or Raptor gene expression differently suppresses PRRSV replication in MARC-145 cells. Gene expression of Rictor and Raptor was down-regulated using the CRISPR/Cas9 system. (a,b) Gene expression analysis to show Rictor or Raptor gene expression was down-regulated nearly 60 % dependent on the transformation efficiency (>90 % of gRNA). (c) MARC-145-sgControl/sgRictor/sgRaptor cells were infected with DsRed-labelled PRRSV (m.o.i. of 1), visualized and imaged with fluorescence microscopy at 36 h p.i. n=3; a, b and c indicate p<0.001, 0.01 and 0.05, respectively, relative to the control.
Fig. 7.
Suppression of Rictor, the key subunit of mTORC2, significantly suppresses PRRSV replication in MARC-145 cells, through regulating type I IFN production and action signalling. MARC-145-sgControl/sgRictor/sgRaptor cells were infected with DsRed-labelled PRRSV (m.o.i. of 1), and quantified using a SpectraMax i3 at 36 h p.i. (a); or infected with DsRed-labelled PRRSV for 24 h. Then cell culture supernatants were collected and PRRSV was inactivated with UV illumination. (b–d) Supernatants were used to detect IFN production and action with a bioassay in MARC-145 cells stably transformed with an IRF3-, IRF7- or Mx1-promoter driven luciferase reporter system; n=3; a, b and c indicate p<0.001, 0.01 and 0.05, respectively, relative to the control.
Methods
Cells and viruses
Experiments involving animals and viruses were approved by the Kansas State University Institutional Animal Care and Use and Biosafety Committees. Animal procedures and isolation of porcine alveolar macrophages and peripheral blood mononuclear cells (PBMCs) were previously described [21, 27]. In brief, 5-week-old clinically healthy pigs from a herd without viral infection history were used for obtaining primary cells. PBMCs were isolated from blood collected by jugular venipuncture from anaesthetized pigs, using Histopaque-1077 (Sigma). Immediately after euthanasia, macrophages were obtained by lavaging lungs with 1× PBS (pH7.4, Sigma), then washing cells three times with RPMI (Roswell Park Memorial Institute)-1640 medium (Gibco). Isolated primary cells were used immediately or cryopreserved in Recovery Cell Culture Freezing Medium (Gibco). African green monkey kidney (MARC-145; ATCC) cells were grown in modified Eagle’s medium (MEM, Gibco) containing 8 % foetal bovine serum (FBS; Gibco) and 1× antibiotic-antimycotic (Gibco). 293FT (Invitrogen) cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10 % FBS (Gibco), 1× MEM non-essential amino acids solution (NEAA; Gibco). Macrophages and PBMCs were maintained in RPMI-1640 medium containing 10 % FBS and 1× antibiotic-antimycotic. mDCs were generated from PBMCs stimulated with IL-4 (2 ng ml−1) and GM-CSF (5 ng ml−1) (R&D Systems) and maintained in RPMI-1640 medium containing 10 % FBS and 1× antibiotic-antimycotic [43]. P129-GFP and DsRed-labelled PRRSV were used in this study [44].
Cell polarization, viral infection and transcriptomic shotgun sequencing
Procedures for macrophage polarization were performed as previously described [21, 27]. Briefly, porcine alveolar macrophages were stimulated with mediators (LPS, IFN-α1, IFN-γ, IL-4 and IL-10; R&D Systems) at 20 ng ml−1 for 30 h, followed by infection of P129-GFP at a multiplicity of infection (m.o.i.) of 0.1 for 5 h. After polarization and infection, cells were washed twice with fresh culture medium, and then total RNA was extracted from 3×107 cells of each treatment using RNA/DNA/protein purification kit (Norgen Biotek). To be qualified for constructing RNA-Seq libraries, RNA concentration and quality were evaluated with a NanoDrop 8000 spectrometer (NanoDrop) and Agilent 2100 Bioanalyzer (Agilent Technologies) to ensure RNA samples with A260/A280 >1.8 and RNA integrity number (RIN) >7.0. All transcriptomic shotgun sequencing was conducted following the procedures of Illumina Pipeline (BGI Americas). Genome-wide transcriptomic analysis was performed using 25–30 M clean reads per sample. Data analyses were conducted as previously described [21, 27].
Antiviral analysis
MARC-145 cells were treated with mTOR inhibitors rapamycin and PP242, or mTOR activator MHY1485 (all Sigma), at different concentrations for 12, 24, 36 or 48 h, and then infected with DsRed-labelled PRRSV at an m.o.i. of 1.0 for 36 h. All images were collected using a Nikon fluorescence microscope at a magnification of ×20, and viral infection was quantified with a SpectraMax i3 (Molecular Devices). Porcine primary cells were infected with DsRed-labelled PRRSV at an m.o.i. of 0.5 with mTOR inhibitors or activator for 20 h, visualized with fluorescence microscopy and quantified using a SpectraMax i3. All chemicals used were dissolved in dimethyl sulfoxide (DMSO, cell culture grade; Sigma).
Bioassays and ELISA
MARC-145 cells were treated with mTOR inhibitors or activator for 24 h, and infected with DsRed-labelled PRRSV at an m.o.i. of 1.0 for another 24 h. Supernatants from each cell culture sample were inactivated with UV light for 1 h, and used to measure IFNs with a bioassay in MARC-145 cells stably transformed with IRF3-, IRF7- or Mx1-promoter driven luciferase reporter systems, or an ELISA kit (R&D System a) for detecting IFN-α subtypes [37]. In brief, MARC-145 (IRF3, IRF7 or Mx1) cells were treated with inactivated supernatants for 24 h, lysed with Glo lysis buffer and quantified by Steady-Glo Luciferase Assay System (Promega).
Real-time PCR and Western blotting assay
The primer information used for PCR assays of porcine type I IFNs was adapted from our previous publications (Tables S1 and S2, Sang et al. [5, 37]). Porcine alveolar macrophages were infected with DsRed-labelled PRRSV at an m.o.i of 0.5 with treatment of mTOR inhibitors or activator for 12 h. Total RNA was extracted from 2×105 cells of each treatment using an RNA/DNA/protein purification kit. Real-Time PCR (RT-PCR) assay was performed using a GoTaq 2-Step RT-PCR System (Promega). Total cDNA was reverse transcribed from RNA pools (2 µg RNA in a 20 µl reaction mixture) using random primers. RT-PCR analysis was conducted using a StepOnePlus RT-PCR system (Applied Biosystems). Reactions were run with 1 µl cDNA in a 20 µl reaction mixture, and set at 95 °C for 2 min followed by 40 amplification cycles of 95 °C for 15 s, 60 °C for 1 min with a melting curve, 95 °C for 15 s, 60 °C for 1 min, a ramp from 60 to 95 °C at an 1 % rate, and 95 °C for 15s. Critical threshold (Ct) values and melt curves were monitored and collected. Relative gene expression data were first normalized against Ct values of the housekeeping gene (β-Actin), and the relative expression index (2-ΔΔCt) was determined compared with expression levels of control sample for stimulated regulation.
Gene silencing based on the CRISPR/Cas9 system
pHR-Cas9-2A-puro and phU6/BB-GFP plasmids were constructed on the basis of pHR-SFFV-dCas9-BFP (Addgene, no. 46910), pSpCas9(BB)-2A-GFP (Addgene, no. 48138) and pgRNA-humanized (Addgene, no. 44248). All sgRNA expression constructs were obtained with BbsI by inserting an annealed oligo pair encoding 20 nt guide sequences. All restriction enzymes, Quick Ligation kit, Quick-Load Taq ×2 Master Mix and Phusion High-Fidelity PCR kit were purchased from New England Biolabs.
Lentiviral constructs for efficient transfection and expression of Cas9 and sgRNA in mammalian cells were produced using a second-generation lentiviral system with pMD2.G (Addgene, no. 12259) and psPAX2 (Addgene, no. 12260). 293FT cells were transfected with envelope plasmid (pMD2.G), packaging plasmid (psPAX2) and transfer plasmid (pHR-Cas9-2A-puro for Cas9 expression or phU6/BB-xx-GFP for sgRNA expression) at a ratio of 0.9 : 1.5 : 2.1 or 0.9 : 1.5 : 1.5 (μg). Briefly, 293FT cells were grown in 24-well plates overnight to reach 70 % confluence and transfected with 1 µg DNA/well in total using XtremeGene9 transfection reagent (Roche) at a transfection reagent : DNA ratio of 2.5 : 1. After incubation for 8 h, the growth medium was refreshed, and lentivirus-containing supernatants were collected for transducing target cells at 48 h post-transfection. The MARC-145 cells that stably express Cas9-2A-puro were enriched by lentiviral transduction, and selected with a complete culture medium including 7 µg ml−1 puromycin (InvivoGen). After obtaining the Cas9-expressing cell line, five different sgRNA-expressing lentiviruses for Rictor and three different sgRNA-expressing lentiviruses for Raptor were combined and transformed into Cas9-expressing cells to knockout/down-regulate target genes as described previously [45].
Statistical analysis
All statistical analyses were performed using Student’s t-test. Data are presented as mean±sem. P<0.05 was considered statistically significant.
Funding information
This work was supported by USDA NIFA AFRI 2013-67015-21236 and USDA NIFA AFRI 2015-665 67015-23216 to Y. S. We thank the Department of Anatomy and Physiology COBRE facility cores (funded by NIH P20-RR017686 and College of Veterinary Medicine of Kansas State University).
Acknowledgements
We thank our laboratory manager, Dr Barbara Lutjemeier, and the undergraduate research assistants, Mr Wyatt Brichalli, Mr Joseph Bergkamp and Ms Jessica Pearson at Kansas State University for their excellent technical support.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: DEG, differentially expressed gene; IRF, interferon regulatory factor; ISG, interferon-stimulated gene; LPS, lipopolysaccharide; mDC, monocyte-derived dendritic cells; mTOR, mechanisitic target of rapamycin; mTORC, mTOR complex; PRR, pattern recognition receptor; PRRSV, porcine reproductive and respiratory syndrome virus; RT-PCR, real-time PCR.
Two supplementary figures and two supplementary tables are available with the online Supplementary Material.
References
- 1.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann HH, Schneider WM, Rice CM. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Weerd NA, Nguyen T. The interferons and their receptors—distribution and regulation. Immunol Cell Biol. 2012;90:483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sang Y, Bergkamp J, Blecha F. Molecular evolution of the porcine type I interferon family: subtype-specific expression and antiviral activity. PLoS One. 2014;9:e112378. doi: 10.1371/journal.pone.0112378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem. 2007;282:20047–20051. doi: 10.1074/jbc.R700004200. [DOI] [PubMed] [Google Scholar]
- 7.Levy DE, Marié IJ, Durbin JE. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol. 2011;1:476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 11.Saleiro D, Platanias LC. Intersection of mTOR and STAT signaling in immunity. Trends Immunol. 2015;36:21–29. doi: 10.1016/j.it.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saleiro D, Platanias LC. ULK1 in type I interferon response. Oncotarget. 2015;6:24586–24587. doi: 10.18632/oncotarget.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarogoulidis P, Lampaki S, Turner JF, Huang H, Kakolyris S, et al. mTOR pathway: a current, up-to-date mini-review. Oncol Lett. 2014;8:2367–2370. doi: 10.3892/ol.2014.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa-Mattioli M, Sonenberg N. RAPping production of type I interferon in pDCs through mTOR. Nat Immunol. 2008;9:1097–1099. doi: 10.1038/ni1008-1097. [DOI] [PubMed] [Google Scholar]
- 15.Livingstone M, Sikström K, Robert PA, Uzé G, Larsson O, et al. Assessment of mTOR-dependent translational regulation of interferon stimulated genes. PLoS One. 2015;10:e0133482. doi: 10.1371/journal.pone.0133482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, et al. Toll-like receptor–mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur S, Lal L, Sassano A, Majchrzak-Kita B, Srikanth M, et al. Regulatory effects of mammalian target of rapamycin-activated pathways in type I and II interferon signaling. J Biol Chem. 2007;282:1757–1768. doi: 10.1074/jbc.M607365200. [DOI] [PubMed] [Google Scholar]
- 18.Kaur S, Sassano A, Dolniak B, Joshi S, Majchrzak-Kita B, et al. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci USA. 2008;105:4808–4813. doi: 10.1073/pnas.0710907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saleiro D, Mehrotra S, Kroczynska B, Beauchamp EM, Lisowski P, et al. Central role of ULK1 in type I interferon signaling. Cell Rep. 2015;11:605–617. doi: 10.1016/j.celrep.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lekmine F, Uddin S, Sassano A, Parmar S, Brachmann SM, et al. Activation of the p70 S6 kinase and phosphorylation of the 4E-BP1 repressor of mRNA translation by type I interferons. J Biol Chem. 2003;278:27772–27780. doi: 10.1074/jbc.M301364200. [DOI] [PubMed] [Google Scholar]
- 21.Sang Y, Brichalli W, Rowland RR, Blecha F. Genome-wide analysis of antiviral signature genes in porcine macrophages at different activation statuses. PLoS One. 2014;9:e87613. doi: 10.1371/journal.pone.0087613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katholnig K, Linke M, Pham H, Hengstschläger M, Weichhart T. Immune responses of macrophages and dendritic cells regulated by mTOR signalling. Biochem Soc Trans. 2013;41:927–933. doi: 10.1042/BST20130032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon S. The role of the macrophage in immune regulation. Res Immunol. 1998;149:685–688. doi: 10.1016/S0923-2494(99)80039-X. [DOI] [PubMed] [Google Scholar]
- 24.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 25.Sang Y, Miller LC, Blecha F. Macrophage polarization in virus-host interactions. J Clin Cell Immunol. 2015;6 doi: 10.4172/2155-9899.1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weichhart T, Hengstschläger M, Linke M. Regulation of innate immune cell function by mTOR. Nat Rev Immunol. 2015;15:599–614. doi: 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sang Y, Rowland RR, Blecha F. Antiviral regulation in porcine monocytic cells at different activation states. J Virol. 2014;88:11395–11410. doi: 10.1128/JVI.01714-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lisi L, Aceto P, Navarra P, dello Russo C. mTOR kinase: a possible pharmacological target in the management of chronic pain. Biomed Res Int. 2015;2015:1–13. doi: 10.1155/2015/394257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 30.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e1000038. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi YJ, Park YJ, Park JY, Jeong HO, Kim DH, et al. Inhibitory effect of mTOR activator MHY1485 on autophagy: suppression of lysosomal fusion. PLoS One. 2012;7:e43418. doi: 10.1371/journal.pone.0043418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun MX, Huang L, Wang R, Yu YL, Li C, et al. Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy. 2012;8:1434–1447. doi: 10.4161/auto.21159. [DOI] [PubMed] [Google Scholar]
- 33.Duan X, Nauwynck HJ, Pensaert MB. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV) Arch Virol. 1997;142:2483–2497. doi: 10.1007/s007050050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sang Y, Rowland RR, Blecha F. Interaction between innate immunity and porcine reproductive and respiratory syndrome virus. Anim Health Res Rev. 2011;12:149–167. doi: 10.1017/S1466252311000144. [DOI] [PubMed] [Google Scholar]
- 35.Huang C, Zhang Q, Feng WH. Regulation and evasion of antiviral immune responses by porcine reproductive and respiratory syndrome virus. Virus Res. 2015;202:101–111. doi: 10.1016/j.virusres.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, et al. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci. 2016;4:129–154. doi: 10.1146/annurev-animal-022114-111025. [DOI] [PubMed] [Google Scholar]
- 37.Sang Y, Rowland RR, Hesse RA, Blecha F. Differential expression and activity of the porcine type I interferon family. Physiol Genomics. 2010;42:248–258. doi: 10.1152/physiolgenomics.00198.2009. [DOI] [PubMed] [Google Scholar]
- 38.Sang Y, Rowland RR, Blecha F. Porcine type I interferons: polymorphic sequences and activity against PRRSV. BMC Proc. 2011;5:S8. doi: 10.1186/1753-6561-5-S4-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnorr JJ, Schneider-Schaulies S, Simon-Jödicke A, Pavlovic J, Horisberger MA, et al. MxA-dependent inhibition of measles virus glycoprotein synthesis in a stably transfected human monocytic cell line. J Virol. 1993;67:4760–4768. doi: 10.1128/jvi.67.8.4760-4768.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 41.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 42.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loving CL, Brockmeier SL, Sacco RE. Differential type I interferon activation and susceptibility of dendritic cell populations to porcine arterivirus. Immunology. 2007;120:217–229. doi: 10.1111/j.1365-2567.2006.02493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sang Y, Shi J, Sang W, Rowland RR, Blecha F. Replication-competent recombinant porcine reproductive and respiratory syndrome (PRRS) viruses expressing indicator proteins and antiviral cytokines. Viruses. 2012;4:102–116. doi: 10.3390/v4010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.