Abstract

Neuropeptides in several animals undergo an unusual post-translational modification, the isomerization of an amino acid residue from the l-stereoisomer to the d-stereoisomer. The resulting d-amino acid-containing peptide (DAACP) often displays biological activity higher than that of its all-l-residue analogue, with the d-residue being critical for function in many cases. However, little is known about the full physiological roles played by DAACPs, and few studies have examined the interaction of DAACPs with their cognate receptors. Here, we characterized the signaling of several DAACPs derived from a single neuropeptide prohormone, the Aplysia californica achatin-like neuropeptide precursor (apALNP), at their putative receptor, the achatin-like neuropeptide receptor (apALNR). We first used quantitative polymerase chain reaction and in situ hybridization experiments to demonstrate receptor (apALNR) expression throughout the central nervous system; on the basis of the expression pattern, we identified novel physiological functions that may be mediated by apALNR. To gain insight into ligand signaling through apALNR, we created a library of native and non-native neuropeptide analogues derived from apALNP (the neuropeptide prohormone) and evaluated them for activity in cells co-transfected with apALNR and the promiscuous Gα subunit Gα-16. Several of these neuropeptide analogues were also evaluated for their ability to induce circuit activity in a well-defined neural network associated with feeding behavior in intact ganglia from Aplysia. Our results reveal the specificity of apALNR and provide strong evidence that this receptor mediates diverse physiological functions throughout the central nervous system. Finally, we show that some native apALNP-derived DAACPs exhibit enhanced stability toward endogenous proteases, suggesting that the d-residues in these DAACPs may increase the peptide lifetime, in addition to influencing receptor specificity, in the nervous system. Ultimately, these studies provide insight into signaling at one of the few known DAACP-specific receptors and advance our understanding of the roles that l- to d-residue isomerization play in neuropeptide signaling.
Polypeptides, including neuropeptides, often undergo post-translational modifications (PTMs) that significantly influence their biological activity.1−3 One poorly understood PTM is the enzyme-catalyzed isomerization of one amino acid residue from the l-stereoisomer to the d-stereoisomer (Figure 1a) to form a d-amino acid-containing peptide (DAACP).4−10 DAACPs have been identified from diverse animals in multiple phyla, where they act as neuropeptides,6−9 hormones,10 and toxins4,11 and, in many cases, are significantly more biologically active than their all-l-residue analogues.7,8,12 DAACPs are difficult to identify by commonly used mass spectrometry (MS)-based peptide characterization techniques because DAACPs and their all-l-residue diastereomers have identical masses. Methods have been recently developed to address this challenge and enhance the identification of DAACPs.8,9,13−16 Nonetheless, the full functions of identified DAACPs are still unknown, and many DAACPs may remain unidentified. In addition, although d-residues have been shown to increase the protease resistance of synthetic peptides12,17 and several DAACP toxins and toxin fragments,4,5 relatively few studies have directly examined the influence of d-residues on the stability of cell–cell signaling DAACPs to central nervous system (CNS) proteases.
Figure 1.
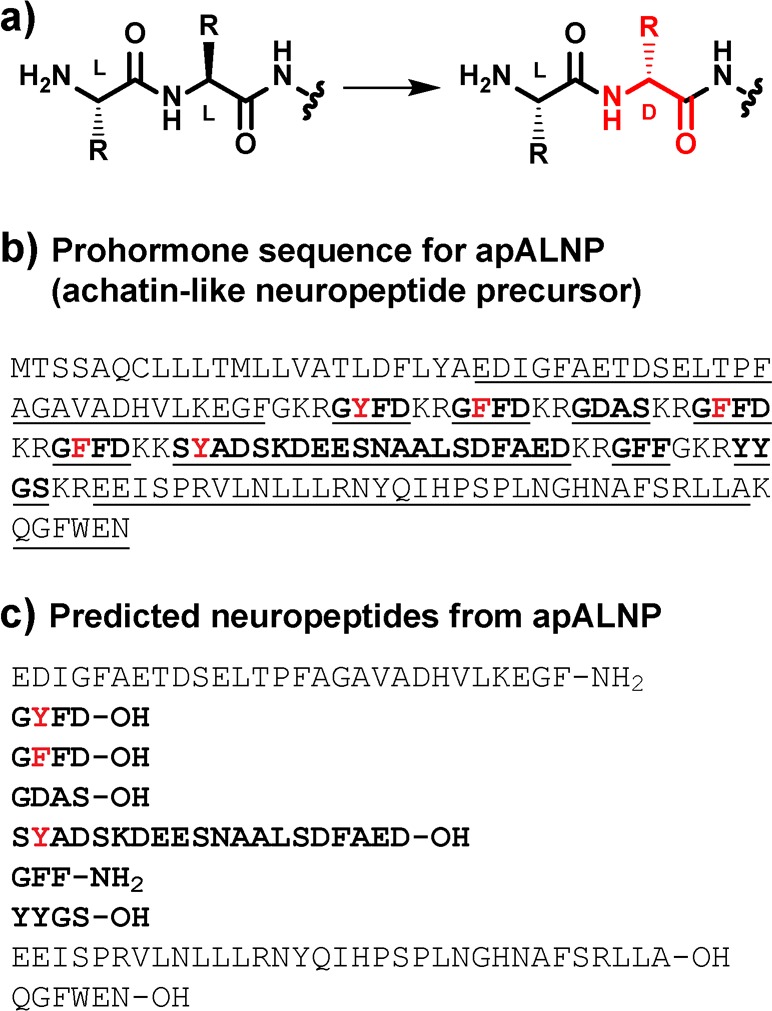
(a) Generation of a DAACP by an l/d-isomerase. (b) Protein sequence of apALNP (GenBank entry AAW30457.1). Predicted peptides generated from prohormone processing25 of apALNP are underlined and listed in panel c. Note that SdYADSKDEESNAALSDFAED-OH was isolated as SdYADSKDEESNAALSDFA-OH in a prior report9 (see the note in the Supporting Information). For panels b and c, peptides in bold were tested for apALNR activation (see Table 1). Residues previously identified to be converted to d-residues are colored red.
The marine mollusk Aplysia californica is an excellent model for investigations of learning and memory, neural circuits, neurochemistry, and neural signaling because it possesses a relatively small number of neurons, many of which are easily identifiable and have well-defined functions.8,9,18−23 Importantly, four DAACPs have been reported in Aplysia: GdFFD-OH,6,8 GdYFD-OH,9 SdYADSKDEESNAALSDFA-OH,9 and NdWF-NH2.7 (In this report, a lowercase “d” indicates the following residue is a d-residue.) GdFFD-OH, GdYFD-OH, and SdYADSKDEESNAALSDFA-OH each arise from post-translational processing of a single precursor, the Aplysia achatin-like neuropeptide precursor [apALNP (Figure 1b,c)]. GdFFD-OH and GdYFD-OH are active in the feeding and locomotor circuits, as assessed by electrophysiology and behavioral experiments,8,9,24 whereas SdYADSKDEESNAALSDFA-OH is inactive in these circuits. NdWF-NH2, which is not derived from apALNP, is cardioactive. For all three active DAACPs in Aplysia, the results of experiments with the all-l-residue analogues demonstrated that the d-residue is essential for bioactivity, highlighting the importance of this PTM for function.
Several peptide toxins and designed therapeutics bearing d-residues interact with mammalian neuropeptide receptors,11,26 but the ways in which endogenous DAACPs enact their functions are less well understood, primarily because few receptors for cell–cell signaling DAACPs are known. As part of a recent study of G protein-coupled receptor (GPCR) deorphanization in Platynereis, Bauknecht and Jékely27 identified related GPCRs from several phyla, here called achatin-like neuropeptide receptors (ALNRs), that were activated by DAACPs derived from ALNP homologues. The authors showed that the Aplysia ALNR [apALNR (Figures S1 and S2)] was activated by GdFFD-OH, but not by GFFD-OH, suggesting that this receptor may be the endogenous receptor for GdFFD-OH. To the best of our knowledge, ALNRs are among the first GPCRs identified that appear to recognize DAACPs as their native ligands. However, outside of identifying this ligand–receptor pair, little is known about the range of functions mediated through ALNRs in the CNS.
A critical examination of an ALNR, including its expression and ligand specificity, would clarify the roles played by ALNP-derived DAACPs in the CNS. Here, we study the molecular and physiological actions of Aplysia apALNP-derived peptides through apALNR. Overall, our results represent one of the first detailed studies of a GPCR that appears to signal primarily through a DAACP and provide evidence that the functions of apALNP-derived DAACPs extend beyond feeding and locomotor circuits to control other behaviors throughout the CNS. Ultimately, our results expand the roles that DAACPs play in the Aplysia CNS and help to clarify the role of l- to d-residue isomerization in both the function and biological stability of these neuropeptides.
Results and Discussion
Expression of apALNR across the CNS
Understanding the tissue localization of receptor expression (e.g., which specific cells transcribe mRNA for the receptor) is a prerequisite for understanding the physiological functions enacted through the receptor.28,29 In Aplysia, regions of the CNS (and even many specific neurons) are mapped to well-known physiological and behavioral functions. Because GdFFD-OH is active in the feeding and locomotor circuits,8,24 controlled primarily by neurons in the buccal and pedal ganglia, respectively, we predicted apALNR would be expressed in these ganglia. The extent of apALNR expression in other tissues is not known.
To gain insight into the relative expression of apALNP and apALNR across different tissues, we performed quantitative polymerase chain reaction (qPCR) experiments to measure gene expression for each major ganglion in the CNS [buccal, cerebral, pedal, pleural, and abdominal (Figure 2a and Figures S3–S5)], as well as other non-CNS tissues (the buccal mass and gill). In contrast to the pedal ganglia-specific expression for apALNP (Figure 2b), which is consistent with previous in situ hybridization (ISH) experiments,8 we observed that apALNR mRNA was present at relatively uniform levels across each ganglion of the CNS (Figure 2c). As expected for a GPCR,30 the expression level of apALNR is low compared to that of the neuropeptide prohormone apALNP, whose products are targeted for release and are generally not recycled.
Figure 2.
(a) Cartoon representation of the Aplysia CNS, highlighting the relative positions of each major ganglion and major connective nerves (cartoon not drawn to scale). Relative expression of (b) apALNP (neuropeptide prohormone) and (c) apALNR (receptor) in various tissues, as determined by qPCR. The buccal mass and gill were included as non-neural tissue. Note the differences in the vertical axes. Individual points represent each of three biological sets of five animals. Bars represent the mean ± the standard deviation (SD) of these three biological sets.
To investigate the cellular localization of apALNR, we performed ISH using digoxigenin-labeled riboprobes that bind apALNR mRNA. ISH is a useful qualitative technique for identifying specific cells that express a given transcript, and the results complement the quantitative information gained from qPCR measurements. Consistent with gene expression measurements by qPCR, our ISH experiments for apALNR mRNA revealed cell-specific staining in the buccal, cerebral, pedal, and pleural ganglia (Figure 3 and Figure S6). In the buccal ganglia, staining was most obvious in several of the large motor neurons located at the ventral surface. This staining in the buccal ganglia is consistent with the activity reported for GdFFD-OH and GdYFD-OH in the feeding network.8,9 In the cerebral ganglia, staining was most pronounced in the metacerebral cells (MCCs) and the region corresponding to the H and right G clusters. Weaker staining was observed on the outer lateral edge in the A/B cluster and in a small number of stained cells in the E clusters. In the pedal ganglia, expression was broadly distributed, with the most intense staining in septa IIIb and IIIc of the dorsal surface, consistent with the effects of GdFFD-OH on locomotor programs and locomotor behavior previously observed.9,24 Staining was weaker in neurons of the pleural ganglia than in the pedal ganglia, except for noticeable staining of LPl1, a giant neuron in the left pleural ganglion. The pattern of staining in the abdominal ganglia was not as consistent between preparations as it was for other ganglia examined (Figure S7), which could reflect technical issues or biological variability in expression between animals.
Figure 3.

Representative images showing the localization of apALNR mRNA by ISH across different ganglia in the Aplysia CNS. Antisense probes revealed apALNR expression (left), while sense probes were included as negative controls (right). (a) Buccal ganglia (caudal surface). (b) Cerebral ganglia (dorsal surface). MCCs highlighted with arrows. (c) Pedal and pleural ganglia (dorsal–lateral view). Legend: LPed, left pedal; RPed, right pedal; LPl, left pleural; RPl, right pleural. LPl1 highlighted with an arrow. Scale bars are 500 μm.
Together, the qPCR and ISH experiments confirmed the predicted expression of apALNR in the buccal and pedal ganglia but also revealed expression of apALNR in the cerebral, pleural, and abdominal ganglia, regions that were not previously anticipated to express receptors for apALNP-derived DAACPs. The cellular expression of apALNR immediately suggests that apALNP-derived DAACPs may play additional roles beyond the specific feeding and locomotor circuits previously identified.8,24 For example, the MCCs of the cerebral ganglia enhance the strength of buccal muscle contractions and modulate the output of the central pattern generator (CPG) for biting movements.31,32 Cell-specific expression of apALNR in the MCCs suggests that apALNP-derived DAACPs may modulate these specific feeding behaviors through these cells. Similarly, apALNR expression in LPl1 suggests that apALNP-derived DAACPs may help regulate defensive mucus release, a behavior mediated by this neuron.33
GdFFD-OH Increases the Excitability of LPl1
The presence of the apALNR transcript in LPl1, an easily identifiable cell asymmetrically present in the left pleural ganglion, made this cell a good candidate for testing the activity of apALNP-derived DAACPs for physiological functions outside of the feeding and locomotor circuits previously tested.8,9,24 Neuropeptides, through activation of cell-surface receptors, can increase or decrease the excitability of a neuron. To evaluate the effect of GdFFD-OH on LPl1, which expresses apALNR, we examined the excitability of the LPl1 neuron under different conditions by electrophysiology. In these experiments, 3 s constant current pulses were applied to LPl1 every 60 s and LPl1 excitability was measured by the number of spikes evoked by the current pulses during the 3 s. Consistent with its positive staining by apALNR ISH, we found that the excitability of LPl1 was increased by a perfusion of GdFFD-OH, but not by GFFD-OH, in electrophysiology experiments on isolated pleural–pedal ganglion preparations (Figure 4 and Figure S8). Recordings on LPl1 were performed in high-divalent saline, which limits polysynaptic influences. Of course, receptors other than apALNR may also be activated by GdFFD-OH, contributing to the observed modulation. Nevertheless, this finding confirms that apALNP-derived peptides can modulate the activity of neurons outside of the feeding and locomotor circuits, including a cell type known to function in defensive mucus release.
Figure 4.
LPl1 excitability (tested with 3 s current pulses every 60 s) after perfusion of isolated pleural and pedal ganglia of Aplysia with (a) GdFFD-OH [F(3,12) = 59.33; p < 0.001; n = 5] or (b) GFFD-OH [F(3,15) = 1; p > 0.05; n = 6] (see Figure S8). Control: activity before peptide perfusion. Wash: activity after washout of peptide. Bars represent the mean ± the standard error of the mean (SEM). Repeated measures analysis of variance and Bonferroni post hoc test: *p < 0.05; ***p < 0.001. Recordings were made in high-divalent saline.
Activation of apALNR and the Feeding Network by apALNP-Derived Peptides
We next sought to determine whether peptides from apALNP other than GdFFD-OH were ligands for apALNR. Post-translational processing of apALNP is predicted to give rise to several peptides, GFFD-OH, GYFD-OH, GDAS-OH, SYADSKDEESNAALSDFAED-OH, GFF-NH2, and YYGS-OH, and N- and C-terminal peptides (Figure 1b,c).25 Of these, we have identified GdYFD-OH and SdYADSKDEESNAALSDFA-OH as endogenous DAACPs in the CNS of Aplysia, in addition to GdFFD-OH.8,9
We synthesized neuropeptides predicted to arise from prohormone processing of apALNP (Figure 1b,c), along with DAACP analogues, to test for direct activation of apALNR in CHO-K1 cells expressing apALNR. For peptides that have not been detected as DAACPs, predicted DAACP analogues were designed with the d-residue at position 2, the location of the d-residue in all known molluscan DAACPs. Three predicted N- and C-terminal peptides, which lack structural similarity to known DAACPs from mollusks, were not evaluated. To determine apALNR activation in response to potential agonists, we transfected CHO-K1 cells with plasmids for the expression of apALNR and also Gα-16, a promiscuous Gα subunit that associates with most GPCRs and activates the phospholipase C signaling pathway.34 Activation of the canonical Gαq signaling was then measured using a commercially available assay that measures the accumulation of IP1 in the presence of LiCl upon activation of phospholipase C.35 Consistent with a prior report,27 we found that GdFFD-OH activates apALNR whereas GFFD-OH does not, using the IP1 accumulation assay (Table 1 and Figure S9). Interestingly, we found that GdYFD-OH was a potent agonist, with potency virtually identical to that of GdFFD-OH. The high apALNR potency for GdYFD-OH is consistent with the fact that this peptide displays high biological activity in physiological networks associated with feeding and locomotor behavior, similar to GdFFD-OH.9 In contrast, GYFD-OH, the all-l-residue analogue, was completely devoid of apALNR activity, indicating that the d-Tyr residue is critical for receptor activation for GdYFD-OH. Neither SdYADSKDEESNAALSDFAED-OH nor SYADSKDEESNAALSDFAED-OH activated apALNR up to 50000 nM (Table 1).
Table 1. apALNR Activation and Feeding Network Activity for apALNP-derived Peptidesa.
| peptide | apALNR EC50 (nM) (IP1 assay) | feeding network (electrophys.) |
|---|---|---|
| GdFFD-OH | 30 | active |
| GFFD-OH | >200000 | not active |
| GdYFD-OH | 30 | active |
| GYFD-OH | >500000 | not active |
| SdYADSKDEESNAALSDFAED-OH | >50000 | not active |
| SYADSKDEESNAALSDFAED-OH | >50000 | not active |
| GdFF-NH2 | >500000 | not active |
| GFF-NH2 | >500000 | not active |
| YdYGS-OH | >500000 | NT |
| YYGS-OH | >500000 | NT |
| GdDAS-OH | >500000 | NT |
| GDAS-OH | >500000 | NT |
Primary sequences of select peptides predicted from processing of apALNP, along with associated apALNR activation EC50 values, as determined by the IP1 accumulation assay in CHO-K1 cells transiently transfected with apALNR and Gα-16 (see Figure S9 for all dose–response curves). For apALNR-active compounds, EC50 values are the mean from at least three independent experiments. See Table S1 for the error associated with these measurements. For apALNR-inactive compounds, the EC50 is listed as being greater than the highest concentration tested, from at least two independent experiments showing no or negligible activity. Activity in the feeding network is determined by electrophysiology measurements on intact buccal and cerebral ganglia (Figure 6, Figure S10, and Figure S11). Active: perfusion of the peptide induced statistically higher circuit activity relative to that of control conditions with no peptide perfusion. Not active: no increase in activity was detected. NT: not tested. Feeding network activity values for GdFFD-OH/GFFD-OH and GdYFD-OH/GYFD-OH are from previous reports.8,9
We also tested the predicted neuropeptide diastereomers GdFF-NH2/GFF-NH2, GdDAS-OH/GDAS-OH, and YdYGS-OH/YYGS-OH for activation of apALNR using the IP1 accumulation assay. We have detected several of these peptides in homogenized ganglion extracts by liquid chromatography (LC)–electrospray ionization mass spectrometry (MS) or with single-cell matrix-assisted laser desorption/ionization (MALDI) MS,8,9 but the chirality of the endogenous peptides is currently unknown. We found that none of the structural variants of GFF-NH2, GDAS-OH, or YYGS-OH that we tested activated apALNR (Table 1), though some variants appeared to show minor activity at the highest concentration tested (500000 nM). Thus, the presence of a d-residue at position 2 in a short peptide, even one as structurally similar to GdFFD-OH as GdFF-NH2, is not sufficient to confer apALNR activity.
We previously found that GdFFD-OH and GdYFD-OH were both potent activators of the CPG associated with feeding behavior in electrophysiology experiments, while their all-l-residue analogues, GFFD-OH and GYFD-OH, were not (Table 1).8,9,24 In the study presented here, we evaluated the ability of SdYADSKDEESNAALSDFAED-OH, SYADSKDEESNAALSDFAED-OH, GdFF-NH2, and GFF-NH2 to induce circuit activity in the feeding network by electrophysiology. These electrophysiology experiments differ from the LPl1 experiments described above in that we measured the cyclic activity bursts of the I2 nerve of the buccal ganglion and corresponding well-defined cycles of activity in several specific buccal neurons that are known to lead to stereotyped feeding responses.36,37 Upon evaluating activity in the feeding network, we found that all four peptides were inactive (Table 1, Figure S10, and Table S1). Thus, for each peptide tested, in vitro activation of apALNR (or lack thereof) determined by the IP1 accumulation assay in cells transfected with apALNR matched their physiological effects in the Aplysia feeding network.
Together, our results support the identification of both GdFFD-OH and GdYFD-OH as key apALNP-derived agonists of apALNR. The matching activity for apALNP peptides in apALNR activation as determined by the IP1 accumulation assay and in the feeding network as determined by electrophysiology measurements supports the hypothesis that apALNR may mediate the effects of apALNP-derived peptides in the feeding circuit. Furthermore, our results suggest that endogenous peptides, such as GYFD-OH, SdYADSKDEESNAALSDFAED-OH, and SYADSKDEESNAALSDFAED-OH, are inactive in the feeding network because of their inability to activate a key receptor, and not because of other factors such as proteolytic degradation.
Activation of apALNR by GdFFD-OH Analogues
To gain more insight into how apALNR recognizes its ligands and to determine if other peptides may be ligands for the receptor, we designed a library of GdFFD-OH analogues bearing single-residue substitutions or terminal modifications and evaluated the ability of each of these peptides to activate apALNR in CHO-K1 cells transiently transfected with both apALNR and Gα-16 using the IP1 accumulation assay described above (Figure 5, Figure S9, and Table S2). The results, summarized in Figure 5, reveal the specificity of the receptor and indicate that each residue of GdFFD-OH and both terminal charges make important contributions to apALNR activity, as judged by apALNR activation potency (EC50 values). For example, position 2 appears to require an extended hydrophobic d-residue, position 3 can tolerate smaller hydrophobic residues but not charged residues, and position 4 requires a residue with a carbonyl group on its side chain (see the Supporting Information for more details). These results, which show that even relatively minor modifications to GdFFD-OH lead to dramatic losses of potency in most cases, suggest that GdFFD-OH and GdYFD-OH are likely the primary agonists for this receptor in vivo, although unidentified or unrelated sequences may also be endogenous agonists.
Figure 5.
Primary sequences of GdFFD-OH analogues made through single-residue substitutions (sub.) or terminal modifications (mod.), along with associated EC50 values for apALNR activation (in parentheses) as determined by the IP1 accumulation assay in CHO-K1 cells transiently transfected with both apALNR and Gα-16 (see Figure S9 for all dose–response curves and Table S2 for errors associated with these measurements). For non-natural analogues of GdFFD-OH, the substituted residue or terminus is colored red. For apALNR-active compounds, EC50 is the mean from at least three independent experiments. For apALNR-inactive compounds, the EC50 is listed as being greater than the highest concentration tested, from at least two independent experiments showing no or negligible activity. EC50 values for GFFD-OH, GYFD-OH, and GdYFD-OH, which are included in Table 1, are repeated here for comparison.
Physiological Activity of GdFFD-OH Analogues
To test the hypothesis that apALNR is a mediator of the physiological effects of GdFFD-OH, we examined whether a subset of the GdFFD-OH analogues described could directly induce feeding circuit activity in isolated buccal and cerebral ganglia. For these studies, we chose GdFFD-OH analogues with high apALNR potency (dAdFFD-OH), intermediate potency (GdLFD-OH, GdFAD-OH, Ac-GdFFD-OH, and GdFFD-NH2), or no activity (AdFFD-OH) in the IP1 accumulation assay. Consistent with its high apALNR potency in the IP1 accumulation assay, we found that dAdFFD-OH activated the Aplysia feeding network in a manner similar to that of GdFFD-OH and GdYFD-OH (Figure 6 and Figure S11).8,9 GdLFD-OH, GdFAD-OH, Ac-GdFFD-OH, and GdFFD-NH2 were each weaker at inducing circuit activation than dAdFFD-OH was, consistent with their reduced apALNR potency relative to that of dAdFFD-OH in the IP1 accumulation assay. Interestingly, we found that AdFFD-OH, which showed no ability to activate apALNR in the IP1 accumulation assay experiments, was modestly active in the feeding network. This contrasts with several other neuropeptides evaluated (e.g., GdFF-NH2 and SdYADSKDEESNAALSDFAED-OH, described above), which were unable to activate apALNR by the IP1 accumulation assay and showed no activity in the feeding circuit. The inconsistency between apALNR activation in cell-based assays with a transiently transfected receptor and electrophysiology activity in intact ganglia for AdFFD-OH might indicate the inability of the apALNR IP1 accumulation assay to completely recapitulate the complex biological interactions present in living neural networks. Indeed, peptide potencies can differ significantly among different expression systems and intact tissues.38,39
Figure 6.
Feeding circuit activity induced by GdFFD-OH analogues (10–6 or 10–5 M), as determined by electrophysiological recordings on intact buccal and cerebral ganglia (see Figure S11). Control: activity before peptide perfusion. Wash: activity after washout of peptide. Bars represent the mean ± the standard error of the mean for dAdFFD [F(3,18) = 12.51; p < 0.001; n = 7], GdLFD-OH [F(3,15) = 8.056; p < 0.01; n = 6], GdFAD-OH [F(3,18) = 30.74; p < 0.001; n = 7], Ac-GdFFD-OH [F(3,21) = 14.68; p < 0.001; n = 8], GdFFD-NH2 [F(3,21) = 26.13; p < 0.001; n = 8], and AdFFD-OH [F(3,15) = 12.5; p < 0.001; n = 6]. Repeated measures analysis of variance and Bonferroni post hoc test: *p < 0.05; **p < 0.01; ***p < 0.001.
Alternatively, activity for AdFFD-OH in the feeding network may indicate that alternative or modified isoforms of apALNR exist in vivo with altered selectivity or that additional unrelated receptors are activated by AdFFD-OH. Overall, the concordant activity for the analogues tested, both for receptor activation by the IP1 accumulation assay and in the feeding network by electrophysiology (including apALNP-derived peptides, described above), is consistent with apALNR as a mediator of the biological activity of this family of DAACPs, although the unexpected activity of AdFFD-OH in the feeding network leaves open the possibility that additional receptors for apALNP-derived DAACPs may also be present.
Stability of DAACPs in CNS Homogenates
d-Residues are known to enhance the stability of engineered peptides to proteases,5,12,17 but little is known about how d-residues influence the lifetime of naturally occurring DAACPs that act as cell–cell signaling peptides. To gain insight into the relative stability of DAACPs from Aplysia to endogenous proteases, we incubated exogenous GdFFD-OH, GdYFD-OH, GFFD-OH, GYFD-OH, and NdWF-NH2 in ganglion homogenate extract and monitored the time course of peptide degradation by LC–MS with multiple-reaction monitoring (MRM) (Figure 7). We found that the all-l-residue peptides GFFD-OH and GYFD-OH were relatively rapidly degraded, with half-lives of 8.4 and 8.7 min, respectively. NdWF-NH2, a cardioactive DAACP, was more stable under these conditions (half-life of 27 min). Previous studies have shown that the stability of NdWF-NH2 is similar to that of NWF-NH2 in ganglion membrane fractions,40 so we did not evaluate NWF-NH2 in this experiment. Interestingly, both GdFFD-OH and GdYFD-OH were even more stable than NdWF-NH2, with half-lives of 530 and 1100 min, respectively, in the ganglion homogenate.
Figure 7.

Stability of 50 μM neuropeptides in Aplysia cerebral and abdominal ganglion homogenate extracts in PBS (pH 7.4) as determined by LC–MRM. Each point represents the mean ± the standard deviation of three biological sets of two animals each. Panels a and b show the same data plotted on different time scales, while panel c shows calculated half-life values for each peptide.
Unlike classical neurotransmitters, neuropeptides are released from both synaptic and nonsynaptic sites and can travel relatively long distances (on the order of micrometers) from their site of release to activate distal receptors.41 This volume transmission mode of signaling within the CNS, and even hormonal-like roles traveling longer distances, are possible for neuropeptides that display long lifetimes in the extracellular space.42,43 Our results are consistent with the hypothesis that the d-residue in apALNP-derived DAACPs increases the lifetime of these peptides to allow signaling across relatively long distances. However, these studies were performed in the environment of soluble ganglion homogenate extract, which likely lacks membrane-bound enzymes that may degrade these compounds and contains intracellular enzymes that would not naturally come into contact with these compounds in vivo. Further studies will be required to assess if GdFFD-OH and GdYFD-OH indeed possess longer lifetimes or travel greater distances in vivo relative to those of all-l-residue peptides of similar length. Nevertheless, our results show that GdFFD-OH and GdYFD-OH are more stable in an environment containing endogenous CNS proteases than their all-l-residue analogues and another known DAACP (NdWF-NH2) from the same animal. This suggests that in some cases the d-residue can greatly extend the lifetime of cell–cell signaling DAACPs, in addition to being critical for receptor activation.
Conclusions
Prior to our work, Bauknecht et al.27 showed d-Phe-specific activation of apALNR by GdFFD-OH, but little else about this ligand–receptor family has been studied. Although previous studies44,45 have explored how DAACP structure relates to specific physiological functions (e.g., neuronal activity or muscle contraction), the receptors for most cell–cell signaling DAACPs have not been identified, and thus, little is known about the interactions of DAACPs with their receptors throughout the CNS. Here, we provide multiple compelling lines of evidence demonstrating that apALNR does indeed function as a receptor for the apALNP-derived DAACPs GdFFD-OH and GdYFD-OH and mediates multiple physiological effects throughout the CNS. First, qPCR and ISH analyses demonstrate, for the first time, that apALNR is expressed in the Aplysia CNS. Interestingly, whereas apALNP expression is largely restricted to the pedal ganglia, apALNR expression is distributed throughout the CNS. These results indicate that apALNP-derived DAACPs may be produced in one central ganglion (i.e., pedal) to affect targets throughout the CNS. Indeed, the apALNR expression data enabled us to identify a novel cellular target of apALNP-derived DAACPs (LPl1), in addition to known targets in feeding and locomotor networks.8,9,24 The broad distribution of apALNR expression across the entire CNS suggests that many other roles could be played by this interesting DAACP receptor family. Receptor activation experiments with non-natural GdFFD-OH analogues (Figure 5) revealed that each residue of this ligand makes important contributions to potency and strongly suggest that GdFFD-OH and GdYFD-OH are likely the primary ligands for apALNR. Finally, GdFFD-OH and GdYFD-OH were more stable than their all-l-residue analogues and NdWF-NH2 in ganglion homogenate extract, providing evidence that the lifetimes of some cell–cell signaling peptides are significantly increased in vivo because of the incorporation of the d-residue, and these peptides may diffuse relatively long distances from their point of release throughout the CNS to activate distal receptors.
The largely concordant activity of peptides in both receptor activation by IP1 accumulation assays and physiological experiments is consistent with the hypothesis that apALNR mediates known physiological activities of GdFFD-OH and GdYFD-OH.8,9 However, there may be other receptors that contribute to the physiological functions of GdFFD-OH and GdYFD-OH. In fact, the physiological activity of AdFFD-OH in the feeding network, despite no apALNR activity in the IP1 accumulation assay, suggests that there are additional receptors (or isoforms of apALNR) with differing ligand specificities that are activated by GdFFD-OH or GdYFD-OH. In the future, it will be of great interest to knock down apALNR expression to determine the specific contributions of this receptor to single-neuron excitability and/or network activity. Regardless of whether apALNR is the sole receptor for GdFFD-OH and GdYFD-OH, our results provide strong evidence that GdFFD-OH and GdYFD-OH are major ligands for apALNR, and that apALNR is expressed across many regions of the CNS, including in regions associated with feeding (buccal ganglia) and locomotion (pedal ganglia),8,9 and in specific cells known to control additional functions, ultimately suggesting that GdFFD-OH and GdYFD-OH play roles in a variety of behaviors.
Despite extensive efforts, there are still a large number of orphan GPCRs in vertebrates, including mammals.46 The identification of ALNRs as receptors whose endogenous ligands require a d-residue for activation suggests that some orphan GPCRs may be selective for DAACPs, and deorphanization efforts that do not consider l- to d-residue isomerization may miss ligand–receptor identifications. Overall, our results represent one of the first in-depth explorations of a receptor that recognizes a DAACP as its native ligand and provide evidence that apALNP-derived DAACPs play a variety of functions throughout the CNS, either by directly activating neurons or by acting as neuromodulators.41,47 Given the growing number of DAACPs that continue to be identified among a wide variety of organisms,4−11 our findings may be of significance for understanding l- to d-residue isomerization in other animals, including vertebrates.
Methods
Detailed procedures can be found in the Supporting Information.
qPCR
qPCR experiments were performed using Power SYBR Green PCR Master Mix (Applied Biosystems, 4368708), according to the manufacturer’s specifications. The relative expression level for each gene was calculated using the ΔCt method.48 For each gene, ΔCt was calculated as the difference between the Ct values (mean of technical triplicates) for the gene of interest and GAPDH. Relative expression values were calculated using 2–ΔCt. See the Supporting Information for more details.
In Situ Hybridization (ISH)
The method for the ISH experiments was adapted from ref (49) using antisense or sense digoxigenin-labeled RNA probes corresponding to an ∼750 bp region of the apALNR mRNA (XM_005106549.2). Riboprobes were synthesized using a SP6/T7 DIG RNA labeling kit (Roche), following the manufacturer’s instructions. See the Supporting Information for expanded details.
Peptide Synthesis and Purification
Peptides GDAS-OH and GdDAS-OH were purchased from CPC Scientific. All other peptides were synthesized by solid-phase peptide synthesis based on Fmoc protection of the main chain amine. Peptides with a C-terminal acid were synthesized on the solid phase using Wang resin preloaded with the C-terminal residue (Novabiochem or Anaspec). Peptides with a C-terminal amide were synthesized on NovaPEG Rink Amide resin (Novabiochem, 855047). Coupling reactions were performed by treating the resin with a solution of ≥4 molar equivalents of Fmoc-protected amino acid with appropriate side chain protecting groups, activated with benzotriazole-1-yl-oxy-tris-pyrrolidino-phosphonium hexafluorophosphate (PyBOP) and N,N-diisopropylethylamine (DIEA) (1:1:2 amino acid:PyBOP:DIEA molar ratio) in a solution of 0.1 M N-hydroxybenzotriazole in N-methyl-2-pyrrolidone. Coupling reactions were allowed to proceed at room temperature (RT) for >40 min with stirring or gentle shaking, after which the resin was rinsed with three to five washes of dimethylformamide (DMF). Deprotection of the Fmoc protecting group was performed in a solution of 20% piperidine in DMF for 20 min at RT with gentle stirring or shaking, after which the resin was rinsed with three to five washes of DMF. Acetylation of the N-terminus for Ac-GdFFD-OH was achieved by treating the resin with an 8:2:1 DMF/DIEA/acetic anhydride solution for 10 min. After the completion of the synthesis, peptides were cleaved from the resin and side chain protecting groups were removed using a solution of 95% trifluoroacetic acid, 2.5% H2O, and 2.5% triisopropylsilane for >3 h. After cleavage, most of the cleavage solution was removed by evaporation, and the peptides were dissolved in a water/acetonitrile (ACN) mixture or dimethyl sulfoxide for high-performance liquid chromatography (HPLC) purification. For SdYADSKDEESNAALSDFAED-OH and SYADSKDEESNAALSDFAED-OH, peptides were precipitated by the addition of cold methyl tert-butyl ether. Crude peptide mixtures were purified by reversed-phase HPLC and then dried under vacuum. Peptides were dissolved in a water/ACN solution and concentrations determined by UV absorbance and extinction coefficients calculated from the primary sequence at 214 nm50 or 280 nm.51 On the basis of this calculated stock concentration, peptides were aliquoted and dried under vacuum. The final peptide purity was assessed by reversed-phase HPLC and the identity confirmed by MALDI-TOF MS (see the Supporting Information).
IP1 Accumulation Assays for apALNR Activation
CHO-K1 cells (ATCC, CCL-61) were transiently transfected with apALNR [region corresponding to XP_005106606.1, in pcDNA3.1(+)] and Gα-16 [in pcDNA3.1(+)] using Turbofect transfection reagent (ThermoFisher Scientific, R0531). After exposure to potential agonist peptides for 1 h, activation of apALNR was detected by monitoring IP1 accumulation using an IPOne Detection Kit (Cisbio, 62IPAPEB), following the manufacturer’s instructions with minor modification. While we found that the recommended amount of IP1-d2 and anti-IP1-cryptate (1×) worked well, we also obtained comparable EC50 values using half these amounts (0.5×), so many assays were performed using 0.5× reagents. See the Supporting Information for more details.
Electrophysiology
Intracellular and extracellular recordings of the physiological activity from CNS preparations (either the cerebral and buccal ganglia or the pleural and pedal ganglia) were made as described previously.8,52 Mean values were compared using repeated measures one-way analysis of variance, assuming sphericity, with the Bonferroni post test in GraphPad Prism 7. See the Supporting Information for notes on the statistical analyses and more details.
Peptide Stability Assay
Cerebral and abdominal ganglia were isolated from Aplysia (65–85 g) and homogenized in phosphate-buffered saline (PBS) (pH 7.4). Each biological set included cerebral and abdominal ganglia from two animals. Tissue was centrifuged (14000g and 4 °C for 10 min), and the supernatant was removed and the protein content estimated using a BCA Protein Assay Kit (ThermoFisher Scientific, 23235). The homogenate from each biological set was diluted to 1000 μg/mL protein. A stock solution containing each peptide at 200 μM was prepared in PBS. The peptide stock in PBS (125 μL) was added to the ganglion homogenate (375 μL) and the resulting mixture incubated at 37 °C (final conditions being 50 μM peptide and 750 μg/mL homogenate protein). At each time point, 10 μL of the reaction mixture was removed, the reaction quenched with 20 μL of a solution containing 25 μM GdFF-NH2 (as an internal control) and 1% formic acid (FA) in a 50% ACN/water mixture, and the quenched reaction stored at −20 °C until analysis. For analysis, each quenched solution was thawed, diluted to 0.1% FA in a 4% ACN/water mixture, and desalted with C18 solid-phase exchange centrifuge spin columns (ThermoFisher Scientific, 89870). Desalted and dried samples were dissolved in 0.1% FA in water and analyzed using a Bruker EVOQ Elite triple-quadrupole mass spectrometer coupled to a Bruker Advance UHPLC instrument, in MRM mode in positive ion mode. Channels of parent and fragment ion pairs were identified using the MRM Builder in the MS Workstation software for the EVOQ instrument or were manually entered on the basis of the predicted fragment ions. The parent ion (m/z, mass window of 0.7)/fragment ion (m/z, mass window of 2) pairs for each peptide were as follows: GFFD-OH, GdFFD-OH = 485.2/120.2; GYFD-OH, GdYFD-OH = 501.2/120.2; NdWF-NH2 = 465.2/448.6; GdFF-NH2 (synthesized with [13C]Gly) = 370.2/120.1. For each time point, the peak area in the resulting LC–MRM chromatogram for the peptide of interest was first normalized to the peak area for GdFF-NH2, and the percent peptide remaining was subsequently calculated relative to the “1 min” time point for each biological set. Data were plotted and half-life values calculated using a one-phase exponential decay model in GraphPad Prism 7.
Acknowledgments
The authors thank P. Bauknecht and G. Jékely for the generous gift of plasmids containing apALNR and Gα-16 for mammalian expression and the UIUC Roy J. Carver Biotechnology Center for providing resources, instrument time, and helpful advice. This work was supported by the National Institutes of Health, via Grant P30 DA018310 from the National Institute on Drug Abuse, and Grant R01 NS031609 from the National Institute of Neurological Disorders and Stroke (J.V.S.), the National Resource for Aplysia, funded by PHS grant P40 OD010952, and the National Natural Science Foundation of China, via Grants 31671097, 31371104 (J.J.), and J1103512 and J1210026 (School of Life Sciences). J.W.C. was supported in part by a Beckman Institute Postdoctoral Fellowship, funded by a Beckman Foundation gift to the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.8b00167.
Detailed materials and methods, notes on apALNR characterization, statistical analyses, and peptide selection/activity/inactivity, Tables S1 and S2, Figures S1–S11 (as noted in the text), Figures S12–S14, peptide characterization data, and supporting references (PDF)
Author Present Address
# R.H.R.-G.: Department of Cellular Biology, University of Georgia, Athens, GA 30602.
The authors declare no competing financial interest.
Supplementary Material
References
- Fricker L. D.; Lim J.; Pan H.; Che F. Y. (2006) Peptidomics: identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom. Rev. 25, 327–344. 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- Mzhavia N.; Berman Y.; Che F. Y.; Fricker L. D.; Devi L. A. (2001) ProSAAS processing in mouse brain and pituitary. J. Biol. Chem. 276, 6207–6213. 10.1074/jbc.M009067200. [DOI] [PubMed] [Google Scholar]
- Hook V.; Funkelstein L.; Lu D.; Bark S.; Wegrzyn J.; Hwang S. R. (2008) Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu. Rev. Pharmacol. Toxicol. 48, 393–423. 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck S. D.; Siok C. J.; Krapcho K. J.; Kelbaugh P. R.; Thadeio P. F.; Welch M. J.; Williams R. D.; Ganong A. H.; Kelly M. E.; Lanzetti A. J.; Gray W. R.; Phillips D.; Parks T. N.; Jackson H.; Ahlijanian M. K.; Saccomano N. A.; Volkmann R. A. (1994) Functional consequences of posttranslational isomerization of Ser(46) in a calcium channel toxin. Science 266, 1065–1068. 10.1126/science.7973665. [DOI] [PubMed] [Google Scholar]
- Torres A. M.; Menz I.; Alewood P. F.; Bansal P.; Lahnstein J.; Gallagher C. H.; Kuchel P. W. (2002) D-amino acid residue in the C-type natriuretic peptide from the venom of the mammal, Ornithorhynchus anatinus, the Australian platypus. FEBS Lett. 524, 172–176. 10.1016/S0014-5793(02)03050-8. [DOI] [PubMed] [Google Scholar]
- Kamatani Y.; Minakata H.; Kenny P. T. M.; Iwashita T.; Watanabe K.; Funase K.; Sun X. P.; Yongsiri A.; Kim K. H.; Novales-Li P.; Novales E. T.; Kanapi C. G.; Takeuchi H.; Nomoto K. (1989) Achatin-I, an endogenous neuroexcitatory tetrapeptide from Achatina fulica Férussac containing a D-amino acid residue. Biochem. Biophys. Res. Commun. 160, 1015–1020. 10.1016/S0006-291X(89)80103-2. [DOI] [PubMed] [Google Scholar]
- Morishita F.; Nakanishi Y.; Kaku S.; Furukawa Y.; Ohta S.; Hirata T.; Ohtani M.; Fujisawa Y.; Muneoka Y.; Matsushima O. (1997) A novel D-amino-acid-containing peptide isolated from Aplysia heart. Biochem. Biophys. Res. Commun. 240, 354–358. 10.1006/bbrc.1997.7659. [DOI] [PubMed] [Google Scholar]
- Bai L.; Livnat I.; Romanova E. V.; Alexeeva V.; Yau P. M.; Vilim F. S.; Weiss K. R.; Jing J.; Sweedler J. V. (2013) Characterization of GdFFD, a D-amino acid-containing neuropeptide that functions as an extrinsic modulator of the Aplysia feeding circuit. J. Biol. Chem. 288, 32837–32851. 10.1074/jbc.M113.486670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livnat I.; Tai H. C.; Jansson E. T.; Bai L.; Romanova E. V.; Chen T. T.; Yu K.; Chen S. A.; Zhang Y.; Wang Z. Y.; Liu D. D.; Weiss K. R.; Jing J.; Sweedler J. V. (2016) A D-amino acid-containing neuropeptide discovery funnel. Anal. Chem. 88, 11868–11876. 10.1021/acs.analchem.6b03658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyez D.; Van Herp F.; Rossier J.; Le Caer J. P.; Tensen C. P.; Lafont R. (1994) Evidence for a conformational polymorphism of invertebrate neurohormones: D-amino acid residue in crustacean hyperglycemic peptides. J. Biol. Chem. 269, 18295–18298. [PubMed] [Google Scholar]
- Richter K.; Egger R.; Kreil G. (1987) D-alanine in the frog skin peptide dermorphin is derived from L-alanine in the precursor. Science 238, 200–202. 10.1126/science.3659910. [DOI] [PubMed] [Google Scholar]
- Miller S. M.; Simon R. J.; Ng S.; Zuckermann R. N.; Kerr J. M.; Moos W. H. (1995) Comparison of the proteolytic susceptibilities of homologous L-amino acid, D-amino acid, and N-substituted glycine peptide and peptoid oligomers. Drug Dev. Res. 35, 20–32. 10.1002/ddr.430350105. [DOI] [Google Scholar]
- Tao Y.; Quebbemann N. R.; Julian R. R. (2012) Discriminating D-amino acid-containing peptide epimers by radical-directed dissociation mass spectrometry. Anal. Chem. 84, 6814–6820. 10.1021/ac3013434. [DOI] [PubMed] [Google Scholar]
- Tao Y.; Julian R. R. (2014) Identification of amino acid epimerization and isomerization in Crystallin proteins by tandem LC-MS. Anal. Chem. 86, 9733–9741. 10.1021/ac502296c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams C. M.; Zubarev R. A. (2005) Distinguishing and quantifying peptides and proteins containing D-amino acids by tandem mass spectrometry. Anal. Chem. 77, 4571–4580. 10.1021/ac0503963. [DOI] [PubMed] [Google Scholar]
- Jia C.; Lietz C. B.; Yu Q.; Li L. (2014) Site-specific characterization of D-amino acid containing peptide epimers by ion mobility spectrometry. Anal. Chem. 86, 2972–2981. 10.1021/ac4033824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton R. C. D.; Milton S. C. F.; Kent S. B. H. (1992) Total chemical synthesis of a D-enzyme: The enantiomers of HIV-1 protease show reciprocal chiral substrate specificity. Science 256, 1445–1448. 10.1126/science.1604320. [DOI] [PubMed] [Google Scholar]
- Hawkins R. D.; Kandel E. R.; Bailey C. H. (2006) Molecular mechanisms of memory storage in Aplysia. Biol. Bull. 210, 174–191. 10.2307/4134556. [DOI] [PubMed] [Google Scholar]
- Gardner D. (1971) Bilateral symmetry and interneuronal organization in the buccal ganglia of Aplysia. Science 173, 550–553. 10.1126/science.173.3996.550. [DOI] [PubMed] [Google Scholar]
- Jacob M. H. (1984) Neurogenesis in Aplysia californica resembles nervous system formation in vertebrates. J. Neurosci. 4, 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church P. J.; Lloyd P. E. (1991) Expression of diverse neuropeptide cotransmitters by identified motor neurons in Aplysia. J. Neurosci. 11, 618–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing J.; Gillette R.; Weiss K. R. (2009) Evolving concepts of arousal: Insights from simple model systems. Rev. Neurosci. 20, 405–427. 10.1515/REVNEURO.2009.20.5-6.405. [DOI] [PubMed] [Google Scholar]
- Kandel E. R. (2001) The molecular biology of memory storage: A dialogue between genes and synapses. Science 294, 1030–1038. 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Yang C. Y.; Yu K.; Wang Y.; Chen S. A.; Liu D. D.; Wang Z. Y.; Su Y. N.; Yang S. Z.; Chen T. T.; Livnat I.; Vilim F. S.; Cropper E. C.; Weiss K. R.; Sweedler J. V.; Jing J. (2016) Aplysia locomotion: Network and behavioral actions of GdFFD, a D-amino acid-containing neuropeptide. PLoS One 11, e0147335 10.1371/journal.pone.0147335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey B. R.; Amare A.; Zimmerman T. A.; Rodriguez-Zas S. L.; Sweedler J. V. (2006) NeuroPred: A tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 34, W267–272. 10.1093/nar/gkl161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer R.; Gaehwiler B. H.; Buescher H. H.; Hill R. C.; Roemer D. (1982) Opiate antagonistic properties of an octapeptide somatostatin analog. Proc. Natl. Acad. Sci. U. S. A. 79, 4815–4817. 10.1073/pnas.79.15.4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauknecht P.; Jékely G. (2015) Large-scale combinatorial deorphanization of Platynereis neuropeptide GPCRs. Cell Rep. 12, 684–693. 10.1016/j.celrep.2015.06.052. [DOI] [PubMed] [Google Scholar]
- Gomes I.; Bobeck E. N.; Margolis E. B.; Gupta A.; Sierra S.; Fakira A. K.; Fujita W.; Müller T. D.; Müller A.; Tschöp M. H.; Kleinau G.; Fricker L. D.; Devi L. A. (2016) Identification of GPR83 as the receptor for the neuroendocrine peptide PEN. Sci. Signaling 9, ra43. 10.1126/scisignal.aad0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes I.; Aryal D. K.; Wardman J. H.; Gupta A.; Gagnidze K.; Rodriguiz R. M.; Kumar S.; Wetsel W. C.; Pintar J. E.; Fricker L. D.; Devi L. A. (2013) GPR171 is a hypothalamic G protein-coupled receptor for BigLEN, a neuropeptide involved in feeding. Proc. Natl. Acad. Sci. U. S. A. 110, 16211–16216. 10.1073/pnas.1312938110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R.; Schiöth H. B. (2005) The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol. Pharmacol. 67, 1414–1425. 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- Rosen S. C.; Weiss K. R.; Goldstein R. S.; Kupfermann I. (1989) The role of a modulatory neuron in feeding and satiation in Aplysia: Effects of lesioning of the serotonergic metacerebral cells,. J. Neurosci. 9, 1562–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K. R.; Cohen J. L.; Kupfermann I. (1978) Modulatory control of buccal musculature by a serotonergic neuron (metacerebral cell) in Aplysia. J. Neurophysiol. 41, 181–203. 10.1152/jn.1978.41.1.181. [DOI] [PubMed] [Google Scholar]
- Rayport S. G.; Ambron R. T.; Babiarz J. (1983) Identified cholinergic neurons R2 and LPl1 control mucus release in Aplysia. J. Neurophysiol. 49, 864–876. 10.1152/jn.1983.49.4.864. [DOI] [PubMed] [Google Scholar]
- Offermanns S.; Simon M. I. (1995) G alpha 15 and G alpha 16 couple a wide variety of receptors to phospholipase C. J. Biol. Chem. 270, 15175–15180. 10.1074/jbc.270.25.15175. [DOI] [PubMed] [Google Scholar]
- Trinquet E.; Fink M.; Bazin H.; Grillet F.; Maurin F.; Bourrier E.; Ansanay H.; Leroy C.; Michaud A.; Durroux T.; Maurel D.; Malhaire F.; Goudet C.; Pin J. P.; Naval M.; Hernout O.; Chrétien F.; Chapleur Y.; Mathis G. (2006) D-myo-inositol 1-phosphate as a surrogate of D-myo-inositol 1,4,5-tris phosphate to monitor G protein-coupled receptor activation. Anal. Biochem. 358, 126–135. 10.1016/j.ab.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Due M. R.; Jing J.; Weiss K. R. (2004) Dopaminergic contributions to modulatory functions of a dual-transmitter interneuron in Aplysia. Neurosci. Lett. 358, 53–57. 10.1016/j.neulet.2003.12.058. [DOI] [PubMed] [Google Scholar]
- Jing J.; Weiss K. R. (2005) Generation of variants of a motor act in a modular and hierarchical motor network. Curr. Biol. 15, 1712–1721. 10.1016/j.cub.2005.08.051. [DOI] [PubMed] [Google Scholar]
- Broichhagen J.; Podewin T.; Meyer-Berg H.; von Ohlen Y.; Johnston N. R.; Jones B. J.; Bloom S. R.; Rutter G. A.; Hoffmann-Röder A.; Hodson D. J.; Trauner D. (2015) Optical control of insulin secretion using an incretin switch. Angew. Chem., Int. Ed. 54, 15565–15569. 10.1002/anie.201506384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly D. (2012) The structure and function of the glucagon-like peptide-1 receptor and its ligands,. Br. J. Pharmacol. 166, 27–41. 10.1111/j.1476-5381.2011.01687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita F.; Matsushima O.; Furukawa Y.; Minakata H. (2003) Deamidase inactivates a D-amino acid-containing Aplysia neuropeptide. Peptides 24, 45–51. 10.1016/S0196-9781(02)00275-9. [DOI] [PubMed] [Google Scholar]
- van den Pol A. N. (2012) Neuropeptide transmission in brain circuits. Neuron 76, 98–115. 10.1016/j.neuron.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M.; Leng G. (2006) Dendritic peptide release and peptide-dependent behaviours. Nat. Rev. Neurosci. 7, 126–136. 10.1038/nrn1845. [DOI] [PubMed] [Google Scholar]
- Branton W. D.; Mayeri E.; Brownell P.; Simon S. B. (1978) Evidence for local hormonal communication between neurons in Aplysia. Nature 274, 70–72. 10.1038/274070a0. [DOI] [PubMed] [Google Scholar]
- Kim K. H.; Takeuchi H.; Kamatani Y.; Minakata H.; Nomoto K. (1991) Structure-activity relationship studies on the endogenous neuroactive tetrapeptide Achatin-I on giant neurons of Achatina fulica Férussac. Life Sci. 48, PL91–PL96. 10.1016/0024-3205(91)90131-T. [DOI] [PubMed] [Google Scholar]
- Fujita K.; Minakata H.; Nomoto K.; Furukawa Y.; Kobayashi M. (1995) Structure-activity relations of Fulicin, a peptide containing a D-amino acid residue. Peptides 16, 565–568. 10.1016/0196-9781(95)00022-C. [DOI] [PubMed] [Google Scholar]
- Vassilatis D. K.; Hohmann J. G.; Zeng H.; Li F.; Ranchalis J. E.; Mortrud M. T.; Brown A.; Rodriguez S. S.; Weller J. R.; Wright A. C.; Bergmann J. E.; Gaitanaris G. A. (2003) The G protein-coupled receptor repertoires of human and mouse. Proc. Natl. Acad. Sci. U. S. A. 100, 4903–4908. 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E. (2012) Neuromodulation of neuronal circuits: Back to the future. Neuron 76, 1–11. 10.1016/j.neuron.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D.; Livak K. J. (2008) Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3, 1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Jezzini S. H.; Bodnarova M.; Moroz L. L. (2005) Two-color in situ hybridization in the CNS of Aplysia californica. J. Neurosci. Methods 149, 15–25. 10.1016/j.jneumeth.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Conibear A. C.; Daly N. L.; Craik D. J. (2012) Quantification of small cyclic disulfide-rich peptides. Biopolymers 98, 518–524. 10.1002/bip.22121. [DOI] [PubMed] [Google Scholar]
- Gill S. C.; von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326. 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Jing J.; Alexeeva V.; Chen S. A.; Yu K.; Due M. R.; Tan L. N.; Chen T. T.; Liu D. D.; Cropper E. C.; Vilim F. S.; Weiss K. R. (2015) Functional characterization of a vesicular glutamate transporter in an interneuron that makes excitatory and inhibitory synaptic connections in a molluscan neural circuit. J. Neurosci. 35, 9137–9149. 10.1523/JNEUROSCI.0180-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






