Abstract
Background
Despite the health concerns and nutritional importance of fatty acids, there is a relative paucity of studies in the literature that report genetic or genomic parameters, especially in the case of sheep populations. To investigate the genetic architecture of fatty acid composition of sheep, we conducted genome-wide association studies (GWAS) and estimated genomic heritabilities for fatty acid profile in Longissimus dorsi muscle of 216 male sheep.
Results
Genomic heritability estimates for fatty acid content ranged from 0.25 to 0.46, indicating that substantial genetic variation exists for the evaluated traits. Therefore, it is possible to alter fatty acid profiles through selection. Twenty-seven genomic regions of 10 adjacent SNPs associated with fatty acids composition were identified on chromosomes 1, 2, 3, 5, 8, 12, 14, 15, 16, 17, and 18, each explaining ≥0.30% of the additive genetic variance. Twenty-three genes supporting the understanding of genetic mechanisms of fat composition in sheep were identified in these regions, such as DGAT2, TRHDE, TPH2, ME1, C6, C7, UBE3D, PARP14, and MRPS30.
Conclusions
Estimates of genomic heritabilities and elucidating important genomic regions can contribute to a better understanding of the genetic control of fatty acid deposition and improve the selection strategies to enhance meat quality and health attributes.
Keywords: Candidate genes, Fatty acid composition, Variance components, Ovine
Background
In recent years there has been a growing concern relative to the health attributes of foods that are consumed by increasingly health conscious consumers [1]. In particular, consumers are becoming gradually concerned relative to the amount of fatty acids in red meat [2]. Meat produced by ruminants is generally related to higher levels of saturated fatty acids (SFA), which are widely associated with the development of heart disease, stroke, diabetes, and obesity [3–5].
On the other hand, moderate levels of consumption of monounsaturated fatty acids (MUFA) are related to a decrease in serum cholesterol, consequently reducing the risk of heart diseases and strokes [4, 6–8]. Although found in a smaller proportion, red meat is also composed of polyunsaturated fatty acids (PUFA), which are strictly essential because they are not synthesized by humans and thus must be consumed daily to maintain proper body function [9]. These fatty acids influence several metabolic functions such as cell signaling, enzymatic regulation, eicosanoid synthesis, regulation of neuronal migration, neuromodulatory activity, and neurotransmitter activity [10, 11].
Despite the health concerns and nutritional importance of fatty acids, there is a relative paucity of studies in the literature that report estimates of genetic parameters, especially in the case of sheep populations. Estimating genetic parameters such as heritability, understanding the mechanisms underlying genetic variation in phenotypes, and predicting the genetic merit of an animal as a parent are fundamental pieces of information to enable genetic and ultimately phenotypic changes for designing successful animal breeding programs.
Fatty acids are complex traits, with several factors affecting their composition, such as sex, diet, age, and genetics [12]. Furthermore, routine phenotypic data collection is not practical given live-animal proxies do not exist for these traits and the expenses associated with collecting them. Thus, genomic information could play a critical role in enabling selection to improve them by allowing the design of animal breeding programs to increase the frequency of favorable alleles in the population [13, 14]. Concomitantly, the use of genomic information can increase the accuracy of estimated breeding values (EBV), thus increasing the rate of genetic change [15–17].
An important first step for the genetic evaluation of fatty acids content in meat is to investigate the genetic architecture of these complex traits and to identify variants associated with genes or regulatory elements through a genome-wide association study (GWAS) [18]. By estimating the degree to which these traits are heritable, and elucidating candidate genes, it could be possible to enable selection that will add commercial value to the sheep meat by providing consumers with a quality product that is also beneficial to health. Thus, the objectives of this study were to estimate genomic heritabilities and, for the first time, identify regions and candidate genes associated with fatty acid profiles in the Longissimus dorsi muscle of sheep using bivariate models.
Results and discussion
Fatty acid profiles
Descriptive statistics of the traits evaluated in this study are described in Table 1. The percentage of intramuscular fat (IMF) ranged between 1.62 and 4.93. These values were similar to those reported in sheep by [19–21], but lower than the values reported by [22, 23].
Table 1.
Descriptive statistics for intramuscular fat and fatty acid (FA) profile traits of Longissimus dorsi muscle in Santa Inês sheep
| Trait | Nomenclature | aN | Mean | bSD | cMin | dMax |
|---|---|---|---|---|---|---|
| Intramuscular fat (%) | IMF | 216 | 3.46 | 0.769 | 1.62 | 4.93 |
| Myristic acid (mg/mg) | C14:0 | 216 | 2.21 | 0.752 | 0.96 | 4.35 |
| Palmitic acid | C16:0 | 216 | 22.17 | 1.949 | 10.51 | 29.43 |
| Stearic acid | C18:0 | 216 | 20.29 | 3.342 | 12.53 | 33.26 |
| Palmitoleic acid | C16:1 | 216 | 1.54 | 0.245 | 0.98 | 2.16 |
| Oleic acid | C18:1 | 216 | 36.04 | 3.04 | 23.92 | 45.99 |
| Linoleic acid | C18:2 ω6 | 216 | 3.99 | 1.151 | 1.47 | 8.05 |
| Conjugated linoleic acid | CLA c9t11 | 216 | 0.43 | 0.184 | 0.076 | 0.924 |
| Alpha-Linolenic acid | C18:3 ω3 | 216 | 0.30 | 0.150 | 0.00 | 0.797 |
| Sum of saturated FA | SFAt | 216 | 46.92 | 4.399 | 28.38 | 59.88 |
| Sum of monounsaturated FA | MUFAt | 216 | 45.17 | 3.919 | 32.02 | 62.21 |
| Sum of polyunsaturated FA | PUFAt | 216 | 6.55 | 1.927 | 2.37 | 12.39 |
| Sum of omega-3 FA | ω3t | 216 | 0.89 | 0.532 | 0.111 | 2.99 |
| Sum of omega-6 FA | ω6t | 216 | 5.23 | 1.618 | 1.82 | 10.6 |
| Ratio of PUFA to SFA | PUFA/SFA | 216 | 0.14 | 0.049 | 0.048 | 0.322 |
| Ratio of ω6 to ω3 | ω6/ω3 | 216 | 7.62 | 5.33 | 2.82 | 55.32 |
anumber of phenotyped animals; bstandard deviation; cminimum; dmaximum
The predominant individual SFAs were palmitic (C16:0, 22.17%), stearic (C18:0, 20.29%), and myristic (C14:0, 2.21%) corresponding to about 95% of the SFA present in the Longissimus dorsi muscle. These results agreed with those reported in previous studies [19, 23–25], where these SFAs were predominant in sheep. The C14:0 and C16:0 acids have been associated with increased serum level of cholesterol and LDL (low-density lipoproteins) and decreased levels of HDL (high density lipoproteins) in the blood, major factors related to obesity, atherosclerosis, hypertension, heart, and coronary diseases [3–5]. Although C18:0 is abundant in sheep meat, studies suggest that it has no impact on the increase of serum cholesterol level, being considered a neutral fatty acid relative to human health [26–28]. Additionally, a large part of the C18:1 content is produced by the desaturation of C18:0 [12].
In this study, the most abundant individual fatty acid was oleic (C18:1, 36.04%), corresponding to about 90% of MUFA, whereas the palmitoleic acid (C16:1, 2.21%) corresponded approximately only 5% of MUFA. These results are similar to those reported in previous studies [19, 20, 22, 24, 25]. C18:1 is one of the main fatty acids in the Mediterranean diet, where several studies report a low incidence of heart disease, despite high-fat consumption [29–31]. Similar to C18:1, C16:1 has been also associated with several health benefits, however, a recent study performed by Hoffmann et al. [8] suggested that increased consumption of C16:1 is associated with some diseases such as the development of cardiomyopathy.
In this study, PUFA concentrations were only 6.55%, the lowest among fatty acid groups. Consequently, a lower ratio of PUFA/SFA (0.14) was observed in this study, consistent with the range between 0.04 and 0.14 reported in the literature [20–24]. A diet with a low PUFA/SFA ratio is associated with a high level of serum cholesterol and strongly correlated with coronary diseases [32, 33]. The value recommended by the World Health Organization [34] is above 0.45, which is related to a lower incidence of coronary heart disease [35].
In this study, the parameters ω3t (sum of omega-3 fatty acids) and ω6t (sum of omega-6 fatty acids) corresponded to approximately 14 and 86% of the PUFA, respectively. The percentage for ω3t were lower than those reported in earlier studies [19, 21, 25], whereas for ω6t the percentage described by the same authors ranged from 3.49 to 12.65. For individual PUFA, the most abundant was linoleic acid (C18:2 ω6, 3.99%), which corresponded to approximately 61% of PUFA and made up about 77% of ω6 fatty acids. The concentrations of ω3t fatty acids for this study were low, less than 1% of the total fatty acids. Alpha-linolenic acid (C18:3 ω3) had the highest concentration, making up 37% of the ω3t fatty acids; however, only 0.37% of the total fatty acids. In previous studies the values for C18:2 ω6 ranged between 0.28 and 9.32%, while for C18:3 ω3 ranged between 0.42 and 2.03% [19–25].
Conjugated linoleic acid (CLA c9t11) corresponded to about 7% of the PUFA, and only 0.43% of the total fatty acids. Results ranging from 0.44 to 1.37 were observed in other ovine studies [19, 21, 23]. CLA c9t11 is a PUFA considered to be beneficial to human health given there is evidence that it has immunostimulatory, antioxidant, antimutagenic, anticarcinogenic, and anti-inflammatory action [36–38]. Due to the results observed in this study, a high ω6/ω3 ratio (7.62) was observed. A very high ω6/ω3 ratio stimulates the pathogenesis of many illnesses, including cardiovascular disease, cancer, and inflammatory and autoimmune diseases, whereas a lower ω6/ω3 ratio, below 4.0, exerts suppressive effects [39].
Genomic heritability
Estimates of genomic heritability for fatty acids traits from the current study were between 0.25 and 0.46 (Table 2). Estimated genomic heritabilities for the individual SFA were high for C14:0 (0.44) and C18:0 (0.30), and moderate for C16:0 (0.25). For Scottish Blackface sheep, Karamichou et al. [40] using the traditional relationship matrix derived from pedigree information, reported slightly lower heritability estimates when compared to the current study for C16:0 (0.29), C18:0 (0.24), and C14:0 (0.14). Bolormaa et al. [41] also found lower genomic heritabilities for C14:0 (0.15), C16:0 (0.11), and C18:0 (0.19) in nine sheep breeds.
Table 2.
Genomic heritabilities (h2) for fatty acid (FA) profile of Longissimus dorsi muscle in Santa Inês sheep
| Trait | Nomenclature | aVg | bVe | cVp | h2 (dSE) |
|---|---|---|---|---|---|
| Myristic acid | C14:0 | 0.11 | 0.13 | 0.24 | 0.44 (0.045) |
| Palmitic acid | C16:0 | 0.95 | 2.84 | 3.79 | 0.25 (0.033) |
| Stearic acid | C18:0 | 2.63 | 6.18 | 8.81 | 0.30 (0.037) |
| Palmitoleic acid | C16:1 | 0.014 | 0.034 | 0.048 | 0.30 (0.035) |
| Oleic acid | C18:1 | 2.23 | 5.77 | 8.00 | 0.28 (0.035) |
| Linoleic acid | C18:2 ω6 | 0.32 | 0.88 | 1.20 | 0.27 (0.034) |
| Conjugated linoleic acid | CLA c9t11 | 0.005 | 0.010 | 0.015 | 0.34 (0.045) |
| Alpha-Linolenic acid | C18:3 ω3 | 0.006 | 0.007 | 0.013 | 0.46 (0.045) |
| Sum of saturated FA | SFAt | 4.45 | 9.44 | 13.89 | 0.32 (0.039) |
| Sum of monounsaturated FA | MUFAt | 3.58 | 7.98 | 11.56 | 0.31 (0.038) |
| Sum of polyunsaturated FA | PUFAt | 0.90 | 2.35 | 3.25 | 0.28 (0.034) |
| Sum of omega-3 FA | ω3t | 0.05 | 0.09 | 0.15 | 0.37 (0.045) |
| Sum of omega-6 FA | ω6t | 0.64 | 1.77 | 2.41 | 0.27 (0.034) |
| Ratio of PUFA to SFA | PUFA/SFA | 0.0006 | 0.0015 | 0.0021 | 0.28 (0.034) |
| Ratio of ω6 to ω3 | ω6/ω3 | 6.48 | 13.27 | 19.75 | 0.33 (0.042) |
aadditive genetic variance; b residual variance; c total phenotypic variance; d standard error
Moderate genomic heritability estimates for C16:1 (0.30) and C18:1 (0.28) acids were obtained in the present study. These results are almost identical to those reported by Karamichou et al. [40], 0.31 and 0.27, for the respective MUFA. Estimates of genomic heritability were high for CLA c9t11 (0.46), and moderate for C18:3 ω3 (0.34) and C18:2 ω6 (0.27). The heritabilities estimated by Karamichou et al. [40] was identical for C18:3 ω3 (0.34), however, were lower for CLA c9t11 (0.30) and C18:2 ω6 (0.10). Bolormaa et al. [41] estimated genomic heritability of 0.15 for C18:2 ω6 and Mortimer et al. [42], using the traditional relationship matrix, reported a heritability estimate of 0.22 for the same acid for crossbred and Merino sheep.
Estimates of genomic heritability reported herein for SFAt (0.32), MUFAt (0.31), and PUFAt (0.28) were all moderate. These results are lower than those reported by Karamichou et al. [40] who reported heritabilities above 0.40 for the same traits. Generally, these results suggest that there is an important genetic effect associated with phenotypic variation in these traits, although two different breeds were used between the current study and that of Karamichou et al. [40].
Estimates of genomic heritability for ω6t (0.27) and ω3t (0.37) were moderate. Genomic heritability estimates for these traits have not been previously reported in sheep populations. However, in beef cattle, genomic heritability estimates for ω3t and ω6t ranged from 0.08 to 0.34, slightly lower in general than the present study [2, 43–45].
The genomic heritability estimates for ω6/ω3 (0.33) and PUFA/SFA (0.28) ratios were moderate. These genetic parameter estimates have not been reported for a sheep population in the literature until now. In cattle for the ω6/ω3 and PUFA/SFA ratios, smaller genomic heritabilities were estimated for Bos indicus cattle, ranging from 0.07 to 0.14 [43, 44], and higher for Bos taurus, ranging from 0.12 to 0.42 [2, 45].
In general, genomic heritability estimates for the fatty acid content indicate that substantial genetic variation exists for the evaluated traits, thus, there is a possibility of altering the fatty acid profiles through selection. These results are important for sheep breeding programs that aim at improving the meat fatty acid composition. Although the genomic heritabilities of fatty acids are of different magnitude compared to the literature, it was possible to observe some consistency in the results, where higher estimates of genomic heritabilities were always obtained for C14:0, C16:0, C18:0, C16:1 and C18:1.
The moderate to high genomic heritabilities observed in this study can be attributed to the fact that there has been no direct selection for these traits in the current population, and thus substantial genetic variation has been maintained [46].
Genome-wide association studies
The GWAS was performed to identify genomic regions (genomic windows) that explained the highest proportion of genetic variance. Each window was further divided into 10 continuous SNPs. A total of 27 different QTL regions ranging from 321,411 to 860,291 Kb were distributed in 11 different chromosomes: 1, 2, 3, 5, 8, 12, 14, 15, 16, 17, and 18. Twenty-three different putative candidate genes (PCG) harbored into these 11 associated genomic regions were identified, which are related to biological processes associated with fatty acid content in skeletal muscle.
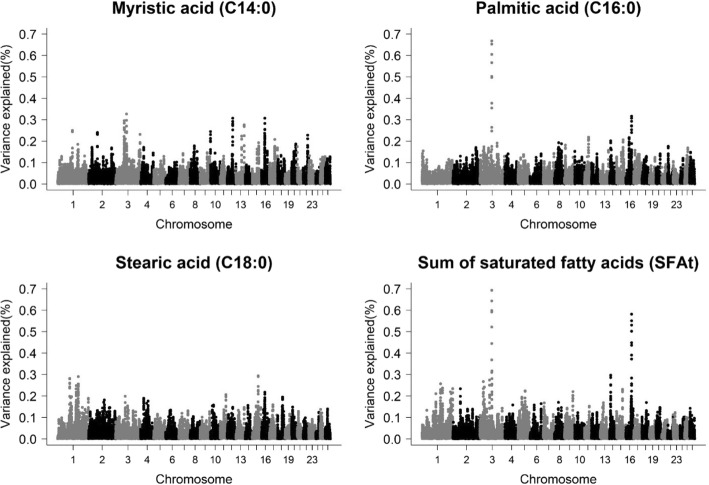
Saturated fatty acids
Ten different QTL regions were associated with C14:0, C16:0, C18:0 and SFAt, which explained between 0.30 and 0.74% of the additive genetic variance (Table 3 and Fig. 1). QTL regions on chromosomes 3 and 16 were associated with C14:0, harboring NPAS2 (neuronal PAS domain protein 2) and MRPS30 (mitochondrial ribosomal protein S30) genes, respectively. NPAS2 gene can be considered as PCG due to the important role in metabolic pathways for regulating lipid metabolism. These pathways include peroxisome proliferator-activated receptor alpha (PPARα), which acts in the control of beta-oxidation of fatty acids [47]. PPARs are receptors for nuclear hormones and transcription factors that regulate the expression of genes involved in lipid and glucose metabolism [48].
Table 3.
Genomic regions and candidate genes associated with the saturated fatty acid (SFA) profile of the Longissimus dorsi muscle of Santa Inês sheep
| Trait | Nomenclature | Genomic Window | Length (Kb) | aVg (%) | Candidate genes |
|---|---|---|---|---|---|
| Myristic acid | C14:0 | 3:78397685–78915234 | 517,549 | 0.30 | – |
| 3:99901070–100285876 | 384,806 | 0.33 | NPAS2 | ||
| 16:30002131–30349083 | 346,952 | 0.31 | MRPS30 | ||
| Palmitic acid | C16:0 | 3:107748212–108088562 | 340,350 | 0.67 | TPH2, TRHDE |
| 16:55961351–56458897 | 497,546 | 0.32 | CDH12 | ||
| Stearic acid | C18:0 | 1:185195800–185614760 | 418,960 | 0.30 | PARP14 |
| 15:52854077–53676145 | 822,068 | 0.39 | DGAT2, WNT11 | ||
| Sum of SFA | SFAt | 3:107809276–108146152 | 336,876 | 0.74 | TPH2, TRHDE |
| 3:109215221–109694971 | 479,750 | 0.32 | – | ||
| 14:11458148–11862331 | 404,183 | 0.30 | – | ||
| 15:52854077–53676145 | 822,068 | 0.58 | DGAT2, WNT11 |
aadditive genetic variance explained by each window
Fig. 1.
Manhattan plot for the proportion of genetic variance explained by windows of 10 adjacent SNPs for saturated fatty acids in the Longissimus dorsi muscle of Santa Inês sheep
MRPS30 is related to the synthesis of proteins inside the mitochondria, being one of more than 70 mitochondrial ribosomal protein components that are encoded by the nuclear genome [49]. One of the Gene Ontology (GO) terms related to this gene is cellular apoptosis (GO:0006915), playing an important role in the induction of apoptosis by saturated fatty acids in several cells [6, 8, 50–52].
Two genomic regions were associated with C16:0 content, on chromosome 3 at 107.7 Mb. In the first region two PCG were identified, TPH2 (tryptophan hydroxylase 2) and TRHDE (thyrotropin releasing hormone degrading enzyme). TPH2 is associated with the serotonergic system, involved in a large number of physiological functions including lipolysis [53, 54] and oxidoreductase activity (GO:0004510), which is involved in oxidation reactions of a variety of substrates including retinoids and steroids [55].
TRHDE gene is related to thyrotropin-releasing hormone in humans [56]. In general, thyroid hormone plays a critical role in mediating changes in development and metabolism in humans [57, 58]. GO annotations relate this gene to the integral component of the plasma membrane (GO:0005887). In sheep, the TRHDE gene is an important candidate gene, because it was related to the amount of internal fat in Merino sheep [59], and post-weaning gain in three sheep populations [60].
In the region on chromosome 16 at 55 Mb, the CDH12 (cadherin 12) gene was found, which was associated with the C14:0 and C18:0 content in milk of Holstein cows and with the Wnt signaling pathway [61]. The Wnt signaling pathway has been shown to have inhibitory effects of adipogenesis [62, 63]. Wnt signaling pathways dysfunctions are associated with obesity and lipodystrophy [64, 65]. Additionally, GO annotations relate the CDH12 as an integral component of the cell membrane (GO:0016021).
Two regions on different chromosomes were associated with C18:0, on chromosome 1 at 185 Mb and chromosome 15 at 52.85 Mb. Chromosome 1 harbors the PARP14 gene (poly (ADP-ribose) polymerase family member 14) and chromosome 15 includes DGAT2 (diacylglycerol o-acyltransferase 2) and WNT11 (Wnt family member 11) genes. PARP14 is involved in the aerobic glycolysis process and promotes the survival of cancer cells by regulating the transcription of template DNA (GO:0006355).
DGAT2 (diacylglycerol o-acyltransferase 2), which is an important gene associated with the catalysis of the final stage of triacylglycerol biosynthesis [66] was identified in the region of chromosome 15 at 52.85 Mb. As the CDH12 gene, WNT11 gene is related to the Wnt signaling pathway and it is also linked to adipogenesis processes [67].
For SFAt, four genomic regions were found. Chromosome 3 at 107.8 Mb overlapped with the region associated with C16:0 (chromosome 3 at 107.7 Mb) and the same candidate genes (TPH2 and TRHDE) were found. The region of chromosome 15 at 52.85 Mb was common to C18:0, therefore, the same candidate genes were observed (DGAT2 and WNT11). This is expected since a high proportion of SFAt is composed by C16:0 and C18:0. In the regions of chromosome 3 at 109 Mb and chromosome 14 at 11 Mb, no candidate genes were identified.
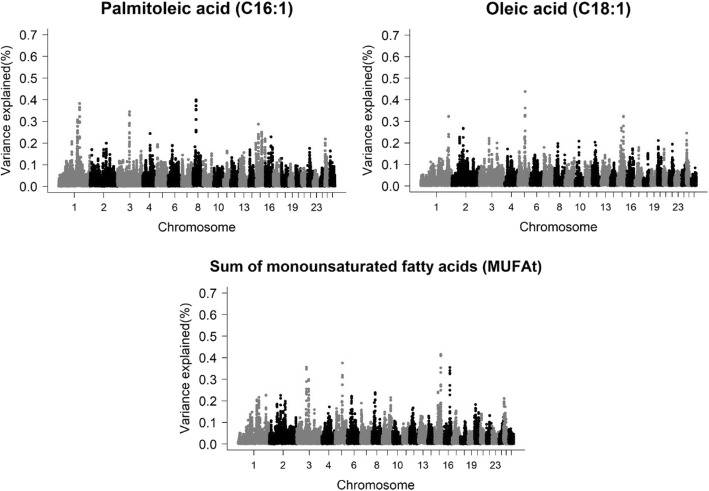
Monounsaturated fatty acids
For MUFA, ten different genomic regions were found (Table 4 and Fig. 2). For C18:1, three genomic regions encompassing PCG were found. The first region was observed on chromosome 1 at 247 Mb and the PCG detected was COPB2 (coatomer protein complex subunit beta 2). The COPB2 gene plays an essential role in the Golgi membrane (GO:0000139) and metabolic pathways related to the transport of cholesterol and sphingolipid of the Golgi complex and the endoplasmic reticulum to the plasma membrane in the region (GO:0006888).
Table 4.
Genomic regions and candidate genes associated with the monounsaturated fatty acids (MUFA) profile of the Longissimus dorsi muscle of Santa Inês sheep
| Trait | Nomenclature | Genomic Window | Length (Kb) | aVg (%) | Candidate genes |
|---|---|---|---|---|---|
| Oleic acid | C18:1 | 1:247008204–247578979 | 570,775 | 0.32 | COPB2 |
| 5:69713533–70306809 | 593,276 | 0.44 | – | ||
| 15:52894279–53729913 | 835,634 | 0.32 | DGAT2 | ||
| Palmitoleic acid | C16:1 | 1:168328606–168777508 | 448,902 | 0.31 | ALCAM |
| 1:185195800–185614760 | 418,960 | 0.38 | PARP14 | ||
| 3:107809276–108146152 | 336,876 | 0.33 | TPH2, TRHDE | ||
| 3:109215221–109694971 | 479,750 | 0.34 | – | ||
| 8:28651040–29263838 | 612,798 | 0.40 | FOXO3, OSTM1 | ||
| Sum of MUFA | MUFAt | 3:109215221–109694971 | 479,750 | 0.30 | – |
| 3:88827198–89234916 | 407,718 | 0.36 | – | ||
| 5:69713533–70306809 | 593,276 | 0.38 | – | ||
| 15:52894279–53729913 | 835,634 | 0.42 | DGAT2 |
aadditive genetic variance explained by each window
Fig. 2.
Manhattan plot for the proportion of genetic variance explained by windows of 10 adjacent SNPs for monounsaturated fatty acids in the Longissimus dorsi muscle of Santa Inês sheep
In the second region located on chromosome 5 at 69 Mb, no candidate genes were found. The region on chromosome 15 at 52.89 Mb overlapped with the C18:0 region (chromosome 15 at 52.85 Mb) including a common PCG (DGAT2). This genetic correlation can be attributed to the fact that C18:0 is a precursor of C18:1, by the desaturation process [68].
We found five genomic regions associated with C16:1 acid. On chromosome 1 at 168 Mb the ALCAM (activated leukocyte cell adhesion molecule) gene was found, which encodes the activated leukocyte adhesion molecule, linked to cell adhesion and migration (GO:0007155), adaptive immune response (GO:0002250), and immunological synapse (GO:0001772). Another region on chromosome 1 at 185 Mb was common between C18:0 and C16:1, so the same candidate gene was detected (PARP14). Two regions of chromosome 3 at 107.8 and 109 Mb associated with C16:1 were found, however in the second, no candidate genes were identified. The first region was also associated with SFAt and C16:0, thus the same candidate genes were identified (TPH2 and TRHDE). Because C16:0 acid is a precursor of C16:1 (desaturation) and C18:0 (elongation) [68] and is related to the SFAt, it is expected that there are common genetic mechanisms for these fatty acids.
In the region of chromosome 8 at 28 Mb two candidate genes FOXO3 (forkhead box O3) and OSTM1 (osteopetrosis associated transmembrane protein 1) were associated with C16:1. The FOXO3 gene is involved in functions related to apoptosis through the expression of genes necessary for cell death [69]. Unsaturated or saturated fatty acids play an essential role in cell apoptosis process, such as hepatocytes [70] and β-cells [71].
OSTM1 encodes a protein involved in the degradation of G proteins via the ubiquitin-dependent proteasome pathway (RefSeq, Jul 2008 [72]). These proteins have been reported to be related to the regulation of body weight and metabolic function, hyperinsulinemia, glucose tolerance, and insulin resistance [73].
Four genomic regions associated with MUFAt were found, with two being in common to C18:1 (chromosome 5 at 69 Mb and chromosome 15 at 52.89 Mb), and one common to C16:1 (chromosome 3 at 109 Mb). Thus, PCG was only found on chromosome 1 (DGAT2). The levels of lipids in the blood plasma may have a direct influence on adaptive immunity [74]. Another region was observed on chromosome 3 at 88 Mb without any associated candidate gene.
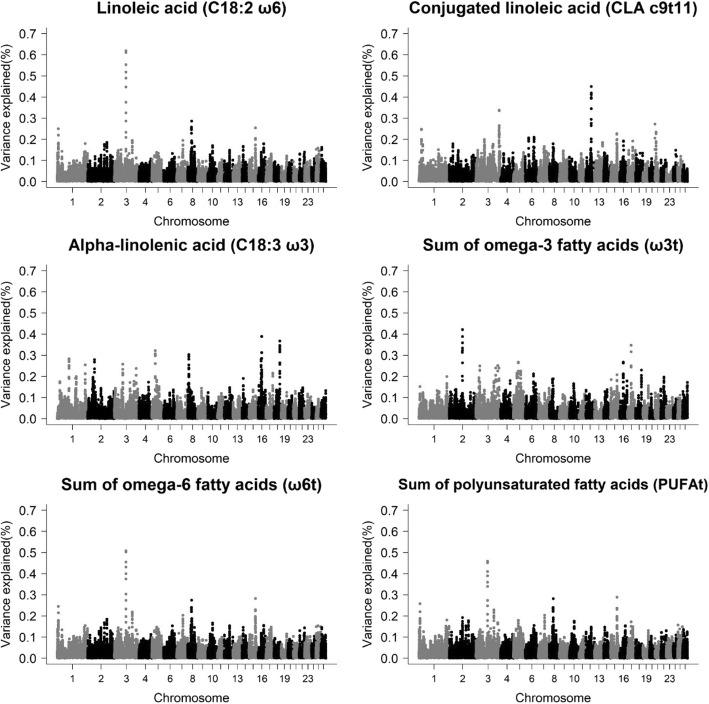
Polyunsaturated fatty acids
Eleven different genomic regions were observed for C18:3 ω3, C18:2 ω6, CLA c9t11, ω6t, ω3t, and PUFAt, distributed over eight chromosomes (Table 5 and Fig. 3). For C18:2 ω6, two regions were found on different chromosomes. One of these regions was common to C16:1 and SFAt (chromosome 3 at 107.8 Mb) and overlapped to C16:0 (107.7). Thus, the same PCG were identified (TPH2 and THRDE). In the region of chromosome 8 at 35 Mb, no PCG were found.
Table 5.
Genomic regions and candidate genes associated with the polyunsaturated fatty (PUFA) acids profile of the Longissimus dorsi muscle of Santa Inês sheep
| Trait | Nomenclature | Genomic Window | Length (Kb) | aVg (%) | Candidate genes |
|---|---|---|---|---|---|
| Linoleic acid | C18:2 ω6 | 3:107809276–108146152 | 336,876 | 0.62 | TPH2, TRHDE |
| 8:35511823–35833234 | 321,411 | 0.30 | – | ||
| alpha-Linolenic acid | C18:3 ω3 | 5:35136857–35559907 | 423,050 | 0.32 | TNFAIP8 |
| 8:10032564–10601751 | 569,187 | 0.30 | UBE3D, ME1 | ||
| 16:32987534–33510366 | 522,832 | 0.39 | PLCXD3, C6, C7 | ||
| 18:55048766–55542400 | 493,634 | 0.37 | CCDC88C, FBLN5 | ||
| Conjugated linoleic acid | CLA c9t11 | 3:212363790–212692333 | 328,543 | 0.34 | CACNA1C |
| 12:49048350–49840917 | 792,567 | 0.45 | – | ||
| Sum of omega-3 FA | ω3t | 2:122297721–123158012 | 860,291 | 0.42 | – |
| 16:33207525–33550464 | 342,939 | 0.30 | C6, C7 | ||
| Sum of omega-6 FA | ω6t | 3:107809276–108146152 | 336,876 | 0.51 | TPH2, TRHDE |
| 15:58185924–58511281 | 325,357 | 0.30 | – | ||
| Sum of PUFA | PUFAt | 3:107809276–108146152 | 336,876 | 0.46 | TPH2, TRHDE |
| 8:35511823–35833234 | 321,411 | 0.30 | – | ||
| 15:58185924–58511281 | 325,357 | 0.30 | – |
aadditive genetic variance explained by each window
Fig. 3.
Manhattan plot for the proportion of genetic variance explained by windows of 10 adjacent SNPs for polyunsaturated fatty acids in the Longissimus dorsi muscle of Santa Inês sheep
For C18:3 ω3, four genomic regions were found on chromosomes 5, 8, 16, and 18. In the region of chromosome 5 at 35 Mb, the candidate gene identified was the TNFAIP8 (tumor necrosis factor, alpha-induced protein 8). In humans, this gene has been confirmed to be important for the maintenance of immune homeostasis and a regulator of apoptosis, and plays a main role in the oncogenesis of several cancer types [75].
In the region located on chromosome 8 at 10 Mb the ME1 (malic enzyme 1) and UBE3D (ubiquitin-protein ligase E3D) genes were identified. ME1 is related to the tricarboxylate transport system that produces NADPH and acetyl-CoA, necessary components in fatty acid biosynthesis [76]. On the other hand, the UBE3D gene is related to E3 ubiquitin-protein ligase which accepts ubiquitin from specific E2 ubiquitin-conjugating enzymes, and transfers it to substrates, usually supporting their degradation by the proteasome [77, 78]. In a general context, the function of the ubiquitin-proteasome pathway can be controlled physiologically, in part, by fatty acids within cellular membranes [79].
On chromosome 16 at 32 Mb the following candidate genes associated with C18:3 ω3 were detected: PLCXD3 (phosphatidylinositol-specific phospholipase C X domain containing 3), C6 (complement C6), and C7 (complement C7) genes. PLCXD3 gene was related to phospholipases, a group of enzymes that hydrolyze the phospholipids in fatty acids and other lipophilic molecules (GO:0016042). C6 and C7 are genes related to the membrane attack complex (GO:0005579), playing key roles in the innate and acquired immune response (GO:0045087) by assisting in inflammatory responses against infections [80]. The ω3 acids influence the activation of inflammatory cells processes from signal transduction to protein expression, even involving effects at the genomic level [81].
In the region of chromosome 18 at 55 Mb, related to C18:3 ω3, the PCG were CCDC88C (coiled-coil domain containing 88C) and FBLN5 (fibulin-5). CCDC88C has been described as a negative regulator of the Wnt signaling pathway (GO: 0016055). FBLN5 plays an essential role in the assembly of elastic fibers that provide strength and flexibility to the connective tissue. This gene also exerts an important pleiotropic effect together with the DGAT1 gene in Holstein dairy cows [82].
For CLA c9t11, two genomic regions were found in the region on chromosome 3 at 212 Mb, the PCG identified was the CACNA1C (calcium voltage-gated channel subunit alpha1 C). CACNA1C belongs to a family of genes that provide instructions for constructing calcium channels (GO:0005891). Long chain fatty acids are involved in the calcium channel activation processes, possibly acting at some nearby lipid binding sites on these channels or directly over the channel protein itself [83–85]. In the region of chromosome 12 at 49 Mb, no PCG related to CLA c9t11 was observed.
Two genomic regions were associated with ω3t on two different chromosomes. The region of chromosome 16 at 33 Mb overlapped with the genomic region associated with C18:3ω3 and the same genes were observed (C6 and C7). For ω6t two genomic regions were observed and explained greater than 0.30% of the additive genetic variance. One of these regions was common to C18:2 ω6 on chromosome 3 at 107 Mb, and the same candidate genes were found (TPH2 and TRHDE). In the region of chromosome 15 at 58 Mb, none candidate gene was found. For PUFAt, three genomic regions in common to 18:2 ω6 and ω6t were found, consistent with the fact that the same regions were associated with other PUFA traits.
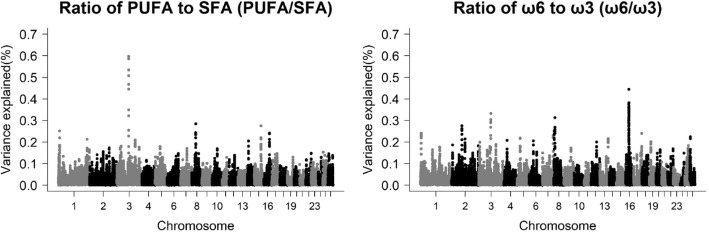
ω6/ω3 and PUFA/SFA ratios
Six genomic regions were observed for the ω6/ω3 ratio, whereas only one genomic region was observed on chromosome 3 for the PUFA/SFA ratio (Table 6 and Fig. 4). For the ω6/ω3 ratio, only the regions of chromosome 8 at 10 Mb and chromosome 16 at 30 Mb harbored PCG. The region of chromosome 8 was common to C18:3 ω3, thus, the same candidate genes were observed (UBE3D and M1). This result is expected since C18:3 ω3 is one of the most abundant ω3 acids, and consequently, it is directly involved in the ω6/ω3 ratio. The region on chromosome 16 was common to C14:0, so the same candidate gene was observed (MRPS30) suggesting that there may be a genetic correlation with some other polyunsaturated fatty acid that has not been addressed in this study. For the PUFA/SFA ratio, only one region on chromosome 3 at 107.8 Mb, common to several individual fatty acids and groups of the fatty acids (C16:0, C16:1, PUFAt, SFAt, C18:2 ω6, and ω6t) was found. This was expected given the PUFA/SFA ratio uses all these fatty acids for its calculation.
Table 6.
Genomic regions and candidate genes associated with the ω6/ω3 and PUFA/SFA ratios of the Longissimus dorsi muscle of Santa Inês sheep
| Trait | Nomenclature | Genomic Window | Length (Kb) | aVg (%) | Candidate genes |
|---|---|---|---|---|---|
| Ratio of ω6 to ω3 | ω6/ω3 | 3:109215221–109694971 | 479,750 | 0.33 | – |
| 8:10032564–10601751 | 569,187 | 0.31 | UBE3D, ME1 | ||
| 16:28050501–28436992 | 386,491 | 0.38 | – | ||
| 16:29245079–29691641 | 446,562 | 0.44 | – | ||
| 16:30002131–30349083 | 346,952 | 0.38 | MRPS30 | ||
| Ratio of PUFA to SFA | PUFA/SFA | 3:107809276–108146152 | 336,876 | 0.60 | TPH2, TRHDE |
aadditive genetic variance explained by each window
Fig. 4.
Manhattan plot for the proportion of genetic variance explained by windows of 10 adjacent SNPs for PUFA/SFA and ω6/ω3 ratios in the Longissimus dorsi muscle of Santa Inês sheep
In the present study, important genomic regions associated with fatty acids profile were identified, providing an improved biological understanding of fatty composition in ovine. The identification of these genes in sheep using genomic approach is the first step to search for causal variations of large effect and can contribute to genetic evaluations of relevant traits in the future. Despite the observed moderate genomic heritabilities, our results suggest the meat fatty acid profile of Santa Inês sheep is controlled mainly by many QTL of small effects. This was expected since from the quantitative genetics point of view, meat fatty acids profile is a complex trait controlled by multiple genes and is influenced by several loci throughout the genome [86]. Additionally, genomic regions identified explained most, but not all, of the additive genetic variance for traits, possibly because there are causal mutations with low minor allele frequency and consequently in incomplete linkage disequilibrium with the SNPs [86]. Therefore, the identification of genetic variants of large effects can be difficult because the contribution of each genomic region to additive genetic variance is small. This implies that genomic selection may be an essential tool for the improvement of these traits, since it captures the effects of all genetic markers simultaneously. In addition, these genomic regions can be used in fine mapping studies, which will be useful to search for causative variations.
Conclusion
Moderate to high genomic heritabilities were estimated for fatty acid profiles in this study, suggesting that these traits can be altered by selection. Several genomic regions and PCG associated with fatty acid profiles were identified, which can be used in later studies to fine-mapping the causal variations. The results described in this study have not been previously reported in sheep. Thus, this research is the first step toward understanding the genetic and metabolic mechanisms involved in the phenotypic determination of fatty acids in sheep meat; information that can be useful to define the selection strategies for these traits with the aim to obtain a benefic product to human health.
Methods
Animals and phenotypes
The study was conducted using phenotypes of 216 non-castrated male Santa Inês sheep, selected randomly, with unknown pedigree. Genomic relationships revealed full- and half-sibling relationships as well as unrelated individuals, with an average relationship among individuals of − 0.005 (standard deviation = 0.0648; minimum = − 0.12; maximum = 0.97). The experiment was conducted at the Experimental Farm of the School of Veterinary Medicine and Animal Science belonging to the Federal University of Bahia, located in the municipality of São Gonçalo dos Campos – Bahia. Animals were raised in individual pens (1.0 m2) from 110 days of age on average and slaughtered around 90 days later. Meat samples were collected from the Longissimus dorsi muscle, between the 12 and 13th ribs of each animal, 24 h after slaughter and stored at − 20 °C until further analysis to determine the fatty acid profile and intramuscular fat percentage (IMF). The animals were raised in three different periods between 2015 and 2016. The concentrate for these 216 ovine consisted of ground corn grain, soybean meal, urea, mineral supplement specific for sheep and the forage was Tifton 85 hay. All diets consisted of 2.45 Mcal/kg DM of metabolisable energy, all of them formulated according to the NRC (2007), in order to meet the nutritional requirements for lambs with estimated weight gains of 200 g/day. Of these, 100 individuals were separated into 5 experimental groups with 20 animals each and received diets comprised of cottonseed (ground or whole) with presence or absence of chitosan (in five different levels). Animals were fed with a forage:concentrate ratio of 50:50.
Other group of 72 Santa Inês lambs were separated into four sets with 18 individuals each. The diets corresponded to two sizes of hay particles of Tifton-85 (6 and 13 mm) and two forage:concentrate ratios (50:50 and 70:30). The diets for these 172 animals were formulated to be isonitrogeneous with 16% crude protein.
Another set of animals consisted of 38 animals separated by four groups ranging from 9 to 10 animals each. Each group received diets with a forage:concentrate ratio of 50:50 with different levels of crude protein: fixed level with 11% crude protein; fixed level with 13% crude protein; oscillating level 11 and 13% crude protein; oscillating level 13 and 11% crude protein. The remaining six animals received the same diet based on hay and concentrate (50:50), but with 16% crude protein.
IMF content was determined at the Meat Science Laboratory in the Department of Animal Science at UFLA (Lavras, Minas Gerais, Brasil), performed by near-infrared spectrophotometer according to the AOAC: 2007–04 method [87] in Longissimus dorsi muscle (~ 180 g, without fat cover) using FoodScan™ (FOSS, Hillerød, Denmark) with an artificial neural network calibration model and database for the determination of intramuscular fat [88].
The extraction, methylation and reading steps for the determination of fatty acid composition in the Longissimus dorsi (30 g) muscle were conducted at the Animal Nutrition and Growth Laboratory at ESALQ (Piracicaba, São Paulo, Brazil). The fatty acid extraction was conducted as described by Hara and Radin [89]. Subsequently, extracted lipids were hydrolyzed and methylated as described by Christie [90]. Fatty acid methyl esters were quantified with a gas chromatograph (ThermoFinnigan, Thermo Electron Corp., MA, USA) equipped with a flame ionization detector and a 100 m Supelco SP-2560 (Supelco Inc., PA, USA) fused silica capillary column (100 m, 0.25 mm and 0.2 μ m film thickness). The column oven temperature was held at 70 °C for 4 min, then increased to 170 °C at a rate of 13 °C min − 1, and subsequently increased to 250 °C at a rate of 35 °C min − 1, and held at 250 °C for 5 min. The gas fluxes were 1.8 mL min − 1 for carrier gas (He), 45 mL min − 1 for make-up gas (N2), 40 mL min − 1 for hydrogen, and 450 mL min − 1 for synthetic flame gas. One μL of the esterified extract was injected in the chromatographer. Injector and detector temperatures were 250 and 300 °C, respectively.
The fatty acids were identified by comparison of the retention times of methyl esters in the samples with standards of fatty acids from butter reference BCR-CRM 164, Anhydrous Milk Fat-Producer (BCR Institute for Materials and Reference Measurements) and also with commercial standard for 37 fatty acids Supelco TM Component FAME Mix (cat 18,919, Supelco, Bellefonte, PA). Fatty acids were quantified by normalizing the areas of methyl esters. Fatty acids were expressed as a weight percentage (mg/mg), obtained using ChromQuest 4.1 software (Thermo Electron, Milan, Italy).
After extraction, 47 different fatty acids were obtained from the Logissimus dorsi muscle of Santa Inês sheep. Of these, the following fatty acids were selected: myristic (C14:0), palmitic (C16:0), stearic (C18:0), palmitoleic (C16:1), oleic (C18:1, cis-9), linoleic (C18: 2 ω6), conjugated linoleic acid (CLA c9t11) and alpha-linolenic (C18: 3 ω3). These individual fatty acids were selected considering their importance for human health and abundance in the samples evaluated. The sum of saturated acids (C6:0 + C8:0 + C10:0 + C11:0 + C12:0 + C13:0 + C13:0 anteiso + C13:0 iso + C14:0 + C14:0iso + C15:0 + C15:0 anteiso + C15:0 iso + C16:0 + C16:0 iso + C17:0 + C17:0 iso + C18:0 + C20:0 + C22:0 + C23:0 + C24:0), sum of monounsaturated acids (C10:1 + C14:1 c9 + C16:1 + C17:1 + C18:1 c11 + C18:1 c12 + C18:1 c13 + C18:1 c15 + C18:1 + C18:1 t16 + C18:1 t9 + C20:1 + C22:1 + C24:1), sum of poly-unsaturated fatty acids (C18:2 ω6 + CLA c9t11 + C18:3 ω3 + C18:3 ω6 + C20:3 ω6 + C20:2 ω6 + C20:3 ω3 + C20:4 ω6 + C20:5 ω3 + C22:5 ω3 + C22:6 ω3), sum of omega 6 acids (C18:2 ω6 + C18:3 ω6 + C20:3 ω6 + C20:2 ω6 + C20:4 ω6), and sum of omega 6 acids (C18:3 ω3 + C20:3 ω3 + C20:5 ω3 + C22:5 ω3 + C22:6 ω3) were calculated. We also calculated the polyunsaturated/saturated fatty acid and omega 6/omega 3 ratios.
Genotyping of animals and quality control
Genotyping was performed at the Biotechnology Laboratory at ESALQ, Piracicaba, São Paulo, Brazil. A total of 216 animals were genotyped using 54,241 SNPs from the Ovine SNP50 BeadChip (Illumina Inc., San Diego, CA). Quality control of the SNPs consisted of excluding those located on sex chromosomes; monomorphic; minor allele frequency lower than 0.05; call rate lower than 90%; and Hardy-Weinberg equilibrium deviations (difference between expected and observed frequency of heterozygous) higher than 0.15. All genotypes achieved a call rate greater than 90%, and thus no genotypes were removed due to this threshold. After quality control, 42,363 SNPs and 216 animals were retained for further analyses.
Genetic analysis
Genetic (co)variance components were estimated by restricted maximum likelihood (REML) method with an average information algorithm using a genomic relationship matrix, which was calculated as described by Van Raden [16]. Models that account for genomic information are powerful tools for capturing a large proportion of the additive genetic variation, increasing the estimation accuracy of the genetic parameters [2, 91]. The analyses were performed using the BLUPF90 family programs [92]. Genomic heritabilities and genomic breeding values (GEBV) for fatty acid profiles were estimated by bivariate analyses with IMF as an anchor trait. The variance components were fixed to the estimates obtained from the univariate analysis for IMF only, and the realized genomic relationship matrix calculated from the SNP marker information was fitted [16]. The anchor trait approach was used to minimize the bias associated with a sample of selected individuals [93] in the event that selection had occurred for some measure of fatness in this population. Additionally, it is expected that the use of multivariate models will have larger or at least similar power compared to univariate models [94, 95]. Use of related phenotypic traits can improve the power of candidate gene detection [96]. Furthermore, the pattern of pleiotropy could support the detection of candidate genes underlying an association and the genetic mechanisms responsible [41]. For the bivariate analyses the following genomic best linear unbiased prediction (GBLUP) model was used:
where the vectors y1 and y2 refer to the observations of the IMF and fatty acid traits, respectively; X1 and X2 are the design matrices and b1 and b2 are the vectors of the fixed effects for the first and second trait, respectively; Z1 and Z2 are the design matrices and u1 and u2 are the vector of genomic breeding values of the two traits; and e1 and e2 are the vectors of the residual effects. For univariate and bivariate analyses the vector b included the fixed effect of contemporary groups (n = 14, comprised of 6 to 20 individuals) where formed by diet and period of confinement as previously reported. Also, it was assumed that , where G is the realized genomic relationship matrix and is the variance and covariance matrix of the genomic breeding values for the two traits; and , where is the residual variance and covariance matrix for the two traits.
For GWAS the effects of the SNPs () were obtained from GEBV using the following equation described by [97]:
where is the vector of SNP effects; is vector of the GEBV obtained for the genotyped animals; W is a genotype matrix containing the numbers of reference alleles; D is a diagonal matrix of the weights of SNP variances, however, in this study the weights of SNP were not used, thus D = I (identity matrix). It is suggested that the use of SNP windows captures the QTL effects more efficiently than the use of a single SNP, and is relevant to distinguish effects from statistical noise [98]. Thus, the results of GWAS were reported as the proportion of variance explained by a window of 10 adjacent SNPs. The percentage (%) of genetic variance explained by each region was caling equation:
where ui is the genetic value of the i-th region that consisted of 10 consecutive SNPs, is the total genetic variance, Wj is the vector of gene content of the j-th SNP for all individuals, and is the marker effect of the j-th SNP within the i-th region.
Searching for genes
Fatty acid profile traits are polygenic, being affected by many markers with small effect and non-genetic factors. Since the fatty acids traits evaluated in this study seem to be controlled mainly by many QTL of small effect, identifying large effect genes will be difficult, given each marker has a minor contribution to the total genetic variation. Consequently, only genomic regions explaining the largest proportion of additive genetic variation, above 0.30%, were considered to determine the possible QTL regions associated with fatty acids profile traits. The selected regions were used to identify positional candidate genes based on the starting and ending coordinates of each window by surveying the database available in the NCBI (National Center for Biotechnology Information) in OAR3.1 version of the ovine genome and Ensembl Genome Browser [99]. The description of genes regarding their biological function was performed by the DAVID [100] and BioGPS [101] online annotation databases. As needed, human genes were used as background in pathway and gene network investigation.
Acknowledgements
The authors are grateful for the support from the University of São Paulo (ESALQ/USP) and Federal University of Bahia (UFBA).
Funding
This study was conducted with funding from São Paulo Research Foundation (FAPESP) (Proc 13/04504–3) and National Council for Scientific Technological Development (CNPq).
Availability of data and materials
The raw data used during the current study are available from the corresponding author on request.
Abbreviations
- CLA
Conjugated linoleic acid
- FA
Fatty acid
- GEBV
Genomic breeding values
- GO
Gene ontology
- GWAS
Genome-wide association study
- IMF
Intramuscular fat percentage
- MUFA
Monounsaturated fatty acid
- MUFAt
Total monounsaturated fatty acid
- PCG
Putative candidate gene
- PUFA
Polyunsaturated fatty acid
- PUFAt
Total polyunsaturated fatty acid
- QTL
Quantitative trait loci
- SFA
Saturated fatty acid
- SFAt
Total saturated fatty acid
- SNP
Single nucleotide polymorphism
- ω3t
Total of omega 3 fatty acids
- ω6t
Total of omega 6 fatty acids
Authors’ contributions
GAR, SFNP, and GBM conceived and designed the study. ABA, JP, GAR and GBM were crucial in phenotypic and genotypic data editing and statistical analysis. ABA, WVBS, SFNP and GAR participated in the collection of samples for DNA extraction, phenotype collection and statistical analysis. LLC and GBM provided the genotypes. GAR drafted the manuscript. LFBP, GGPC, DPDL, and GBM assisted with phenotypic data. ASMC, FP, VF, JP, GBM, GM, WVBS and MLS edited and contributed important modifications in the manuscript. All authors contributed, read, and agreed on the final manuscript.
Ethics approval and consent to participate
This study was approved by Animal Ethics Committee of the Federal University of Bahia (UFBA), n° 07/2016.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
G. A. Rovadoscki, Email: gregori.rov@usp.br
S. F. N. Pertile, Email: spertile@usp.br
A. B. Alvarenga, Email: abalvarenga@usp.br
A. S. M. Cesar, Email: alinecesar@usp.br
F. Pértille, Email: pertille@usp.br
J. Petrini, Email: juliana.petrini@usp.br
V. Franzo, Email: franzo@usp.br
W. V. B. Soares, Email: webervbs@yahoo.com.br
G. Morota, Email: morota@unl.edu
M. L. Spangler, Email: mspangler2@unl.edu
L. F. B. Pinto, Email: luisfernando@ufba.br
G. G. P. Carvalho, Email: gleidsongiordano@ufba.br
D. P. D. Lanna, Email: dplanna@usp.br
L. L. Coutinho, Email: llcoutinho@usp.br
G. B. Mourão, Email: gbmourao@usp.br
References
- 1.Valencak TG, Gamsjäger L, Ohrnberger S, Culbert NJ, Ruf T. Healthy n-6/n-3 fatty acid composition from five European game meat species remains after cooking. BMC Res Notes. 2015;8:273. doi: 10.1186/s13104-015-1254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saatchi M, Garrick DJ, Tait RG, Mayes MS, Drewnoski M, Schoonmaker J, et al. Genome-wide association and prediction of direct genomic breeding values for composition of fatty acids in Angus beef cattle. BMC Genomics. 2013;14:730. doi: 10.1186/1471-2164-14-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biesalski HK. Meat as a component of a healthy diet - are there any risks or benefits if meat is avoided in the diet? Meat Sci. 2005;70:509–524. doi: 10.1016/j.meatsci.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Haug A, Høstmark AT, Harstad OM. Bovine milk in human nutrition – a review. Lipids Health Dis. 2007;6:25. doi: 10.1186/1476-511X-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu FB, Manson JE, Willett WC. Types of dietary fat and risk of coronary heart disease: a critical review. J Am Coll Nutr. 2001;20:5–19. doi: 10.1080/07315724.2001.10719008. [DOI] [PubMed] [Google Scholar]
- 6.Kasai T, Ohguchi K, Nakashima S, Ito Y, Naganawa T, Kondo N, et al. Increased activity of oleate-dependent type phospholipase D during actinomycin D-induced apoptosis in Jurkat T cells. J Immunol. 1998;161:6469–6474. [PubMed] [Google Scholar]
- 7.Sartorius T, Ketterer C, Kullmann S, Balzer M, Rotermund C, Binder S, et al. Monounsaturated fatty acids prevent the aversive effects of obesity on locomotion, brain activity, and sleep behavior. Diabetes. 2012;61:1669–1679. doi: 10.2337/db11-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann L, Seibt A, Herebian D, Spiekerkoetter U. Monounsaturated 14:1n-9 and 16:1n-9 fatty acids but not 18:1n-9 induce apoptosis and necrosis in murine HL-1 cardiomyocytes. Lipids. 2014;49:25–37. doi: 10.1007/s11745-013-3865-4. [DOI] [PubMed] [Google Scholar]
- 9.Yehuda S, Rabinovitz S, Carasso RL, Mostofsky DI. The role of polyunsaturated fatty acids in restoring the aging neuronal membrane. Neurobiol Aging. 2002;23:843–853. doi: 10.1016/S0197-4580(02)00074-X. [DOI] [PubMed] [Google Scholar]
- 10.Miles EA, Calder PC. Influence of marine n-3 polyunsaturated fatty acids on immune function and a systematic review of their effects on clinical outcomes in rheumatoid arthritis. Br J Nutr. 2012;107 Suppl:S171–S184. doi: 10.1017/S0007114512001560. [DOI] [PubMed] [Google Scholar]
- 11.Bazinet RP, Layé S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat Rev Neurosci. 2014;15:771–785. doi: 10.1038/nrn3820. [DOI] [PubMed] [Google Scholar]
- 12.Wood JD, Enser M, Fisher AV, Nute GR, Sheard PR, Richardson RI, et al. Fat deposition, fatty acid composition and meat quality: a review. Meat Sci. 2008;78:343–358. doi: 10.1016/j.meatsci.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Goddard ME, Hayes BJ. Genomic selection. J Anim Breed Genet. 2007;124:323–330. doi: 10.1111/j.1439-0388.2007.00702.x. [DOI] [PubMed] [Google Scholar]
- 14.Tiezzi F, Parker-Gaddis KL, Cole JB, Clay JS, Maltecca C. A genome-wide association study for clinical mastitis in first parity US Holstein cows using single-step approach and genomic matrix re-weighting procedure. PLoS One. 2015;10:e0114919. doi: 10.1371/journal.pone.0114919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meuwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- 17.Bolormaa S, Pryce JE, Kemper K, Savin K, Hayes BJ, Barendse W, et al. Accuracy of prediction of genomic breeding values for residual feed intake and carcass and meat quality traits in Bos taurus, Bos indicus, and composite beef cattle. J Anim Sci. 2013;91:3088–3104. doi: 10.2527/jas.2012-5827. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Wang Z, Wang S, Li H. Progress of genome wide association study in domestic animals. J Anim Sci Biotechnol. 2012;3:26. doi: 10.1186/2049-1891-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Nudda A, McGuire MK, Battacone G, Manca MG, Boe R, Pulina G. Documentation of fatty acid profiles in lamb meat and lamb-based infant foods. J Food Sci. 2011;76:43–47. doi: 10.1111/j.1750-3841.2010.02027.x. [DOI] [PubMed] [Google Scholar]
- 20.Costa RG, Dos Santos NM, RDCR DEQ, De Sousa WH, Madruga MS, Cartaxo EQ. Physicochemical characteristics and fatty acid profile of meat from lambs with different genotypes and diets. Rev Bras Zootec. 2015;44:248–254. doi: 10.1590/S1806-92902015000700003. [DOI] [Google Scholar]
- 21.Carneiro MMY, Goes RH de T e B de, Silva LHX da, Fernandes ARM, Oliveira RT De, Cardoso CAL, et al. Quality traits and lipid composition of meat from crossbreed Santa Ines ewes fed diets including crushed crambe. Rev Bras Zootec. 2016;45:319–27.
- 22.Diaz M, Velasco S, Cañeque V, Lauzurica S, Ruiz de Huidobro F, Pérez C, et al. Use of concentrate or pasture for fattening lambs and its effect on carcass and meat quality. Small Rumin Res. 2002;43:257–268. doi: 10.1016/S0921-4488(02)00016-0. [DOI] [Google Scholar]
- 23.da Cruz CAC, dos Santos-Cruz CL, Castillo CJC, de Souza AO, da Silva LB, Brito PN. Lipidic characterization of Santa Inês lamb shoulder. Ciência e Tecnol Aliment. 2011;31:508–516. doi: 10.1590/S0101-20612011000200036. [DOI] [Google Scholar]
- 24.Landim AV, Cardoso MTM, Castanheira M, Fioravanti MCS, Louvandini H, McManus C. Fatty acid profile of hair lambs and their crossbreds slaughtered at different weights. Trop Anim Health Prod. 2011;43:1561–1566. doi: 10.1007/s11250-011-9842-0. [DOI] [PubMed] [Google Scholar]
- 25.Sañudo C, Enser ME, Campo MM, Nute GR, Maria G, Sierra I, et al. Fatty acid composition and sensory characteristics of lamb carcasses from Britain and Spain. Meat Sci. 2000;54:339–346. doi: 10.1016/S0309-1740(99)00108-4. [DOI] [PubMed] [Google Scholar]
- 26.Bonanome A, Grundy SM. Effect of Dietary Stearic Acid on Plasma Cholesterol and Lipoprotein Levels. N Engl J Med. 1988;318:1244–8. [DOI] [PubMed]
- 27.Grundy SM. Influence of stearic acid on cholesterol metabolism relative to other long chain fatty acids. Am J Clin Nutr. 1994;60:986S–990S. doi: 10.1093/ajcn/60.6.986S. [DOI] [PubMed] [Google Scholar]
- 28.Tholstrup T, Vessby B, Sandstrom B. Difference in effect of myristic and stearic acid on plasma HDL cholesterol within 24 h in young men. Eur J Clin Nutr. 2003;57:735–742. doi: 10.1038/sj.ejcn.1601605. [DOI] [PubMed] [Google Scholar]
- 29.Mente A, De Koning L, Shannon HS, Anand SS. A systematic review of the evidence supporting a causal link between dietary factors and coronary heart disease. Arch Intern Med. 2009;169:659–669. doi: 10.1001/archinternmed.2009.38. [DOI] [PubMed] [Google Scholar]
- 30.Gillingham LG, Harris-Janz S, Jones PJH. Dietary monounsaturated fatty acids are protective against metabolic syndrome and cardiovascular disease risk factors. Lipids. 2011;46:209–228. doi: 10.1007/s11745-010-3524-y. [DOI] [PubMed] [Google Scholar]
- 31.Joris PJ, Mensink RP. Role of cis-monounsaturated fatty acids in the prevention of coronary heart disease. Curr Atheroscler Rep Current Atherosclerosis Reports. 2016;18:1–7. [DOI] [PMC free article] [PubMed]
- 32.Kang MJ, Shin MS, Park JN, Lee SS. The effects of polyunsaturated:saturated fatty acids ratios and peroxidisability index values of dietary fats on serum lipid profiles and hepatic enzyme activities in rats. Br J Nutr. 2005;94:526–532. doi: 10.1079/BJN20051523. [DOI] [PubMed] [Google Scholar]
- 33.Guo Z, Miura K, Turin TC, Hozawa A, Okuda N, Okamura T, et al. Relationship of the polyunsaturated to saturated fatty acid ratio to cardiovascular risk factors and metabolic syndrome in Japanese: the INTERLIPID study. J Atheroscler Thromb. 2010;17:777–784. doi: 10.5551/jat.4135. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization et al. FAO Food Nutr Pap. 2010;91:1–166. [Google Scholar]
- 35.Harika RK, Eilander A, Alssema M, Osendarp SJM, Zock PL. Intake of fatty acids in general populations worldwide does not meet dietary recommendations to prevent coronary heart disease: a systematic review of data from 40 countries. Ann Nutr Metab. 2013;63:229–238. doi: 10.1159/000355437. [DOI] [PubMed] [Google Scholar]
- 36.Ip C, Dong Y, Ip MM, Banni S, Carta G, Angioni E, et al. Conjugated linoleic acid isomers and mammary Cancer prevention. Nutr Cancer. 2002;43:52–58. doi: 10.1207/S15327914NC431_6. [DOI] [PubMed] [Google Scholar]
- 37.Kelley NS, Hubbard NE, Erickson KL. Conjugated linoleic acid isomers and cancer. J Nutr. 2007;137:2599–2607. doi: 10.1093/jn/137.12.2599. [DOI] [PubMed] [Google Scholar]
- 38.Viladomiu M, Hontecillas R, Bassaganya-Riera J. Modulation of inflammation and immunity by dietary conjugated linoleic acid. Eur J Pharmacol. 2016;785:87–95. [DOI] [PubMed]
- 39.Simopoulos AP. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp Biol Med (Maywood) 2008;233:674–688. doi: 10.3181/0711-MR-311. [DOI] [PubMed] [Google Scholar]
- 40.Karamichou E, Richardson RI, Nute GR, Gibson KP, Bishop SC. Genetic analyses and quantitative trait loci detection, using a partial genome scan, for intramuscular fatty acid composition in Scottish blackface sheep. J Anim Sci. 2006;84:3228–3238. doi: 10.2527/jas.2006-204. [DOI] [PubMed] [Google Scholar]
- 41.Bolormaa S, Hayes BJ, van der Werf JHJ, Pethick D, Goddard ME, Daetwyler HD. Detailed phenotyping identifies genes with pleiotropic effects on body composition. BMC Genomics. 2016;17:224. doi: 10.1186/s12864-016-2538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mortimer SI, van der Werf JHJ, Jacob RH, Hopkins DL, Pannier L, Pearce KL, et al. Genetic parameters for meat quality traits of Australian lamb meat. Meat Sci. 2014;96:1016–1024. doi: 10.1016/j.meatsci.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Cesar AS, Regitano LC, Tullio RR, Lanna DP, Nassu RT, Mudado MA, et al. Genome-wide association study for intramuscular fat deposition and composition in Nellore cattle. BMC Genet. 2014;15:39. doi: 10.1186/1471-2156-15-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemos MVA, Chiaia HLJ, Berton MP, Feitosa FLB, Aboujaoud C, Camargo GMF, et al. Genome-wide association between single nucleotide polymorphisms with beef fatty acid profile in Nellore cattle using the single step procedure. BMC Genomics. 2016;17:213. doi: 10.1186/s12864-016-2511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L, Ekine-Dzivenu C, Vinsky M, Basarab J, Aalhus J, Dugan MER, et al. Genome-wide association and genomic prediction of breeding values for fatty acid composition in subcutaneous adipose and longissimus lumborum muscle of beef cattle. BMC Genet. 2015;16:135. doi: 10.1186/s12863-015-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nogi T, Honda T, Mukai F, Okagaki T, Oyama K. Heritabilities and genetic correlations of fatty acid compositions in longissimus muscle lipid with carcass traits in Japanese black cattle. J Anim Sci. 2011;89:615–621. doi: 10.2527/jas.2009-2300. [DOI] [PubMed] [Google Scholar]
- 47.Muoio DM, MacLean PS, Lang DB, Li S, Houmard JA, Way JM, et al. Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator-activated receptor (PPAR) alpha knock-out mice. Evidence for compensatory regulation by PPAR delta. J Biol Chem. 2002;277:26089–26097. doi: 10.1074/jbc.M203997200. [DOI] [PubMed] [Google Scholar]
- 48.Hwang D. Fatty acids and immune responses--a new perspective in searching for clues to mechanism. Annu Rev Nutr. 2000;20:431–456. doi: 10.1146/annurev.nutr.20.1.431. [DOI] [PubMed] [Google Scholar]
- 49.Kenmochi N, Suzuki T, Uechi T, Magoori M, Kuniba M, Higa S, et al. The human mitochondrial ribosomal protein genes: mapping of 54 genes to the chromosomes and implications for human disorders. Genomics. 2001;77:65–70. doi: 10.1006/geno.2001.6622. [DOI] [PubMed] [Google Scholar]
- 50.Tang DG, La E, Kern J, Kehrer JP. Fatty acid oxidation and signaling in apoptosis. Biol Chem. 2002;383:425–442. doi: 10.1515/BC.2002.046. [DOI] [PubMed] [Google Scholar]
- 51.Staiger K, Staiger H, Weigert C, Haas C, Haring H-U, Kellerer M. Saturated, but not unsaturated, fatty acids induce apoptosis of human coronary artery endothelial cells via nuclear factor- B activation. Diabetes. 2006;55:3121–3126. doi: 10.2337/db06-0188. [DOI] [PubMed] [Google Scholar]
- 52.Wei Y, Wang D, Topczewski F, Pagliassotti MJ. Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am J Physiol Endocrinol Metab. 2006;291:E275–E281. doi: 10.1152/ajpendo.00644.2005. [DOI] [PubMed] [Google Scholar]
- 53.Sumara G, Sumara O, Kim JK, Karsenty G. Gut-derived serotonin is a multifunctional determinant to fasting adaptation. Cell Metab. 2012;16:588–600. doi: 10.1016/j.cmet.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laporta J, Hernandez LL. Serotonin receptor expression is dynamic in the liver during the transition period in Holstein dairy cows. Domest Anim Endocrinol. 2015;51:65–73. doi: 10.1016/j.domaniend.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Haeseleer F, Palczewski K. Short-chain dehydrogenases/reductases in retina. Methods Enzymol. 2000;316:372–83. [DOI] [PubMed]
- 56.Aliesky HA, Pichurin PN, Chen C-R, Williams RW, Rapoport B, McLachlan SM. Probing the genetic basis for thyrotropin receptor antibodies and hyperthyroidism in immunized CXB recombinant inbred mice. Endocrinology. 2006;147:2789–2800. doi: 10.1210/en.2006-0160. [DOI] [PubMed] [Google Scholar]
- 57.Hollenberg AN. The role of the thyrotropin-releasing hormone (TRH) neuron as a metabolic sensor. Thyroid. 2008;18:131–139. doi: 10.1089/thy.2007.0251. [DOI] [PubMed] [Google Scholar]
- 58.Luo L, Jackson I. Thyrotropin releasing hormone ( TRH ) may preserve pancreatic islet cell function : potential role in the treatment of diabetes mellitus. Control. 2007;14:216–221. [PubMed] [Google Scholar]
- 59.Cavanagh CR, Jonas E, Hobbs M, Thomson PC, Tammen I, Raadsma HW. Mapping quantitative trait loci (QTL) in sheep. III. QTL for carcass composition traits derived from CT scans and aligned with a meta-assembly for sheep and cattle carcass QTL. Genet Sel Evol. 2010;42:36. doi: 10.1186/1297-9686-42-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L, Liu J, Zhao F, Ren H, Xu L, Lu J, et al. Genome-wide association studies for growth and meat production traits in sheep. PLoS One. 2013;8:1–12. [DOI] [PMC free article] [PubMed]
- 61.Li C, Sun D, Zhang S, Wang S, Wu X, Zhang Q, et al. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. PLoS One. 2014;9:1–21. [DOI] [PMC free article] [PubMed]
- 62.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 63.Takada I, Kouzmenko AP, Kato S. Wnt and PPARγ signaling in osteoblastogenesis and adipogenesis. Nat Rev Rheumatol. 2009;5:442–447. doi: 10.1038/nrrheum.2009.137. [DOI] [PubMed] [Google Scholar]
- 64.Galic S, Oakhill JS, Steinberg GR. Adipose tissue as an endocrine organ. Mol Cell Endocrinol. 2010;316:129–139. doi: 10.1016/j.mce.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 65.Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yen C-LE, Stone SJ, Koliwad S, Harris C, Farese RV. Thematic review series: Glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keats EC, Dominguez JM, Grant MB, Khan ZA. Switch from canonical to noncanonical Wnt signaling mediates high glucose-induced Adipogenesis. Stem Cells. 2014;32:1649–1660. doi: 10.1002/stem.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drackley JK. Lipid metabolism. In: JPF D’M, editor. Metab. Farm Anim. Metab. Nutr. Anim. 1. New York: CABI Publishing; 2000. p. 438. [Google Scholar]
- 69.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mei S, Ni H-M, Manley S, Bockus A, Kassel KM, Luyendyk JP, et al. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther. 2011;339:487–498. doi: 10.1124/jpet.111.184341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shimabukuro M, Wang MY, Zhou YT, Newgard CB, Unger RH. Protection against lipoapoptosis of beta cells through leptin-dependent maintenance of Bcl-2 expression. Proc Natl Acad Sci U S A. 1998;95:9558–9561. doi: 10.1073/pnas.95.16.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:32–36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deng W, Wang X, Xiao J, Chen K, Zhou H, Shen D, et al. Loss of regulator of G protein signaling 5 exacerbates obesity, hepatic steatosis, inflammation and insulin resistance. PLoS One. 2012;7:e30256. doi: 10.1371/journal.pone.0030256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang Q, Abedi-Valugerdi M, Xie Y, Zhao X-Y, Möller G, Nelson BD, et al. Potent suppression of the adaptive immune response in mice upon dietary exposure to the potent peroxisome proliferator, perfluorooctanoic acid. Int Immunopharmacol. 2002;2:389–397. doi: 10.1016/S1567-5769(01)00164-3. [DOI] [PubMed] [Google Scholar]
- 75.Lou Y, Liu S. The TIPE (TNFAIP8) family in inflammation, immunity, and cancer. Mol Immunol. 2011;49:4–7. doi: 10.1016/j.molimm.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 76.Kuhajda FP, Jennert K, Wood FD, Hennigart RA, Jacobs LB, Dick JD, et al. Fatty acid synthesis : a potential selective target for antineoplastic therapy. Proc Natl Acad Sci. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kobirumaki F, Miyauchi Y, Fukami K, Tanaka H. A novel UbcH10-binding protein facilitates the ubiquitinylation of cyclin B in vitro. J Biochem. 2005;137:133–139. doi: 10.1093/jb/mvi026. [DOI] [PubMed] [Google Scholar]
- 78.Bateman A, Martin MJ, O’Donovan C, Magrane M, Apweiler R, Alpi E, et al. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ando H, Watabe H, Valencia JC, Yasumoto K, Furumura M, Funasaka Y, et al. Fatty acids regulate pigmentation via proteasomal degradation of tyrosinase: a new aspect of ubiquitin-proteasome function. J Biol Chem. 2004;279:15427–15433. doi: 10.1074/jbc.M313701200. [DOI] [PubMed] [Google Scholar]
- 80.Würzner R. Deficiencies of the complement MAC II gene cluster (C6, C7, C9): is subtotal C6 deficiency of particular evolutionary benefit? Clin Exp Immunol. 2003;133:156–159. doi: 10.1046/j.1365-2249.2003.02230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grimm H, Mayer K, Mayser P, Eigenbrodt E. Regulatory potential of n-3 fatty acids in immunological and inflammatory processes. Br J Nutr. 2002;87 Suppl 1:S59–S67. doi: 10.1079/BJN2001457. [DOI] [PubMed] [Google Scholar]
- 82.Mach N, Blum Y, Bannink A, Causeur D, Houee-Bigot M, Lagarrigue S, et al. Pleiotropic effects of polymorphism of the gene diacylglycerol-O-transferase 1 (DGAT1) in the mammary gland tissue of dairy cows. J Dairy Sci. 2012;95:4989–5000. doi: 10.3168/jds.2012-5348. [DOI] [PubMed] [Google Scholar]
- 83.Huang JM, Xian H, Bacaner M. Long-chain fatty acids activate calcium channels in ventricular myocytes. Proc Natl Acad Sci U S A. 1992;89:6452–6456. doi: 10.1073/pnas.89.14.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiao YF, Gomez AM, Morgan JP, Lederer WJ, Leaf A. Suppression of voltage-gated L-type Ca2+ currents by polyunsaturated fatty acids in adult and neonatal rat ventricular myocytes. Proc Natl Acad Sci U S A. 1997;94:4182–4187. doi: 10.1073/pnas.94.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murphy EF, Jewell C, Hooiveld GJ, Muller M, Cashman KD. Conjugated linoleic acid enhances transepithelial calcium transport in human intestinal-like Caco-2 cells: an insight into molecular changes. Prostaglandins Leukot Essent Fatty Acids. 2006;74:295–301. doi: 10.1016/j.plefa.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 86.Kemper KE, Goddard ME. Understanding and predicting complex traits: knowledge from cattle. Hum Mol Genet. 2012;21:45–51. doi: 10.1093/hmg/dds332. [DOI] [PubMed] [Google Scholar]
- 87.AOAC . Official methods of analysis of AOAC INTERNATIONAL. 18. Gaithersburg: Association of Official Analytical chemists; 2007. [Google Scholar]
- 88.Anderson S. Determination of fat, moisture, and protein in meat and meat products by using the FOSS FoodScan near-infrared spectrophotometer with FOSS artificial neural network calibration model and associated database: collaborative study. J AOAC Int. 2007;90:1073–1083. [PubMed] [Google Scholar]
- 89.Hara A, Radin NS. Lipid extraction of tissues with a low-toxicity solvent. Anal Biochem. 1978;90:420–426. doi: 10.1016/0003-2697(78)90046-5. [DOI] [PubMed] [Google Scholar]
- 90.Christie WW. A simple procedure for rapid transmethylation of glycerolipids and cholesteryl esters. J Lipid Res. 1982;23:1072–1075. [PubMed] [Google Scholar]
- 91.Abdollahi-Arpanahi R, Pakdel A, Nejati-Javaremi A, Moradi Shahrbabak M, Morota G, Valente BD, et al. Dissection of additive genetic variability for quantitative traits in chickens using SNP markers. J Anim Breed Genet. 2014;131:183–193. doi: 10.1111/jbg.12079. [DOI] [PubMed] [Google Scholar]
- 92.Misztal I, Tsuruta S, Strabel T, Auvray B, Druet T, Lee DH. BLUPF90 and related programs (BGF90). Proc. 7th world Congr. Genet Appl to Livest Prod. 2002;28:21–22. [Google Scholar]
- 93.Silva MR, Pedrosa VB, Silva JCB, Eler JP, Guimarães JD, Albuquerque LG. Testicular traits as selection criteria for young Nellore bulls. J Anim Sci. 2011;89:2061–2067. doi: 10.2527/jas.2010-3525. [DOI] [PubMed] [Google Scholar]
- 94.Zhu W, Zhang H. Why do we test multiple traits in genetic association studies? J Korean Stat Soc. 2009;38:1–10. doi: 10.1016/j.jkss.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yuan J, Wang K, Yi G, Ma M, Dou T, Sun C, et al. Genome-wide association studies for feed intake and efficiency in two laying periods of chickens. Genet Sel Evol. 2015;47:82. doi: 10.1186/s12711-015-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang JJ, Li J, Williams LK, Buu A. An efficient genome-wide association test for multivariate phenotypes based on the fisher combination function. BMC Bioinformatics. 2016;17:19. doi: 10.1186/s12859-015-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang H, Misztal I, Aguilar I, Legarra A, Muir WM. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet Res (Camb) 2012;94:73–83. doi: 10.1017/S0016672312000274. [DOI] [PubMed] [Google Scholar]
- 98.Habier D, Fernando RL, Kizilkaya K, Garrick DJ. Extension of the bayesian alphabet for genomic selection. BMC Bioinformatics. 2011;12:186. doi: 10.1186/1471-2105-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data used during the current study are available from the corresponding author on request.