Abstract
The secretion of insulin and glucagon from the pancreas and the incretin hormones glucagon-like peptide-1 (GLP-1) and glucose-dependent insulinotropic peptide (GIP) from the gastrointestinal tract is essential for glucose homeostasis. Several novel treatment strategies for type 2 diabetes (T2D) mimic GLP-1 actions or inhibit incretin degradation (DPP4 inhibitors), but none is thus far aimed at increasing the secretion of endogenous incretins. In order to identify new potential therapeutic targets for treatment of T2D, we performed a meta-analysis of a GWAS and an exome-wide association study of circulating insulin, glucagon, GIP, and GLP-1 concentrations measured during an oral glucose tolerance test in up to 7,828 individuals. We identified 6 genome-wide significant functional loci associated with plasma incretin concentrations in or near the SLC5A1 (encoding SGLT1), GIPR, ABO, GLP2R, F13A1, and HOXD1 genes and studied the effect of these variants on mRNA expression in pancreatic islet and on metabolic phenotypes. Immunohistochemistry showed expression of GIPR, ABO, and HOXD1 in human enteroendocrine cells and expression of ABO in pancreatic islets, supporting a role in hormone secretion. This study thus provides candidate genes and insight into mechanisms by which secretion and breakdown of GIP and GLP-1 are regulated.
Keywords: Endocrinology, Genetics
Keywords: Diabetes, Genetic variation, Glucose metabolism
GWAS of circulating fasting and glucose-stimulated concentrations of incretins GIP and GLP-1 identifies new genetic loci involved in hormone secretion and metabolism
Introduction
The hormones secreted in response to nutrient intake are essential for maintaining glucose homeostasis and development of diabetes. Insulin secreted from pancreatic β cells in response to elevated blood glucose acts on peripheral tissues to increase glucose uptake, while glucagon secreted from α cells counteracts low glucose levels by promoting gluconeogenesis and glycogenolysis in the liver (1).
The two incretin hormones, glucose-dependent insulinotropic peptide (GIP) and glucagon-like peptide-1 (GLP-1), are released into the circulation by intestinal enteroendocrine cells (K and L cells, respectively) in response to nutrient stimulation and act on β cells to increase insulin secretion. In addition to their insulinotropic activity both hormones exert a number of other functions, such as promotion of energy storage and bone formation. GIP acts via the GIP receptor (GIPR), which is expressed in numerous tissues, including pancreatic islets, fat, bone, and vascular endothelial cells. GLP-1 acts via the GLP-1 receptor and affects biological functions such as regulation of food intake, stimulation of insulin synthesis, inhibition of glucagon secretion, and gastric emptying (2).
Several therapeutic agents for treatment of type 2 diabetes (T2D) act by increasing GLP-1 action, i.e., GLP-1 receptor agonists or GLP-1 analogs and inhibitors of the dipeptidyl peptidase–4 (DPP4) enzyme, which inactivates both GIP and GLP-1 within minutes after their secretion (3). Furthermore, long-term weight loss, improved glucose tolerance, and remission of T2D following bariatric surgery are suggested to involve an exaggerated GLP-1 response (4).
Numerous GWAS have identified genetic variants that affect plasma concentration of insulin (5–8), but such information is lacking for glucagon, GLP-1, and GIP concentrations.
To identify potential novel targets to therapeutically manipulate circulating levels of these hormones we performed a GWAS of circulating plasma levels of glucagon, GIP, and GLP-1 as well as insulin (as positive control), measured during an oral glucose tolerance test (OGTT) and meta-analyzed with data from an exome-wide study of the same phenotypes in an independent cohort. Thereby, we identified 6 loci associated with incretin levels at genome-wide significance in or near the solute carrier family 5 member 1 (SLC5A1), gastric inhibitory polypeptide receptor (GIPR), ABO blood group (ABO), glucagon-like peptide 2 receptor (GLP2R), coagulation factor XIII A chain (F13A1), and homeobox D1 (HOXD1) genes.
Results
We performed a GWAS of circulating levels of insulin, glucagon, GIP, and GLP-1 measured at 0 and 2 hours of an OGTT (plus 30 minutes for insulin) in a Swedish population-based cohort of 3,344 individuals (Malmö Diet and Cancer study [MDC]) genotyped using the Illumina Infinium OmniExpressExome v1.0 B array. The results were meta-analyzed with data from 4,905 Finnish individuals from the Prevalence, Prediction and Prevention of Diabetes Botnia study (PPP-Botnia) genotyped with the Illumina Infinium Exome v1.0 Beadchip. After quality control (QC), 81,396 exome SNPs were present in analysis of both cohorts. Manhattan and QQ plots are presented in Supplemental Figures 1 and 2 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.93306DS1). The significant associations were used to examine other potential phenotypic associations and the effect on gene expression in target tissues.
Heritability of circulating hormone concentrations.
The “narrow-sense heritability” of hormone concentrations during OGTT, estimated as total variance explained by the genotyped SNPs, i.e., the proportion of variation in the phenotype that can be attributed to effects of the genotyped SNPs, was calculated in the MDC cohort using Genome-Wide Complex Trait Analysis (GCTA) (9). The heritability of insulin ranged from 11% for fasting to 23% for circulating insulin at 30 minutes of the OTGG (30-minute insulin). Fasting GIP, 2-hour GIP, and 2-hour GLP-1 showed heritabilities of approximately 15%, and fasting glucagon showed a heritability of around 20% (Supplemental Table 1). Only the heritabilities of 30-minute insulin and fasting glucagon were significantly higher than 0.
Genetic variants associated with insulin concentration.
As several studies have reported genetic variants associated with insulin concentrations in nondiabetic individuals, we searched for similar associations in our cohorts as positive controls. The strongest association with fasting insulin was seen for the rs2943641 SNP (P = 2.4 × 10–7) near the insulin receptor substrate 1 (IRS1) gene that has previously been shown to be associated with T2D and insulin resistance (10). This locus was also most strongly associated with 2-hour insulin (P = 1.35 × 10–5). As previously reported (5), variants in GIPR were associated with reduced insulin concentration at 30 minutes of an OGTT by 8% per allele (Table 1 and Supplemental Figure 3A). The second strongest association was seen for rs5015480 (P = 4.9 × 10–7) in a previously reported T2D locus near the hematopoietically expressed homeobox (HHEX) gene (11). SNPs at least suggestively associated (P < 10–5) with insulin are presented in Supplemental Table 2.
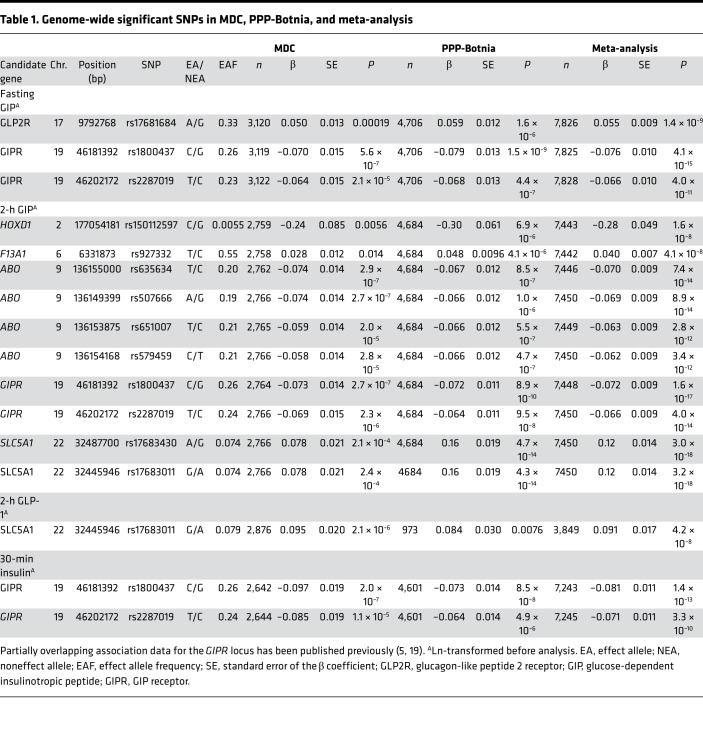
Table 1. Genome-wide significant SNPs in MDC, PPP-Botnia, and meta-analysis.
Genetic variants associated with glucagon concentrations.
The strongest association of fasting glucagon concentrations was seen for an intronic SNP, rs7102710, in the gene encoding spondin 1 (SPON1, P = 8.2 × 10–7). The SPON1 gene was observed to be highly expressed in pancreatic islets from 191 human cadaver donors (mRNA higher than 73.3% of all genes), and the expression correlated positively with hemoglobin A1c (HbA1c) levels (n = 116, r2 = 0.13, P = 5.2 × 10–5). However, rs7102710 was not an expression QTL (eQTL) for any gene within 1 Mb (P > 0.01) in the pancreatic islets. All SNPs significantly or suggestively associated (P < 10–5) with glucagon levels are presented in Supplemental Table 2.
Genetic variants associated with GLP-1 concentration.
We observed a strong association with GLP-1 concentration after OGTT for 2 missense variants in SLC5A1: rs17683011 (Asn51Ser, P = 4.2 × 10–8) and rs17683430 (Ala411Thr, P = 5.2 × 10–8). These 2 variants are in complete linkage disequilibrium (LD) (r2 = 1, D′ = 1) and thus represent the same locus. Each G allele of rs17683011 increased the 2-hour GLP-1 concentration by 9.1% (P = 4.2 × 10–8). SLC5A1 encodes the sodium-dependent glucose transporter 1 (SGLT1), the main mediator of glucose uptake in the gut, which has been shown to be expressed in the apical membrane of both K and L cells and to be essential for incretin secretion in both humans and animal models (12–16). All genome-wide significant associations are presented in Table 1 and Supplemental Figure 3.
Genetic variants associated with GIP concentrations.
Two independent loci were significantly associated with fasting GIP concentration: GIPR and GLP2R (Table 1). All SNPs at least suggestively associated (P < 10–5) with GIP and GLP-1 are presented in Supplemental Table 3.
GIPR.
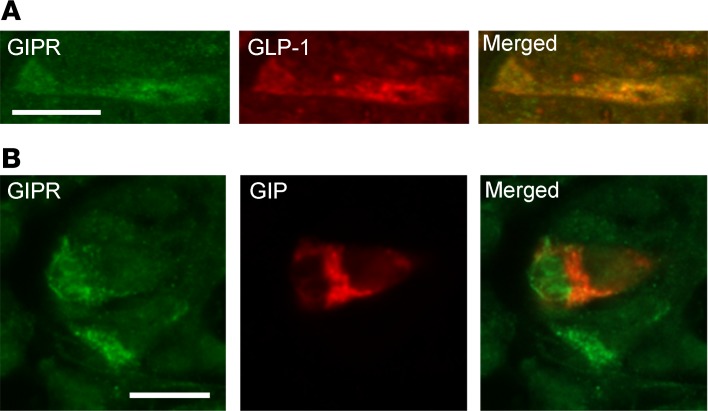
The minor alleles of rs1800437 and rs2287019 (P = 4.0 × 10–11) in the GIPR locus were associated with lower fasting (P = 4.1 × 10–15) and 2-hour (P = 1.6 × 10–17) GIP concentrations. The rs1800437 SNP is in strong LD (r2 = 0.94, D′ = 1) with the rs10423928 variant that has previously been associated with GIP concentrations in the PPP-Botnia cohorts as well as with several diabetes-related phenotypes, including insulin secretion, BMI, and expression of GIPR mRNA in islets (5, 17–19). The rs1800437 and rs2287019 variants are also in relatively strong linkage equilibrium (r2 = 0.7, D′ = 1) with each other. Analysis conditioned on rs1800437 showed no independent association for rs2287019 (P = 0.8), suggesting that they represent the same locus. The minor C allele of rs1800437 was also nominally associated with increased fasting (P = 5.3 × 10–3) but not 2-hour GLP-1 (Table 2). In accordance with previous publications, the same allele was associated with decreased fasting insulin (P = 0.015), 30-minute insulin secretion (P = 1.4 × 10–13), 2-hour insulin concentrations (P = 0.011), BMI (P = 6.0 × 10–7), and increased 2-hour glucose levels (P = 0.011, Table 2) (5, 17). However, in contrast to previously published results, the locus was not an eQTL for the GIPR gene (Supplemental Table 4 and Supplemental Figure 4). We also analyzed GIPR expression in K and L cells by immunohistochemical staining of human colon and jejunum specimens and observed that the GIPR was expressed in subsets of both K and L cells (Figure 1).
Table 2. Association between genome-wide loci and other metabolic phenotypes in meta-analysis of MDC and PPP-Botnia.
Figure 1. GIP receptor expression in enteroendocrine cells.
Human intestinal sections. (A) Colonal sections double immunostained for glucose-dependent insulinotropic peptide receptor (GIPR) and glucagon-like peptide-1 (GLP-1). (B) Jejunal sections double immunostained for GIPR and glucose-dependent insulinotropic peptide (GIP). Scale bar: 10 μm.
GLP2R.
The second locus associated with fasting GIP was rs17681684 (P = 1.4 × 10–9) in GLP2R, which encodes the GLP-2 receptor. rs17681684 was associated with GIP specifically in the fasting state where each A allele associated with 5.5% increased GIP levels. No association with 2-hour GIP could be detected. The same SNP was nominally associated with decreased 2-hour levels of GLP-1 (P = 5 × 10–4), each allele reducing the serum concentration by 3.4%, whereas no effect on fasting levels was found (Table 2). This genetic variant causes an amino acid change from an aspartic acid to asparagine (Asp470Asn), but it is not known if this has a functional effect. The amino acid substitution was predicted to be benign (risk score = 0) by the Polyphen-2 database (20). Because the variant was also nominally associated with decreased glucagon concentrations (P = 0.04), we examined GLP2R mRNA expression in human pancreatic islets from cadaver donors. We found no significant association with GLP2R mRNA expression (P = 0.05) in human pancreatic islets (Supplemental Table 4 and Supplemental Figure 4), and no effect on in vitro insulin secretion (n = 197) measured as stimulatory index or glucagon secretion was detectable.
The rs17681684 SNP was also nominally associated with fasting glucose (P = 0.03), and in the DIAGRAM database (21), including more than 60,000 individuals, the same allele was associated with increased risk of T2D (P = 8.2 × 10–4). Together, the identified variants in the GIPR and GLP2R loci explained 1.2 % of the fasting GIP variation.
In addition to the GIPR locus, we found 4 loci significantly associated with GIP after OGTT (Table 1).
SLC5A1.
The SLC5A1 locus was also associated with 2-hour GIP. Each G allele of rs17683011 associated with 12.2% increased 2-hour GIP levels, and, concomitantly, the corrected insulin response (CIR) was increased by 10.0% (P = 0.002). Fasting glucose was slightly lower among the G allele carriers of rs17683011 (P = 0.03), but no significant association with risk of T2D was detected in either of our cohorts or in the DIAGRAM database (OR = 0.95, P = 0.12). Neither did we observe any significant association with BMI in our cohorts or in the GIANT meta-analysis (P = 0.46) (22).
The G allele of rs17683011 also associated with 3.5% increased 2-hour glucagon concentration (P = 7.3 × 10–5). SLC5A1 mRNA was found to be expressed in pancreatic islets from human donors, where its expression correlated with expression of the glucagon (GCG) gene (r2 = 0.31, P = 2 × 10–17). This supports previous studies showing expression of SLC5A1 in α cells (23). In islets, SLC5A1 expression was significantly higher in carriers of the minor G allele (P = 3.1 × 10–6, Supplemental Table 4 and Supplemental Figure 4), but the locus was not associated with expression of the GCG gene (P = 0.55). We did not observe any significant association with glucagon secretion in human islets in vitro (P = 0.8); however, this could be due to lack of power, given the limited number of individuals included in the analysis (n = 95).
ABO.
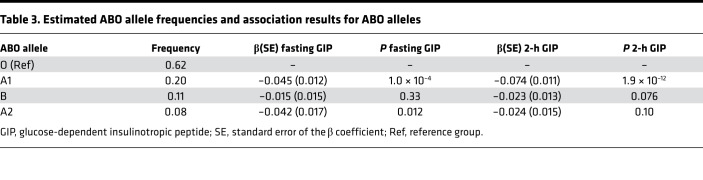
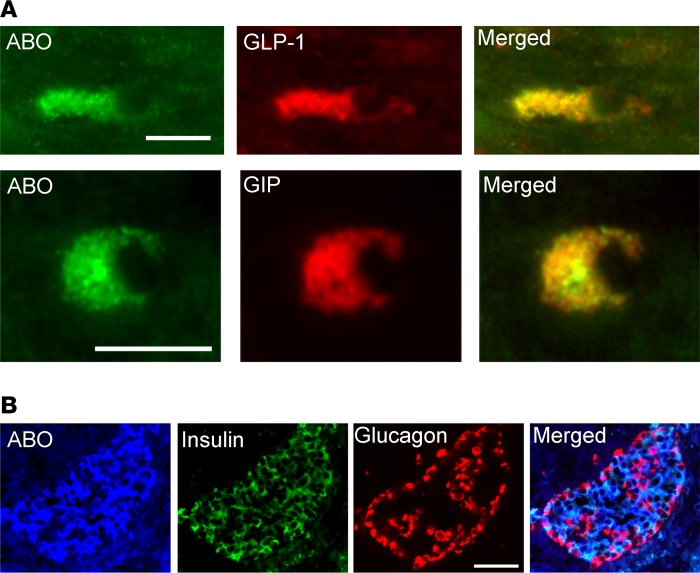
Four SNPs in strong LD (r2 = 0.95-1, D′ = 1), near the ABO gene, were associated with both fasting (P = 6.5 × 10–4) and 2-hour (P = 9.0 × 10–14) GIP concentration but not with GLP-1. The same locus has previously been associated with numerous phenotypes, including fasting glucose (24), soluble E-selectin (25), ICAM-1 (26), and P-selectin (26). The associated SNPs are in low-to-moderate LD with SNPs in ABO that determine blood group (r2 = 0.01––0.51, D′ = 0.19-1). Previous studies have shown that the associated SNPs are proxies for the ABO*A101 allele responsible for blood group A1. We estimated the ABO alleles of individuals in the MDC cohort using haplotypes formed of 3 ABO SNPs: rs8176749, rs8176704, and rs687289 (26). The estimated ABO allele frequencies corresponded well with their expected frequencies (Table 3). The overall P value for the association with 2-hour GIP was 5.1 × 10–8, but after conditioning on rs635634, or the A1-haplotype, no remaining association was detected (P > 0.8) (Supplemental Table 5). This was further confirmed in the PPP-Botnia cohort (Supplemental Table 6) and meta-analysis (Table 3). Immunohistochemistry showed that the ABO protein is expressed in K cells and L cells (Figure 2A).
Table 3. Estimated ABO allele frequencies and association results for ABO alleles.
Figure 2. ABO expression in enteroendocrine cells and pancreatic islets.
(A) Human colonic sections double immunostained for ABO and glucagon-like peptide-1 (GLP-1) as well as ABO and glucose-dependent insulinotropic peptide (GIP), respectively. Scale bar: 10 μm. (B) Human pancreatic sections triple immunostained for ABO, insulin, and glucagon. Scale bar: 50 μm.
The ABO locus was also associated with reduced insulin concentrations at 30 minutes (P = 5.7 × 10–4) and with increased fasting glucose (P = 0.038) and 2-hour glucose (P = 0.0014). In the MDC cohort, the variant associated with increased risk of T2D incidence during a follow-up period of 20–23 years (HR 1.16; CI 95% 1.01–1.31; P = 0.04). In pancreatic islets, rs507666 was strongly associated with ABO mRNA expression (P = 1.0 × 10–7, Supplemental Table 4 and Supplemental Figure 4), but this was mainly due to lower expression among individuals with the blood group O, and the association was no longer significant after exclusion of these rs687289 homozygous individuals. Immunohistochemistry in human pancreatic islets confirmed expression of ABO protein in β cells (Figure 2B), but we found no association with glucose-stimulated insulin secretion (stimulatory index) in vitro (P = 0.8), in islets from 214 human cadaver donors, suggesting that the association with 30-minute insulin could be secondary to the effect on GIP secretion.
The ABO locus was also associated with increased BMI (P = 0.005). The same nominal association has been reported by the CHARGE consortium (24), and, in accordance with findings from this consortium, the association with BMI was primarily seen in females (P = 0.013). The ABO locus has previously been associated with cholesterol uptake and LDL cholesterol concentrations (27). In addition, individuals with blood group A express lower levels of intestinal alkaline phosphatase, which has been shown to affect body weight in interaction with high-fat diet in mice (28). We therefore explored whether a similar interactions between the locus and dietary habits would be seen in humans. Interestingly, we observed an interaction between rs635634 and fat intake (Pinteraction = 0.001), especially saturated fat (Pinteraction = 0.0002) and monounsaturated fat (Pinteraction = 0.004), on BMI (Supplemental Table 7). The positive association between the minor allele of rs635634 and BMI was mainly observed with higher intakes of fat, while it was associated with lower BMI in the tertile with the lowest intake of saturated (P = 0.003) and monosaturated fat (P = 0.02) (Supplemental Table 8). Interestingly, a nominally significant association with cholesterol intake was also observed (P = 0.02).
F13A1.
The T allele of the rs927332 variant was suggestively associated with increased fasting GIP (P = 2.5 × 10–5) and significantly associated with increased 2-hour GIP (P = 4.1 × 10–8) (Table 1) but was not associated with GLP-1. The associated SNP is an eQTL (P = 1.6 × 10–6) for F13A1 in tibial arteries according to the GTEx database (29), indicating that the locus has a functional effect on this gene and, thus, suggesting that this could be the gene responsible for the effect. However, immunohistochemical staining showed no specific expression of F13A1 in the intestinal epithelium. Likewise, we did not observe any associated effect on insulin or glucose concentrations at any time point (Table 2) or any association with risk of T2D.
HOXD1.
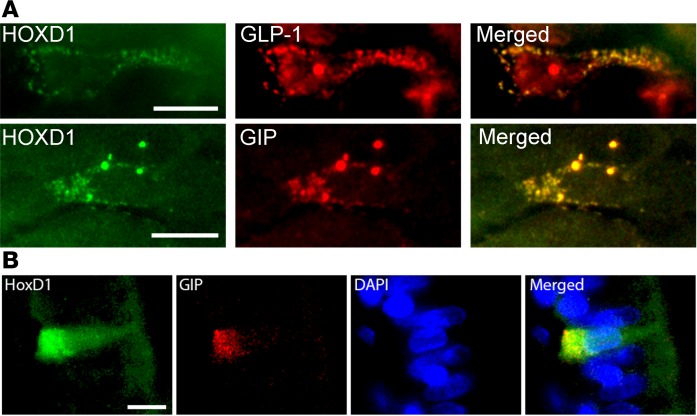
Finally, a rare C allele of rs150112597 (minor allele frequency = 0.005) was found to be associated with 28% reduced 2-hour GIP concentration (P = 1.6 × 10–8). The same C allele was nominally associated with fasting GIP (P = 0.02), decreased 2-hour glucose (P = 0.002), and BMI (P = 0.034). Because of the low frequency, results for this SNP are not reported in the published large meta-analyses of T2D or diabetes-related variables. Immunohistochemical staining of human intestine showed expression of HOXD1 in GIP-expressing K cells in the jejunum, thus providing some further support for the association (Figure 3).
Figure 3. HOXD1 expression in enteroendocrine cells.
(A) Human colonal sections double immunostained for homeobox D1 (HOXD1) and glucagon-like peptide-1 (GLP-1) as well as HOXD1 and glucose-dependent insulinotropic peptide (GIP), respectively. Scale bar: 10 μm. (B) Triple staining, including DAPI, also shows expression in the nucleus. Scale bar: 10 μm.
Together, the 5 loci associated with 2-hour GIP explained 2.6% of the 2-hour GIP variation.
Association of variants in known T2D loci with hormone concentrations during OGTT.
Finally, we did an exploratory analysis of known T2D loci (Supplemental Table 9) (30) to see if we could find evidence that they were associated with glucagon, GIP, and GLP-1 concentrations in our GWAS. Several of these T2D-associated loci showed nominal associations with GIP, GLP-1, and glucagon concentrations, but only the GIPR locus was significant after correction for multiple testing (Supplemental Tables 10–12).
Discussion
With the goal to identify new mechanisms for regulation of hormone secretion in response to oral glucose and thereby potential new drug targets, we identified 5 loci that were associated with circulating insulin, glucagon, and incretin levels at fasting and 2 hours of OGTT.
Blood concentration of incretins is highest at 30–60 minutes after glucose stimulation but remains high until approximately 2 hours, at which time it starts to decline; after approximately 4 hours, it is back to fasting levels (31). The 2-hour concentrations thus depend on both secretion and clearance, and no definite conclusions can therefore be made about which of these processes is affected by the identified loci. It should also be noted that the pancreas and liver are exposed to portal venous concentrations of incretins and that first-pass clearance could alter the peripheral levels of these hormones.
The heritability/familiality of incretin secretion during OGTT has previously been estimated to be in the 50%–75% range (31, 32), which is markedly higher than the heritability estimates obtained in this study. This difference is most likely a consequence of the GCTA method, which only estimates the proportion of variance explained by additive contributions from the genotyped SNPs, not the total heritability. However, the accuracy of the method has also been questioned due to the underlying assumption that may not always be true (33, 34). Given this, and the large confidence intervals, the results should be interpreted with caution.
Identifying genes underlying an association is an inherent problem of GWAS studies, and we can therefore not claim that the genes linked to the loci are really causal. Four of the associated loci are coding nonsynonymous SNPs; GIPR, SLC5A1, and GLP2R are very strong candidates based on their known function, but other genes and/or genetic variants in LD could still be involved in mediating the effect of the identified loci.
SGLT1, the glucose and galactose transporter encoded by SLC5A1, is expressed in the apical cell domain of L and K cells (12), and studies in both rodents and humans have shown that SGLT1 is pivotal for intestinal glucose absorption and glucose-dependent incretin secretion (12–16). In this study, we show that common genetic variants in the SGLT1 transporter gene have a significant effect on both GIP and GLP-1 secretion in humans and as a consequence on insulin response. Interestingly, the same variants are associated with increased glucagon secretion. While this could be an effect on the α cell, both the increased incretin secretion and the increased mRNA expression in pancreatic islets suggest that the genetic variation is involved in increased glucose transport, which would be expected to reduce glucagon secretion. Inclusion of insulin and glucose measures in the regression model did not attenuate the association (β = 0.036 ± 0.0092, P = 9.0 × 10–5 adjusted, β = 0.035 ± 0.0089, P = 7.3 × 10–5 unadjusted), showing that the increase in glucagon secretion was not secondary to increased insulin secretion. Notably, other variants that associated with increased GIP concentrations did not associate with glucagon concentrations, suggesting that this effect is unique to SGLT1. One possibility is that the variants affect extrapancreatic glucagon secretion, since recent data demonstrate that, at least in pancreatectomized individuals, glucagon is secreted from extrapancreatic sources in response to oral glucose stimulation, the L cells being the likely source (35).
The minor allele of the rs635634-tagged locus near ABO has previously been associated with fasting glucose levels in a GWAS meta-analysis of >60.000 individuals (24). Weak nominal associations with increased fasting insulin (P = 0.02) and risk of T2D (P = 0.01) have also been reported (24). In support of this, we observed an association with increased fasting and 2-hour glucose as well as decreased 30-minute insulin secretion and increased risk of T2D. Given the much stronger association seen with GIP concentration, and lack of detectable effect on in vitro glucose-stimulated insulin secretion, it is likely that the association with insulin and glucose-related traits is secondary to the effect on GIP secretion. We could also replicate the reported nominal association with BMI, including the gender specificity of the effect. This is more difficult to explain by an effect on GIP, as the ABO SNP was associated with increased BMI, whereas lower BMI would be the expected effect of decreased GIP concentrations (36).
The ABO locus has been associated with a large number of phenotypes (106 phenotypes in GWAS Central; http://www.gwascentral.org/phenotypes) including soluble ICAM-1, soluble E-selectin, and soluble P-selectin as well as circulating phytosterols, LDL, and cardiovascular disease (25, 26, 37). However, the mechanisms underlying these associations have not been explored. The ABO gene encodes the glycosyl transferase that adds the A and B antigenic determinants to the surface of erythrocytes. The most common allele of the gene is the O allele that has a frame-shift variant, resulting in a nonfunctional protein. The product of the B allele adds a galactose while the A allele enzyme adds an N-acetylgalactosamine to the outer galactose residue of the H-antigen. A1 and A2 mostly produce the same antigen, but the transferase activity of the A1 protein is 38-fold higher than that of A2, resulting in a much higher cell surface density of A antigen in A1 (38, 39). Since O-linked glycosylation is essential to the function of many proteins, it could theoretically affect GIP levels through a number of mechanisms. However, the region also contains other interesting genes that could mediate the effect, including the carboxyl ester lipase (CEL) gene, which is involved in lipid and cholesterol uptake (40).
The ABO locus has consistently been associated with LDL cholesterol levels. The fact that it is also associated with phytosterol levels suggests that this association can be caused by an increased uptake of dietary cholesterol or a decreased excretion of cholesterol from the liver. Interestingly, we found an interaction on BMI between the genotype and intake of saturated fat, a strong marker of dietary and plasma cholesterol, after adjustment for total caloric consumption. In the tertile with the lowest consumption of saturated fat, the ABO locus was associated with lower BMI and the group with the highest intake had an opposite tendency. Whether this really means that people with different blood groups could benefit from different diets needs to be examined. Of note, blood group A expresses lower levels of intestinal alkaline phosphatase, which has been shown to affect body weight in interaction with high-fat diet in mice (28). While this interaction is novel and interesting, it requires replication in larger cohorts with comprehensive dietary data.
The rare C allele of HOXD1 replaces glycine for arginine at position 218 of the HOXD1 protein, a sequence-specific transcription factor involved in the differentiation of endothelium (41) and limb development (42). While HOXD1 has no know function in incretin biology, it is highly expressed in the small intestine according to the GTEx database (29). In addition, we found expression of HOXD1 protein in K and L cells. The question remains of whether HOXD1 might affect differentiation of the gut and the enteroendocrine cells.
GLP-2 is cosecreted with GLP-1 from L cells and has a multitude of functions in the gastrointestinal system, including stimulation of mucosal growth, glucose transport and glucose transporter 2 expression, inhibition of gastric emptying and gastric acid secretion, and stimulation of intestinal blood flow (43). The GLP-2 receptor is expressed in both the small and large intestine and has been reported in subsets of human enteroendocrine cells (44), but the exact cellular location remains controversial due to low specificity of available antibodies. In rat jejunum, expression is mainly seen in enteric neurons and in subepithelial myofibroblasts (45, 46). GLP-2 has been shown to stimulate pancreatic glucagon secretion, which is also supported here by the observed expression difference and effect on fasting glucagon levels (47). However, this could also be an extrapancreatic effect due to the inhibitory effect of the locus on secretion from the L cells.
While several novel treatment strategies for T2D mimic GLP-1 actions (analogs) or inhibit incretin degradation, none is thus far aimed at increasing the secretion of endogenous incretins. This study provides insight into such mechanisms, by which secretion and breakdown of GIP and GLP-1 are regulated.
Methods
Cohorts
MDC reinvestigation cohort.
MDC is a prospective cohort study in which all men and women born between 1923 and 1950 from the city of Malmö were invited to participate. A total of 30,447 individuals underwent a baseline examination between 1991 and 1996 (48). From this cohort, a random sample of 6,103 individuals screened between 1991 and 1994 were selected to participate in a study aimed at studying cardiovascular risk factors, the MDC Cardiovascular Cohort (MDC-CC). All individuals from the MDC-CC were invited to participate in a reinvestigation starting in 2007 (49). Of 6,103 individuals, 3,734 (age 61–85 years) accepted and were reexamined between May 2007 and January 2012 and are included in this study. Individuals with known diabetes (n = 343) were excluded from the OGTT and were thus not included in the analysis of values after OGTT, while individuals diagnosed with diabetes based on the OGTT were included. In total, 527 individuals had T2D. Population characteristics are presented in Supplemental Table 13. Diet analyses were performed in 4,892 nondiabetic individuals with diet information. No data on hormone concentrations were available in these individuals.
PPP-Botnia cohort.
PPP-Botnia is a population-based study in Western Finland started in 2004 to obtain accurate estimates of prevalence and risk factors for T2D, impaired glucose tolerance, impaired fasting glucose, and the metabolic syndrome in the adult population and to use this information for prediction and prevention of the disease (50). The participants were randomly recruited from the National Finnish Population Registry to represent 6%–7% of the population in the 18- to 75-year age range. Altogether, 5,208 individuals participated in the original study and 3,870 (77%) participated in the follow-up study. The number of individuals with diabetes was 307 at the basal visit and 284 at the follow-up. Population characteristics are presented in Supplemental Table 14.
Laboratory assays and other measurements
MDC.
A 75-g OGTT was performed after an overnight fast in individuals without known diabetes. Blood samples were drawn for analysis of insulin, glucagon, GIP, and GLP-1 concentrations at 0 and 120 minutes. Total plasma GLP-1 concentrations (intact GLP-1 and the metabolite GLP-1 9-36 amide) were determined radioimmunologically using an in-house anti-serum (no. 89390; sensitivity <1 pmol/l) as previously described (51). Serum GIP was analyzed using the Human GIP Total ELISA kit EZHGIP-54K (EMD Millipore), which has 100% cross reactivity to human GIP(aa 1–42) and GIP(aa 3–42), and range for the assay was 9.3–2,250/10.1–2,450 mg/ml. Plasma glucose was analyzed using the Hemocue Glucose System (HemoCue AB). Serum insulin was assayed with an ELISA kit (K6219, Dako) and had no significant cross reactivity to proinsulin (range 0.5–206 U/l). Glucagon was measured using a radioimmunoassay (GL-32K, EMD Millipore) with less than 2% cross reactivity to oxyntomodulin (range 18.5–400 pg/ml) (52). For all assays used for hormone measurements, inter-assay variation was <15% and intra-assay variation was <10%. BMI was calculated from direct measurements of weight and height. Dietary habits were measured with a modified diet history method (53).
PPP-Botnia.
A 75-g OGTT was performed after an overnight fast at both at the basal and follow-up visit. Blood samples were drawn at 0, 30, and 120 minutes. Total GIP, 0 and 120 minutes, was measured at the basal visit, whereas GLP-1, 0 and 120 minutes, was measured at follow-up. GLP-1 was measured using GLP-1 (total) radioimmunoassay (GLP1T-36HK, EMD Millipore) with high specificity to GLP-1 (GLP-2, glucagon, and exendin <0.2%). The range was 3–333 pmol/l. Serum insulin was measured by an AutoDelfia fluoroimmunometric assay (B080-101, PerkinElmer). Plasma glucose, glucagon, and serum GIP concentrations were measured as for MDC. For GLP-1 inter-assay, variation was up to 18%. For all other hormone measurements, inter-assay variation was <15% and intra-assay <10%.
Genotyping
Genotyping of PPP-Botnia and MDC was done at the Broad genotyping facility using the Infinium OmniExpressExome v1.0 B Beadchip array (Illumina) for MDC and the Illumina Infinium Exome v1.0 Beadchip for PPP-Botnia.
Genotypes were called using Illumina’s Gencall algorithm in Genomestudio with refinement of rare variants with missing calls undertaken using zCall (54). Standard QC filters were applied, in accordance with the Exome-chip Quality Control SOP version 5, as developed within the UK exome chip consortium (for more information, see the Supplemental Methods). In brief, SNPs were excluded if they were monomorphic, had a low call rate (<99%), or deviated from the Hardy-Weinberg equilibrium (P < 10−4). Samples were excluded if they were duplicates, unexpected first/second relatives, sex mismatches, heterozygosity outliers (>3 SD from mean), ancestral outliers, or had low call rate (<95%). Mean call rate after QC was >99.5% in both cohorts. Cluster plots for all SNPs of interest were inspected, to ensure accuracy of genotype calling.
Immunohistochemistry
Tissue handling.
Human specimens were fixed in 4% paraformaldehyde, cut in 6-μm sections, and mounted on slides. Sections were deparaffinized and then boiled twice for 7 minutes in citrate buffer (0.01 M, pH 6.0) followed by a 20-minute rinse in running tap water and then 5 minutes in 1× PBS.
Immunohistochemical staining of pancreatic sections.
The following primary antibodies were used: rabbit anti-ABO (code bs-7589R, dilution 1:500, Bioss), rabbit anti-F13A1 (code HPA001804, dilution 1:500, Sigma-Aldrich), goat anti-GIP (code sc-23554, dilution 1:500, Santa Cruz Biotechnology), rabbit anti-GIPR (code RbαGIPr551#4, dilution 1:400, provided by Timothy Kieffer, University of British Columbia, Vancouver, British Columbia, Canada), mouse anti-glucagon (code G-2654, dilution 1:9000, Sigma-Aldrich), guinea pig anti-GLP-1 (code 8707, dilution 1:1280, EuroDiagnostika), mouse anti-HoxD1 (code sc-365853, dilution 1:500, Santa Cruz), and guinea pig anti-insulin (code M9003, dilution 1:2500, EuroDiagnostika). Antibodies were diluted in PBS containing 0.25% BSA and 0.25% Triton X-100. Human pancreas, colon, and jejunum sections (5 μm) were incubated with primary antibodies overnight at 4°C, followed by rinsing in PBS with 0.25% Triton X-100 twice for 10 minutes. Thereafter, secondary antibodies (donkey anti-rabbit AMCA [code 711-155-152, Jackson Immunoresearch] for ABO; donkey anti-rabbit Cy2 [code 711-225-152, Jackson Immunoresearch] for ABO, GIPR, and F13A1; donkey anti-goat Texas Red [code PA1-28662, Thermo Fisher Scientific] for GIP; donkey anti-goat Cy2 for GIP [code 705-225-147, Jackson Immunoresearch]; donkey anti-mouse Texas Red [code SAB3701112, Sigma-Aldrich] for glucagon; donkey anti-guinea pig [code 706-075-148, Jackson Immunoresearch] for GLP-1; donkey anti-mouse Cy2 [code 115-225-166, Jackson Immunoresearch] for HoxD1; and donkey anti-guinea pig Cy2 [code 706-225-148, Jackson Immunoresearch] for insulin) were applied for 1 hour at room temperature. Sections were again rinsed in PBS with 0.25% Triton X-10 and mounted in PBS/glycerol (1:1). Cy2 and Texas Red were used at a dilution of 1:400, while AMCA was diluted 1:100.
Imaging and morphometry.
Immunofluorescence was examined in an epifluorescence microscope (Olympus BX60) with a digital camera (Nikon DS-2Mv).
Human islets
RNA isolation.
Isolation of total RNA was done using the AllPrep DNA/RNA Kit (Qiagen). Quality of isolated RNA was assessed by 2100 Bioanalyzer (Agilent Technologies) or 2200 Tapestation (Agilent Technologies), and quantity was measured on a NanoDrop 1000 (NanoDrop Technologies) or a Qubit 2.0 Fluorometer (Thermo Fisher Scientific).
Sample preparation for sequencing.
1 μg total RNA of sufficient quality (RIN > 8) was used for sample preparation for sequencing with a TruSeq RNA sample preparation kit (Illumina). The size selection was done using Agencourt AMPure XP beads (Beckman Coulter), aiming at a fragment size above 300 bp. The resulting libraries were quality controlled on a 2200 Tapestation (Agilent Technologies) before combining 6 samples into one pool for sequencing on one lane on a flow cell sequenced on a HiSeq 2000 (Illumina).
Data analysis.
The raw data were base called and demultiplexed using CASAVA 1.8.2 (Illumina) before alignment to hg19 with STAR. To count the number of reads aligned to specific transcripts featureCounts (http://bioinf.wehi.edu.au/featureCounts/) was used. Raw data were normalized using trimmed mean of M values and transformed using voom (limma R-package) before linear modelling.
Insulin secretion.
Glucose-stimulated insulin secretion was assessed in a dynamic perfusion system, Suprafusion 1000 (BRANDEL). Twenty handpicked islets were perfused with low glucose (1.67 mM) for 42 minutes, high glucose (16.7 mM) for 48 minutes, and then low glucose again. Fractions were collected at 6-minute intervals, and the secreted insulin was measured by ELISA (Mercodia). The stimulatory index is defined as the ratio between the areas under the curves that were calculated for the low and high glucose concentrations.
Glucagon secretion.
Human islets were handpicked under a stereo microscope. First, 6–8 groups of 12 islets were preincubated in a Krebs-Ringer carbonate buffer containing 1 mM glucose in a CO2 incubator for 30 minutes. Next, the islets were incubated in Krebs-Ringer buffer containing 1 or 16.7 mM glucose for 60 minutes before an aliquot of the medium was removed for analysis with RIA (Millipore).
Genotyping.
Genotyping was performed using the Illumina Infinium OmniExpressExome-8 v1.0B or the Illumina Infinium Exome v1.0 chips. Genotype calling was done with Illumina Genome studio software. All samples passed standard genotype QC metrics: sample call rate >98%, only European ancestry assessed by principal component analysis comparisons with HapMap populations, gender matched, no relatedness, and no genome-wide heterozygosity outliers. SNPs were removed if the SNP call rate <98% and Hardy-Weinberg equilibrium test P < 5.7 × 10–7
Statistics
Association of SNPs with hormone levels was analyzed using linear regression models and, for prediction of risk of T2D in MDC, using Cox regression models. All analyses were adjusted for sex and age. Phenotypes with nonnormal distributions (GIP, GLP-1, insulin, glucagon, BMI, and CIR) were log transformed before analysis. Individuals who were on antidiabetic medication were excluded. The diet-gene interaction analyses in MDC were additionally adjusted for energy intake. The software PLINK (55) was used for all genome-wide analyses. A P value of less than 5 × 10–8 was used to claim genome-wide significance. Manhattan and QQ plots are presented in Supplemental Figures 1 and 2. The GWAMA software (56) was used to perform meta-analysis of the phenotype-SNP association results from PPP-Botnia (exome chip) and MDC reinvestigation (OmniExpress Exome chip). Fixed-effects meta-analysis was performed using estimates of the allelic effect size and standard error. Other statistical analysis was done using IBM SPSS Statistics for Windows v.22 or Stata v. 13.1 (StataCorp).
GTEx analysis release v4 (dbGaP accession phs000424.v4.p1) was used for eQTL lookups (29). LD was calculated using SNAP v2.2 (57), based on the 1000 Genomes pilot 1 SNP data set.
The heritability estimates of hormone concentrations during OGTT were calculated using GCTA software, which estimates the narrow-sense heritability captured by the genotyped SNPs (9).
Study approval
The MDC study protocol was approved by the Lund University Ethics Review Committee (532/2006, 51-90). The PPP-Botnia protocol was approved by the Ethics Committee of Helsinki University (608/2003). The collection of human intestinal biopsies was approved by the Lund University Ethics Review Committee (2013/580). The collection of human islets was approved by the Lund University Ethics Review Committee (2011/263). All participants provided written informed consent.
Author contributions
PA participated in design of the study, analyzed data, and wrote the manuscript. AL and NW performed the immunohistochemistry. EL and JJH were responsible for incretin measurements. UK and EOL generated and analyzed data from human pancreatic islets. OA and RBP analyzed data. ES and MOM analyzed diet interactions. LH, TT, and OM generated data. PMN participated in the design of the study. LG participated in the design and supervision of the study. EA participated in the design and supervision of the study, analyzed data, and wrote the manuscript. All authors critically read and reviewed the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the European Research Council Advanced Researcher Grant (Gene Target T2D, 269045), the Swedish Research Council (2010-3490), and the Academy of Finland (FiDiPro grant 263401 and project grant 267882 (all to LG); the Bo and Kerstin Hjelt Foundation; the Albert Påhlsson Research Foundation; and the Swedish Research Council (2007-3533, 2010-2917). PPP-Botnia has been financially supported by grants from the Sigrid Juselius Foundation, the Folkhalsan Research Foundation, the Ollqvist Foundation, the Nordic Center of Excellence in Disease Genetics, the EU (EXGENESIS, EUFP7-MOSAIC), the Signe and Ane Gyllenberg Foundation, the Swedish Cultural Foundation in Finland, the Finnish Diabetes Research Foundation, the Foundation for Life and Health in Finland, the Finnish Medical Society, the Paavo Nurmi Foundation, Helsinki University Central Hospital Research Foundation, the Perklén Foundation, the Närpes Health Care Foundation, and the Ahokas Foundation. The study has also been supported by the Ministry of Education in Finland, the Municipal Heath Care Center and Hospital in Jakobstad, and Health Care Centers in Vasa, Närpes, and Korsholm. Human islets were provided by the Nordic Network for Clinical Islet Transplantation and the EXODIAB Human Tissue Lab.
Version 1. 11/02/2017
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(21):e93306. https://doi.org/10.1172/jci.insight.93306.
Contributor Information
Peter Almgren, Email: Peter.Almgren@med.lu.se.
Andreas Lindqvist, Email: andreas.lindqvist@med.lu.se.
Ulrika Krus, Email: ulrika.krus@med.lu.se.
Liisa Hakaste, Email: Liisa.hakaste@hus.fi.
Emilia Ottosson-Laakso, Email: emilia.ottosson@scilifelab.se.
Olof Asplund, Email: olof.asplund@med.lu.se.
Emily Sonestedt, Email: Emily.Sonestedt@med.lu.se.
Rashmi B. Prasad, Email: rashmi.prasad@med.lu.se.
Esa Laurila, Email: esa.laurila@med.lu.se.
Marju Orho-Melander, Email: Marju.Orho-Melander@med.lu.se.
Olle Melander, Email: Olle.Melander@med.lu.se.
Tiinamaija Tuomi, Email: Tiinamaija.Tuomi@hus.fi.
Nils Wierup, Email: nils.wierup@med.lu.se.
Leif Groop, Email: leif.groop@med.lu.se.
Emma Ahlqvist, Email: Emma.Ahlqvist@med.lu.se.
References
- 1.Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology. 2007;132(6):2131–2157. doi: 10.1053/j.gastro.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 2.McIntosh CH, Widenmaier S, Kim SJ. Glucose-dependent insulinotropic polypeptide (Gastric Inhibitory Polypeptide; GIP) Vitam Horm. 2009;80:409–471. doi: 10.1016/S0083-6729(08)00615-8. [DOI] [PubMed] [Google Scholar]
- 3.Brubaker PL. Minireview: update on incretin biology: focus on glucagon-like peptide-1. Endocrinology. 2010;151(5):1984–1989. doi: 10.1210/en.2010-0115. [DOI] [PubMed] [Google Scholar]
- 4.Meek CL, Lewis HB, Reimann F, Gribble FM, Park AJ. The effect of bariatric surgery on gastrointestinal and pancreatic peptide hormones. Peptides. 2016;77:28–37. doi: 10.1016/j.peptides.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Saxena R, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42(2):142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manning AK, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen G, et al. Genome-wide association study identifies novel loci association with fasting insulin and insulin resistance in African Americans. Hum Mol Genet. 2012;21(20):4530–4536. doi: 10.1093/hmg/dds282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rung J, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41(10):1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 11.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 12.Vrhovac I, et al. Localizations of Na(+)-D-glucose cotransporters SGLT1 and SGLT2 in human kidney and of SGLT1 in human small intestine, liver, lung, and heart. Pflugers Arch. 2015;467(9):1881–1898. doi: 10.1007/s00424-014-1619-7. [DOI] [PubMed] [Google Scholar]
- 13.Dobbins RL, et al. Selective sodium-dependent glucose transporter 1 inhibitors block glucose absorption and impair glucose-dependent insulinotropic peptide release. Am J Physiol Gastrointest Liver Physiol. 2015;308(11):G946–G954. doi: 10.1152/ajpgi.00286.2014. [DOI] [PubMed] [Google Scholar]
- 14.Gorboulev V, et al. Na(+)-D-glucose cotransporter SGLT1 is pivotal for intestinal glucose absorption and glucose-dependent incretin secretion. Diabetes. 2012;61(1):187–196. doi: 10.2337/db11-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhre RE, Frost CR, Svendsen B, Holst JJ. Molecular mechanisms of glucose-stimulated GLP-1 secretion from perfused rat small intestine. Diabetes. 2015;64(2):370–382. doi: 10.2337/db14-0807. [DOI] [PubMed] [Google Scholar]
- 16.Ogata H, et al. KATP channel as well as SGLT1 participates in GIP secretion in the diabetic state. J Endocrinol. 2014;222(2):191–200. doi: 10.1530/JOE-14-0161. [DOI] [PubMed] [Google Scholar]
- 17.Ahlqvist E, et al. Link between GIP and osteopontin in adipose tissue and insulin resistance. Diabetes. 2013;62(6):2088–2094. doi: 10.2337/db12-0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berglund LM, et al. Glucose-dependent insulinotropic polypeptide stimulates osteopontin expression in the vasculature via endothelin-1 and CREB. Diabetes. 2016;65(1):239–254. doi: 10.2337/db15-0122. [DOI] [PubMed] [Google Scholar]
- 19.Lyssenko V, et al. Pleiotropic effects of GIP on islet function involve osteopontin. Diabetes. 2011;60(9):2424–2433. doi: 10.2337/db10-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena R, et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90(3):410–425. doi: 10.1016/j.ajhg.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonner C, et al. Inhibition of the glucose transporter SGLT2 with dapagliflozin in pancreatic alpha cells triggers glucagon secretion. Nat Med. 2015;21(5):512–517. doi: 10.1038/nm.3828. [DOI] [PubMed] [Google Scholar]
- 24.Wessel J, et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun. 2015;6:5897. doi: 10.1038/ncomms6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paterson AD, et al. Genome-wide association identifies the ABO blood group as a major locus associated with serum levels of soluble E-selectin. Arterioscler Thromb Vasc Biol. 2009;29(11):1958–1967. doi: 10.1161/ATVBAHA.109.192971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbalic M, et al. Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels. Hum Mol Genet. 2010;19(9):1863–1872. doi: 10.1093/hmg/ddq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teupser D, et al. Genetic regulation of serum phytosterol levels and risk of coronary artery disease. Circ Cardiovasc Genet. 2010;3(4):331–339. doi: 10.1161/CIRCGENETICS.109.907873. [DOI] [PubMed] [Google Scholar]
- 28.Kaliannan K, et al. Intestinal alkaline phosphatase prevents metabolic syndrome in mice. Proc Natl Acad Sci USA. 2013;110(17):7003–7008. doi: 10.1073/pnas.1220180110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasad RB, Groop L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 2015;6(1):87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gjesing AP, et al. Fasting and oral glucose-stimulated levels of glucose-dependent insulinotropic polypeptide (GIP) and glucagon-like peptide-1 (GLP-1) are highly familial traits. Diabetologia. 2012;55(5):1338–1345. doi: 10.1007/s00125-012-2484-6. [DOI] [PubMed] [Google Scholar]
- 32.Matikainen N, et al. GLP-1 responses are heritable and blunted in acquired obesity with high liver fat and insulin resistance. Diabetes Care. 2014;37(1):242–251. doi: 10.2337/dc13-1283. [DOI] [PubMed] [Google Scholar]
- 33.Krishna Kumar S, Feldman MW, Rehkopf DH, Tuljapurkar S. Limitations of GCTA as a solution to the missing heritability problem. Proc Natl Acad Sci USA. 2016;113(1):E61–E70. doi: 10.1073/pnas.1520109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speed D, Cai N, UCLEB Consortium. Johnson MR, Nejentsev S, Balding DJ. Reevaluation of SNP heritability in complex human traits. Nat Genet. 2017;49(7):986–992. doi: 10.1038/ng.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lund A, et al. Evidence of extrapancreatic glucagon secretion in man. Diabetes. 2016;65(3):585–597. doi: 10.2337/db15-1541. [DOI] [PubMed] [Google Scholar]
- 36.Karras S, Goulis DG, Mintziori G, Katsiki N, Tzotzas T. The effects of incretins on energy homeostasis: physiology and implications for the treatment of type 2 diabetes mellitus and obesity. Curr Vasc Pharmacol. 2012;10(6):781–791. doi: 10.2174/157016112803520909. [DOI] [PubMed] [Google Scholar]
- 37.Silbernagel G, et al. High intestinal cholesterol absorption is associated with cardiovascular disease and risk alleles in ABCG8 and ABO: evidence from the LURIC and YFS cohorts and from a meta-analysis. J Am Coll Cardiol. 2013;62(4):291–299. doi: 10.1016/j.jacc.2013.01.100. [DOI] [PubMed] [Google Scholar]
- 38.Schenkel-Brunner H. Studies on blood-groups A1 and A2. Further evidence for the predominant influence of quantitative differences in the number of A antigenic sites present on A1 and A2 erythrocytes. Eur J Biochem. 1982;122(3):511–514. [PubMed] [Google Scholar]
- 39.Yamamoto F, McNeill PD, Hakomori S. Human histo-blood group A2 transferase coded by A2 allele, one of the A subtypes, is characterized by a single base deletion in the coding sequence, which results in an additional domain at the carboxyl terminal. Biochem Biophys Res Commun. 1992;187(1):366–374. doi: 10.1016/S0006-291X(05)81502-5. [DOI] [PubMed] [Google Scholar]
- 40.Hui DY, Howles PN. Molecular mechanisms of cholesterol absorption and transport in the intestine. Semin Cell Dev Biol. 2005;16(2):183–192. doi: 10.1016/j.semcdb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Park H, et al. Homeobox D1 regulates angiogenic functions of endothelial cells via integrin β1 expression. Biochem Biophys Res Commun. 2011;408(1):186–192. doi: 10.1016/j.bbrc.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 42.Sheth R, et al. Decoupling the function of Hox and Shh in developing limb reveals multiple inputs of Hox genes on limb growth. Development. 2013;140(10):2130–2138. doi: 10.1242/dev.089409. [DOI] [PubMed] [Google Scholar]
- 43.Amato A, Baldassano S, Mulè F. GLP2: an underestimated signal for improving glycaemic control and insulin sensitivity. J Endocrinol. 2016;229(2):R57–R66. doi: 10.1530/JOE-16-0035. [DOI] [PubMed] [Google Scholar]
- 44.Yusta B, et al. Enteroendocrine localization of GLP-2 receptor expression in humans and rodents. Gastroenterology. 2000;119(3):744–755. doi: 10.1053/gast.2000.16489. [DOI] [PubMed] [Google Scholar]
- 45.Pedersen J, et al. The glucagon-like peptide 2 receptor is expressed in enteric neurons and not in the epithelium of the intestine. Peptides. 2015;67:20–28. doi: 10.1016/j.peptides.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 46.Ørskov C, Hartmann B, Poulsen SS, Thulesen J, Hare KJ, Holst JJ. GLP-2 stimulates colonic growth via KGF, released by subepithelial myofibroblasts with GLP-2 receptors. Regul Pept. 2005;124(1-3):105–112. doi: 10.1016/j.regpep.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 47.de Heer J, Pedersen J, Orskov C, Holst JJ. The alpha cell expresses glucagon-like peptide-2 receptors and glucagon-like peptide-2 stimulates glucagon secretion from the rat pancreas. Diabetologia. 2007;50(10):2135–2142. doi: 10.1007/s00125-007-0761-6. [DOI] [PubMed] [Google Scholar]
- 48.Manjer J, et al. The Malmö Diet and Cancer Study: representativity, cancer incidence and mortality in participants and non-participants. Eur J Cancer Prev. 2001;10(6):489–499. doi: 10.1097/00008469-200112000-00003. [DOI] [PubMed] [Google Scholar]
- 49.Rosvall M, et al. Risk factors for the progression of carotid intima-media thickness over a 16-year follow-up period: the Malmö Diet and Cancer Study. Atherosclerosis. 2015;239(2):615–621. doi: 10.1016/j.atherosclerosis.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 50.Isomaa B, et al. A family history of diabetes is associated with reduced physical fitness in the Prevalence, Prediction and Prevention of Diabetes (PPP)-Botnia study. Diabetologia. 2010;53(8):1709–1713. doi: 10.1007/s00125-010-1776-y. [DOI] [PubMed] [Google Scholar]
- 51.Lindgren O, et al. Incretin hormone and insulin responses to oral versus intravenous lipid administration in humans. J Clin Endocrinol Metab. 2011;96(8):2519–2524. doi: 10.1210/jc.2011-0266. [DOI] [PubMed] [Google Scholar]
- 52.Bak MJ, et al. Specificity and sensitivity of commercially available assays for glucagon and oxyntomodulin measurement in humans. Eur J Endocrinol. 2014;170(4):529–538. doi: 10.1530/EJE-13-0941. [DOI] [PubMed] [Google Scholar]
- 53.Wirfält E, Mattisson I, Johansson U, Gullberg B, Wallström P, Berglund G. A methodological report from the Malmö Diet and Cancer study: development and evaluation of altered routines in dietary data processing. Nutr J. 2002;1:3. doi: 10.1186/1475-2891-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldstein JI, et al. zCall: a rare variant caller for array-based genotyping: genetics and population analysis. Bioinformatics. 2012;28(19):2543–2545. doi: 10.1093/bioinformatics/bts479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.