Abstract
Background
Increasing studies showed that miR-200 family (miR-200s) clusters are aberrantly expressed in multiple human cancers, and miR-200s clusters function as tumor suppressor genes by affecting cell proliferation, self-renewal, differentiation, division and apoptosis. Herein, we aimed to investigate the expression and clinical implication of miR-200s clusters in acute myeloid leukemia (AML).
Methods
RT-qPCR was performed to detect expression of miR-200s clusters in 19 healthy donors, 98 newly diagnosed AML patients, and 35 AML patients achieved complete remission (CR).
Results
Expression of miR-200a/200b/429 cluster but not miR-200c/141 cluster was decreased in newly diagnosed AML patients as compared to healthy donors and AML patients achieved CR. Although no significant differences were observed between miR-200s clusters and most of the features, low expression of miR-200s clusters seems to be associated with higher white blood cells especially for miR-200a/200b. Of the five members of miR-200s clusters, low expression of miR-200b/429/200c was found to be associated with lower CR rate. Logistic regression analysis further revealed that low expression of miR-429 acted as an independent risk factor for CR in AML. Based on Kaplan–Meier analysis, low expression of miR-200b/429/200c was associated with shorter OS, whereas miR-200a/141 had a trend. Moreover, multivariate analysis of Cox regression models confirmed the independently prognostic value of miR-200b expression for OS in AML.
Conclusions
Expression of miR-200a/200b/429 cluster was frequently down-regulated in AML, and low expression of miR-429 as an independent risk factor for CR, whereas low expression of miR-200b as an independent prognostic biomarker for OS.
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1494-7) contains supplementary material, which is available to authorized users.
Keywords: miR-200, Expression, Prognosis, Acute myeloid leukemia
Background
Acute myeloid leukemia (AML) is a highly heterogeneous malignant hematological disorder with complex molecular pathophysiology. Although the treatment strategies against AML have been updated in the past decades, the majority of patients eventually succumb to relapse after induction chemotherapy [1]. Clinical outcome of AML remains unsatisfactory especially in those with specific karyotypes/biomarkers such as inv(3)(q21q26.2), t(6;9)(p23; q34), 11q abnormalities other than t(9;11), -5/del(5q), -7, TP53 mutations, FLT3-ITD mutations, C-KIT mutations, WT1 overexpression, and BAALC overexpression [2–4]. The development of effective therapeutic options against AML relies on mechanistic understanding of AML biology, especially in molecular regulators of AML pathogenesis and molecular predictor of AML prognosis [5].
MicroRNAs, a class of small (19–22 nucleotides) single-stranded RNAs, negatively regulate various genes by targeting 3′-untranslated region (3′-UTR) of mRNAs, thereby facilitating translational silencing or degradation of targeted genes [6]. Mounting evidences have implicated that microRNAs play crucial roles in regulating many fundamental and biological processes including cancer development [7]. Moreover, microRNAs have been reported as novel biomarkers for diagnosis and prognosis, and regarded as potential therapeutic targets in AML [8]. For instance, recent studies implicated that several microRNAs such as miR-216b, miR-362-5p, miR-217, and miR-193b were prognosis-related predictors in AML and may involve in AML biology [9–12].
The miR-200 family (miR-200s) clusters includes five members (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) and can be divided into two clusters (miR-200a/b/429 cluster and miR-200c/141 cluster) based on chromosomal location (chromosome 1p36 and chromosome 12p13) [13]. Numerous studies showed that miR-200s clusters are aberrantly expressed in multiple human cancers, and miR-200s clusters function as tumor suppressor genes by affecting cell proliferation, self-renewal, differentiation, division and apoptosis [14]. Although the tumor-suppressive roles of miR-200s clusters have also been reported in solid tumors with prognostic value [14, 15], the expression and clinical implication of miR-200s clusters in AML remains poorly revealed.
In this study, we investigated expression of miR-200s clusters in AML patients except for acute promyelocytic leukemia (APL), and found that low expression of miR-200s clusters acted as potential prognostic biomarkers in AML.
Methods
Patients and treatment
A total of 98 de novo AML patients except for APL and 19 healthy donors were enrolled in this study. Bone marrow (BM) was collected from all the patients at diagnosis time as well as 35 patients at complete remission (CR) time. AML was diagnosed based on the French–American–British (FAB) and 2016 revised World Health Organization (WHO) criteria [16, 17]. All the patients received chemotherapy as reported [18]. Induction chemotherapy therapy was 1–2 courses of daunorubicin combined with cytarabine. Subsequent consolidation treatment after CR for younger patients included high-dose cytarabine, mitoxantrone with cytarabine, and homoharringtonine combined with cytarabine, whereas for older patients received in an individualized manner decided by the physicians, such as CHG protocol (cytarabine, homoharringtonine, and G-CSF). This study was approved by the Ethics Committee of the Affiliated People’s Hospital of Jiangsu University, and written informed consents were informed and signed by all participants in accordance with the Declaration of Helsinki Principles.
Cytogenetic analysis and mutation detection
BM cells were harvested after 1–3 days of unstimulated culture in RPMI 1640 medium (BOSTER, Wuhan, China) containing 20% fetal calf serum (ExCell Bio, Shanghai, China). Cytogenetics for AML patients were analyzed at the newly diagnosis time by conventional R-banding method and karyotype risk was classified according to reported previously [19, 20]. Hotspot mutations in NPM1, C-KIT, DNMT3A, N/K-RAS, IDH1/2, U2AF1, SRSF2 and SETBP1 were detected by high-resolution melting analysis [21–25], whereas mutations in FLT3-ITD and CEBPA were examined by DNA sequencing [26].
RNA isolation and reverse transcription
BM mononuclear cells (BMMNCs) were extracted as reported using Lymphocyte Separation Medium (Absin, Shanghai, China) [27]. According to the manufacturer’s protocols, RNA was extracted from BMMNCs using the mirVana miRNA isolation kit (Ambion, Austin, TX, USA), and was synthesized to cDNA by reverse transcription using MiScript Reverse Transcription Kit (Qiagen, Duesseldorf, Germany).
Real-time quantitative PCR
The level of miR-200s clusters was detected by real-time quantitative PCR (RT-qPCR) using miScript SYBR green PCR kit (Qiagen, Duesseldorf, Germany). The primers were miR-200s specific (Additional file 1: Table S1) and the manufacturer-provided miScript universal primer (Qiagen, Duesseldorf, Germany). The programs for RT-qPCR reactions were performed as reported [28]. U6 small nuclear RNA was selected as the endogenous normalizer detected by RT-qPCR using 2× SYBR Green PCR Mix (Multisciences, Hangzhou, China). Relative miR-200s level was calculated by 2−ΔΔCT method. The healthy donors that possessed the minimal ΔCT between miR-200s (each member) and U6 expression was selected as control, and was defined as 100% expression.
Statistical analysis
Mann–Whitney’s U test was carried to compare the difference of continuous variables between two groups, whereas Pearson Chi square analysis/Fisher exact test were applied to compare the difference of categorical variables between two groups. The impact of miR-200s clusters expression on overall survival (OS) was analyzed by Kaplan–Meier analysis, and Cox regression models (univariate and multivariate analyses) were further used to determine the independently prognostic value of miR-200s cluster expression. The effect of miR-200s clusters expression on CR was determined by Logistic regression analysis (univariate and multivariate analyses). All tests were two sided, and P < 0.05 was defined as statistically significant. SPSS software 20.0 and GraphPad Prism 5.0 was used to conduct the statistical analyses in this study.
Results
Expression of miR-200s in AML
We analyzed miR-200s clusters expression in BM from 19 healthy donors, 98 AML patients, and 35 AML patients achieved CR by RT-qPCR. As presented in Fig. 1, expression of miR-200a/200b/429 clusters but not miR-200c/141 clusters was significantly decreased in AML patients as compared to healthy donors and AML patients achieved CR.
Fig. 1.
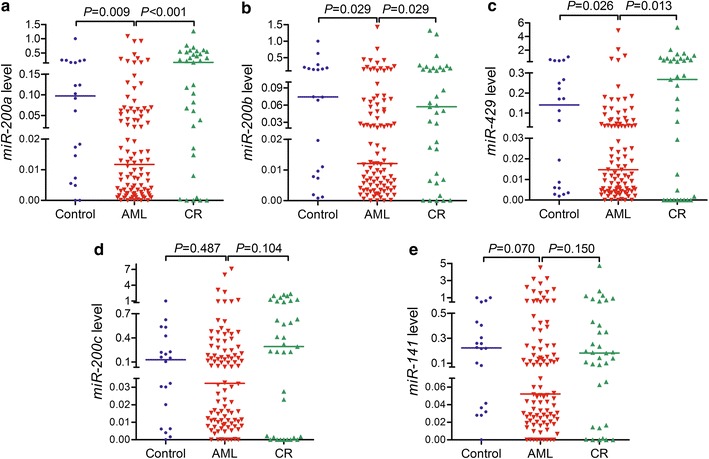
Expression of miR-200s clusters in controls, newly diagnosed AML patients and AML patients achieved CR. a For miR-200a; b For miR-200b; c For miR-429; d For miR-200c; e For miR-141. The distributions of the miR-200s clusters expression in controls, newly diagnosed AML patients and AML patients achieved CR were presented with scatter plots. The median level of miR-200s clusters expression in each group was shown with horizontal line
Relationship between miR-200s and clinical features in AML
To investigate clinical implication of miR-200s clusters expression, the whole-cohort patients were classified into two groups (high and low miR-200s clusters expression) based on the median level of each member of miR-200s clusters, respectively. We analyzed the association between each member of miR-200s clusters expression and clinic-pathologic features including gender, age, white blood cell (WBC) counts, hemoglobin content, platelet counts, blasts (%), FAB subtypes, karyotypes, and common gene mutations. As shown in Table 1, no significant differences were observed between miR-200s clusters expression and most of the features. However, low expression of miR-200s clusters seems to be associated with higher WBC counts especially for miR-200a/200b (P = 0.001 and 0.041, respectively). In addition, low expression of miR-200a was related to male, whereas low expression of miR-141 was correlated with higher hemoglobin content (P = 0.013 and 0.024, respectively).
Table 1.
Correlation of miR-200s cluster expression with clinical/laboratory features in AML patients
| Patient’s features | miR-200a expression | miR-200b expression | miR-429 expression | miR-200c expression | miR-141 expression | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low (n = 49) | High (n = 49) | P | Low (n = 49) | High (n = 49) | P | Low (n = 49) | High (n = 49) | P | Low (n = 49) | High (n = 49) | P | Low (n = 49) | High (n = 49) | P | |
| Sex (male/female) | 36/13 | 23/26 | 0.013 | 32/17 | 27/22 | 0.409 | 31/18 | 28/21 | 0.680 | 32/17 | 27/22 | 0.409 | 31/18 | 28/21 | 0.680 |
| Age (years) | 58 (21–81) | 61 (18–87) | 0.842 | 60 (18–81) | 59 (18–87) | 0.831 | 60 (21–81) | 59 (18–87) | 0.741 | 60 (18–81) | 59 (18–87) | 0.743 | 60 (18–81) | 59 (18–87) | 1.000 |
| WBC (× 109/L) | 38.7 (1.3–528.0) | 9.6 (1.1–130.2) | 0.001 | 35.5 (1.3–528.0) | 9.6 (1.1–130.2) | 0.041 | 34.5 (1.3–528.0) | 13.3 (1.1–130.2) | 0.099 | 34.9 (1.3–528.0) | 13.2 (1.1–130.2) | 0.094 | 35.5 (1.3–528.0) | 10.2 (1.1–116.6) | 0.090 |
| Hemoglobin (g/L) | 78 (53–138) | 76.5 (32–144) | 0.085 | 80 (53–138) | 76.5 (32–144) | 0.330 | 77 (32–138) | 78 (34–134) | 0.940 | 84 (53–138) | 76 (32–144) | 0.124 | 88 (53–138) | 74.5 (32–144) | 0.024 |
| Platelets (× 109/L) | 37 (3–447) | 47 (4–264) | 0.558 | 37 (3–447) | 46 (4–264) | 0.460 | 36 (3–447) | 47 (4–264) | 0.512 | 30 (5–447) | 49 (3–264) | 0.233 | 40 (5–125) | 46 (3–447) | 0.769 |
| BM blasts (%) | 60% (20–99%) | 58% (20–95%) | 0.892 | 63% (20–99%) | 58% (21–95%) | 0.651 | 60% (20–98%) | 59% (20–99%) | 0.757 | 63% (20–99%) | 54% (20–95%) | 0.226 | 62% (20–98%) | 57% (20–99%) | 0.598 |
| FAB subtypes | 0.660 | 0.945 | 0.681 | 0.827 | 0.900 | ||||||||||
| M0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | |||||
| M1 | 4 | 2 | 3 | 3 | 3 | 3 | 2 | 4 | 2 | 4 | |||||
| M2 | 26 | 24 | 23 | 27 | 22 | 28 | 23 | 27 | 24 | 26 | |||||
| M4 | 12 | 15 | 14 | 13 | 15 | 12 | 15 | 12 | 15 | 12 | |||||
| M5 | 6 | 6 | 7 | 5 | 8 | 4 | 7 | 5 | 6 | 6 | |||||
| M6 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| Karyotypes | 0.982 | 0.813 | 0.606 | 0.948 | 0.578 | ||||||||||
| Normal | 25 | 28 | 23 | 30 | 23 | 30 | 24 | 29 | 24 | 29 | |||||
| t(8;21) | 5 | 5 | 5 | 5 | 6 | 4 | 5 | 5 | 6 | 4 | |||||
| + 8 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 3 | 0 | |||||
| − 5/5q- | 1 | 2 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | |||||
| − 7/7q- | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | |||||
| t(9;22) | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | |||||
| Complex | 8 | 7 | 9 | 6 | 10 | 5 | 8 | 7 | 7 | 8 | |||||
| Others | 6 | 5 | 6 | 5 | 6 | 5 | 6 | 5 | 5 | 6 | |||||
| No data | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | |||||
| Gene mutations | |||||||||||||||
| CEBPA (±) | 7/33 | 6/38 | 0.765 | 5/38 | 8/33 | 0.376 | 8/36 | 5/35 | 0.555 | 7/34 | 6/37 | 0.768 | 6/37 | 7/34 | 0.768 |
| NPM1 (±) | 5/35 | 4/40 | 0.730 | 3/40 | 6/35 | 0.307 | 4/40 | 5/35 | 0.730 | 5/36 | 4/39 | 0.735 | 4/39 | 5/36 | 0.735 |
| FLT3-ITD (±) | 6/34 | 3/41 | 0.298 | 6/37 | 3/38 | 0.484 | 6/38 | 3/37 | 0.488 | 5/36 | 4/39 | 0.735 | 6/37 | 3/38 | 0.484 |
| C-KIT (±) | 1/39 | 1/43 | 1.000 | 1/42 | 1/40 | 1.000 | 1/43 | 1/39 | 1.000 | 1/40 | 1/42 | 1.000 | 1/42 | 1/40 | 1.000 |
| N/K-RAS (±) | 4/36 | 6/38 | 0.741 | 3/40 | 7/34 | 0.190 | 5/39 | 5/35 | 1.000 | 6/35 | 4/39 | 0.515 | 6/37 | 4/37 | 0.739 |
| IDH1/2 (±) | 1/39 | 3/41 | 0.618 | 3/40 | 1/40 | 0.616 | 4/40 | 0/40 | 0.118 | 4/37 | 0/43 | 0.052 | 2/41 | 2/39 | 1.000 |
| DNMT3A (±) | 4/36 | 2/42 | 0.418 | 4/39 | 2/39 | 0.676 | 4/40 | 2/38 | 0.678 | 3/38 | 3/40 | 1.000 | 3/40 | 3/38 | 1.000 |
| U2AF1 (±) | 2/38 | 2/42 | 1.000 | 1/42 | 3/38 | 0.354 | 1/43 | 3/37 | 0.343 | 2/39 | 2/41 | 1.000 | 3/40 | 1/40 | 0.616 |
| SRSF2 (±) | 1/39 | 3/41 | 0.618 | 1/42 | 3/38 | 0.354 | 1/43 | 3/37 | 0.343 | 1/40 | 3/40 | 0.616 | 1/42 | 3/38 | 0.354 |
| SETBP1 (±) | 2/38 | 0/44 | 0.224 | 2/41 | 0/41 | 0.494 | 2/42 | 0/40 | 0.495 | 2/39 | 0/43 | 0.235 | 2/41 | 0/41 | 0.494 |
| CR (±) | 16/33 | 23/26 | 0.215 | 14/35 | 25/24 | 0.038 | 14/35 | 25/24 | 0.038 | 14/35 | 25/24 | 0.038 | 15/34 | 24/25 | 0.098 |
WBC white blood cells, BM bone marrow, FAB French–American–British classification, CR complete remission
Prognostic value of miR-200s in AML
To observe the impact of miR-200s clusters expression on clinical outcome in AML, we first determined the association of each member of miR-200s clusters expression with CR. Of the five members of miR-200s clusters, low expression of miR-200b/429/200c was found to be associated with lower CR rate (Table 1, all P = 0.038). Additionally, Logistic regression analysis was further performed to confirm and verify the effect of miR-200s clusters’ expression on CR, and revealed low expression of miR-429 as an independent risk factor for CR in AML (Table 2, P = 0.023).
Table 2.
Univariate and multivariate analyses of variables for overall survival in AML patients
| Variables | Complete remission | Overall survival | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
| OR (95% CI) | P | OR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |
| miR-200a | 0.548 (0.242–1.244) | 0.150 | 1.029 (0.285–3.722) | 0.965 | 0.662 (0.415–1.022) | 0.082 | 1.425 (0.651–3.120) | 0.376 |
| miR-200b | 0.384 (0.167–0.885) | 0.025 | 0.823 (0.199–3.401) | 0.788 | 0.511 (0.319–0.819) | 0.005 | 0.524 (0.305–0.902) | 0.020 |
| miR-429 | 0.384 (0.167–0.885) | 0.025 | 0.331 (0.128–0.858) | 0.023 | 0.558 (0.350–0.891) | 0.015 | 0.820 (0.325–2.073) | 0.675 |
| miR-200c | 0.384 (0.167–0.885) | 0.025 | 0.977 (0.149–6.400) | 0.981 | 0.606 (0.380–0.965) | 0.035 | 0.649 (0.190–2.217) | 0.491 |
| miR-141 | 0.460 (0.201–1.050) | 0.065 | 0.594 (0.192–1.833) | 0.364 | 0.695 (0.437–1.104) | 0.123 | 1.152 (0.582–2.279) | 0.684 |
| Age | 4.229 (1.742–10.266) | 0.001 | 4.555 (1.715–12.095) | 0.002 | 2.046 (1.282–3.266) | 0.003 | 1.732 (1.033–2.902) | 0.037 |
| WBC | 2.367 (1.015–5.520) | 0.046 | 1.846 (0.715–4.767) | 0.206 | 2.002 (1.253–3.199) | 0.004 | 1.560 (0.925–2.629) | 0.095 |
| Karyotype | 3.108 (1.338–7.220) | 0.008 | 2.862 (1.164–7.042) | 0.022 | 1.875 (1.295–2.715) | 0.001 | 1.874 (1.210–2.902) | 0.005 |
| CEBPA mutations | 0.526 (0.160–1.731) | 0.290 | 0.870 (0.413–1.829) | 0.713 | ||||
| NPM1 mutations | 0.833 (0.207–3.358) | 0.798 | 1.200 (0.516–2.793) | 0.672 | ||||
| FLT3-ITD mutations | 0.833 (0.207–3.358) | 0.798 | 0.935 (0.403–2.170) | 0.876 | ||||
| C-KIT mutations | 0.673 (0.041–11.150) | 0.783 | 0.479 (0.066–3.458) | 0.465 | ||||
| N/K-RAS mutations | 3.048 (0.605–15.343) | 0.177 | 1.311 (0.621–2.770) | 0.478 | ||||
| IDH1/2 mutations | Undetermined | 0.999 | 4.671 (1.637–13.326) | 0.004 | 6.662 (1.757–25.268) | 0.005 | ||
| DNMT3A mutations | 1.391 (0.240–8.057) | 0.712 | 1.590 (0.634–3.987) | 0.323 | ||||
| U2AF1 mutations | Undetermined | 0.999 | 2.791 (0.987–7.890) | 0.053 | 5.130 (1.714–15.355) | 0.003 | ||
| SRSF2 mutations | Undetermined | 0.999 | 1.934 (0.693–5.400) | 0.208 | ||||
| SETBP1 mutations | 0.673 (0.041–11.150) | 0.783 | 0.637 (0.088–4.613) | 0.656 | ||||
OR odd ratio, HR hazard ratio, CI confidence interval. Variables including miR-200s cluster expression (Low vs. High), age (≤ 60 vs. > 60 years), WBC (≥ 30 × 109 vs. < 30 × 109/L), karyotype (favorable vs. intermediate vs. poor), and gene mutations (mutant vs. wild-type). Multivariate analysis includes variables with P < 0.200 in univariate analysis
We next evaluated the correlation of each member of miR-200s clusters expression with survival. Based on Kaplan–Meier analysis, low expression of miR-200b/429/200c was associated with shorter OS, whereas miR-200a/141 had a trend (Fig. 2). In addition, we also analyzed the impact of composite members of miR-200s clusters expression on OS by Kaplan–Meier analysis as shown in Fig. 3.
Fig. 2.
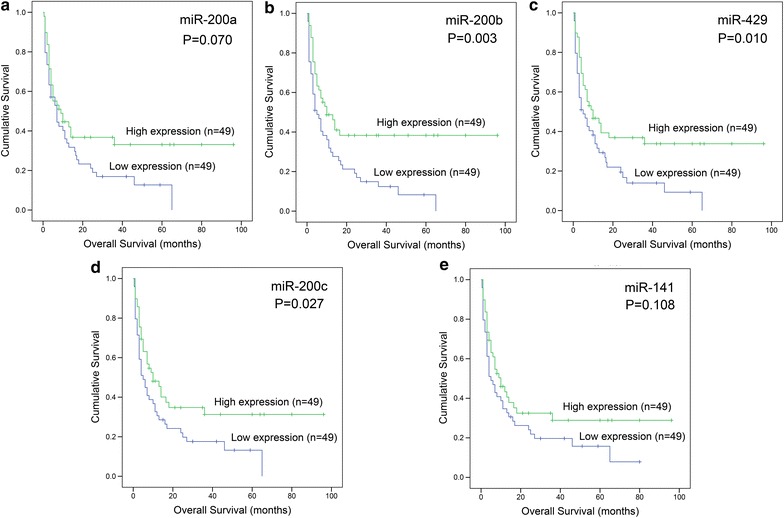
Prognostic value of each member of miR-200s clusters expression in AML. a For miR-200a. b For miR-200b. c For miR-429. d For miR-200c. e For miR-141. Overall survival analyzed between two groups based on median level of each member of miR-200s clusters, and performed by Kaplan–Meier methods
Fig. 3.
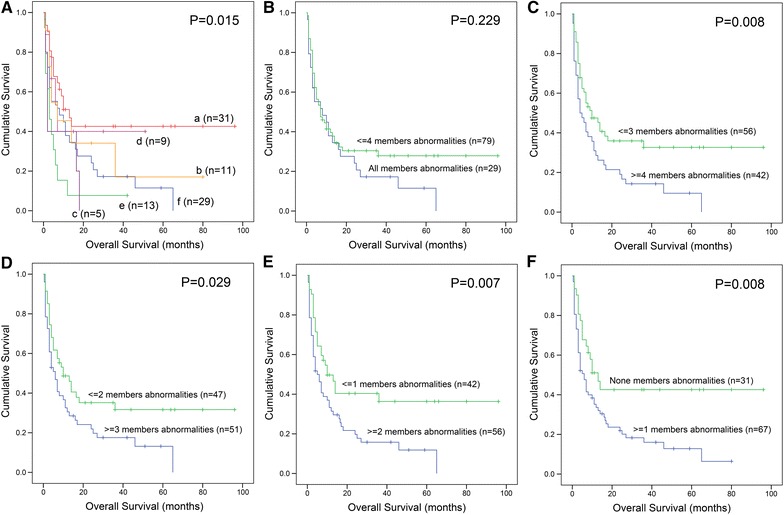
Prognostic value of composite members of miR-200s clusters expression in AML. A Overall survival (OS) analyzed among AML patients with different numbers of abnormal miR-200s clusters expression; a: without miR-200s clusters abnormalities; b: with only one member of miR-200s clusters abnormalities; c: with two members of miR-200s clusters abnormalities; d: with three members of miR-200s clusters abnormalities; e: with four members of miR-200s clusters abnormalities; f: with all members of miR-200s clusters abnormalities. B OS analyzed between two groups (all members of miR-200s clusters abnormalities vs. equal or less than four members of miR-200s clusters abnormalities). C OS analyzed between two groups (equal or more than four members of miR-200s clusters abnormalities vs. equal or less than three members of miR-200s clusters abnormalities). D OS analyzed between two groups (equal or more than three members of miR-200s clusters abnormalities vs. equal or less than two members of miR-200s clusters abnormalities). E OS analyzed between two groups (equal or more than two members of miR-200s clusters abnormalities vs. equal or less than one member of miR-200s clusters abnormalities). F OS analyzed between two groups (equal or more than one member of miR-200s clusters abnormalities vs. without any members of miR-200s clusters abnormalities)
Since miR-200s clusters expression was associated with well-established prognostic factor such as WBC counts, we further conducted a Cox regression model adjusting for prognosis-related factors (age, WBC counts, karyotypic classifications, and gene mutations) for OS. Results showed that low expression of miR-200b acted as an independent prognostic biomarker for OS (P = 0.020, Table 2).
Discussion
In the current study, we for the first time investigated expression of miR-200s clusters in AML, and revealed that most of the members of miR-200s clusters were down-regulated in de novo AML patients. Recently, Li et al. revealed that introduction of a pre-miR-200c reduced the expression of ZEB2 protein and inhibited the proliferation of human leukemia cell lines (HL-60, MOLM-13, and THP-1), and mouse miR-200c significantly impaired the proliferation of mouse leukemia cells [29]. Taken together, these results emphasized the crucial role of miR-200s clusters in leukemogenesis. Although the biological role of miR-200s clusters in AML was less studied, tumor suppressor roles of miR-200s clusters have been identified in a variety of human solid cancers, such as bladder cancer, gastric cancer, colorectal cancer, breast cancer, ovarian cancer, endometrial cancer, pancreatic cancer, gliomas, hepatocellular carcinoma, and lung cancer [14, 30]. The miR-200s clusters were reported as key inhibitors of epithelial-to-mesenchymal transition by directly targeting transcriptional repressors of E-cadherin, ZEB1, and ZEB2 [13]. Moreover, miR-200s clusters also played crucial roles in the repression of cancer stem cells self-renewal and differentiation, modulation of cell division and apoptosis, and reversal of chemoresistance [14, 30]. Notably, in some other hematological malignancies, expression or biological role of miR-200s clusters has been preliminary studied. For instance, Choi et al. reported that miR-200c was decreased in patients with myelodysplastic syndrome (MDS) [31]. González-Gugel et al. revealed that down-regulation of mmu-miR-30a and mmu-miR-141 as well as hsa-miR-193b clearly contributed to enhance the expression of Smoothened (SMO) gene in mouse and human lymphomas and, subsequently, to activate the GLI/Hh signalling [32].
In addition to basic research before, it has been noted that low expression of miR-200s clusters could correlate with adverse clinical outcome and serve as a prognostic biomarker for various cancer patients [15]. Although the potential prognostic value of miR-200s clusters in several human cancers remains controversial, a recent meta-analysis demonstrated that lower tissue expression of miR-200s clusters’ members were associated with poor OS and progression-free survival, whereas lower expression of circulating miR-200s clusters’ members were correlated with favorable prognosis [15]. From our study, we showed the negative effect of low expression of miR-200s clusters on AML chemotherapy response and survival. Moreover, multivariate analysis showed that low expression of miR-429 as an independent risk factor for CR, whereas low expression of miR-200b as an independent prognostic biomarker for OS in AML. Due to some limitations in this study (such as patients numbers, treatment regimens, and single center), prospective studies are needed to verify our results before miR-200s clusters expression could be used routinely as a promising biomarker for risk stratification in AML.
Conclusion
Expression of miR-200a/200b/429 cluster was frequently down-regulated in AML, and low expression of miR-429 as an independent risk factor for CR, whereas low expression of miR-200b as an independent prognostic biomarker for OS.
Additional file
Additional file 1: Table S1. The primer sequences for miR-200s clusters.
Authors’ contributions
JQ and JL conceived and designed the experiments; JZ and LZ performed the experiments; JZ and TZ analyzed the data; YG, WZ and DW collected the clinical data; JM, XW and HG offered technique support; JZ wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
None.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Written informed consents were obtained from all enrolled individuals prior to their participation.
Ethical approval and consent to participate
The present study approved by the Ethics Committee and Institutional Review Board of the Affiliated People’s Hospital of Jiangsu University.
Funding
This work was supported by National Natural Science foundation of China (81270630), Medical Innovation Team of Jiangsu Province (CXTDB2017002), 333 Project of Jiangsu Province (BRA2016131), Six Talent Peaks Project in Jiangsu Province (2015-WSN-115), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX17_1821), Youth Medical Talents Project of “Ke Jiao Qiang Wei” project of Jiangsu province (QNRC2016450, QNRC2016449), China Postdoctoral Science Foundation funded project (2016M601748), Clinical Medical Science and Development Foundation of Jiangsu University (JLY20160011), Social Development Foundation of Kunshan (KS1624), Social Development Foundation of Zhenjiang (SH2016045, SH2016046, SH2017040), Key Medical Talent Program of Zhenjiang City.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- AML
acute myeloid leukemia
- 3′-UTR
3′-untranslated region
- APL
acute promyelocytic leukemia
- BM
bone marrow
- CR
complete remission
- BMMNCs
BM mononuclear cells
- FAB
French–American–British
- WHO
World Health Organization
- RT-qPCR
real-time quantitative PCR
- OS
overall survival
- WBC
white blood cell
Footnotes
Jing-dong Zhou and Liu-chao Zhang contributed equally to this work
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1494-7) contains supplementary material, which is available to authorized users.
Contributor Information
Jing-dong Zhou, Email: zhoujingdong1989@qq.com.
Liu-chao Zhang, Email: 1012517087@qq.com.
Ting-juan Zhang, Email: 1162004751@qq.com.
Yu Gu, Email: 547046087@qq.com.
De-hong Wu, Email: 40534048@qq.com.
Wei Zhang, Email: zhangweizwmz@126.com.
Ji-chun Ma, Email: majichun606@sohu.com.
Xiang-mei Wen, Email: wenxiangmei@126.com.
Hong Guo, Email: guohong1009@163.com.
Jiang Lin, Email: linjiangmail@sina.com.
Jun Qian, Email: qianjun0007@hotmail.com.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Rowley JD. Chromosomal translocations: revisited yet again. Blood. 2008;112(6):2183–2189. doi: 10.1182/blood-2008-04-097931. [DOI] [PubMed] [Google Scholar]
- 3.Mrózek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, Metzeler KH, Schwind S, Wu YZ, Kohlschmidt J, Pettenati MJ, Heerema NA, Block AW, Patil SR, Baer MR, Kolitz JE, Moore JO, Carroll AJ, Stone RM, Larson RA, Bloomfield CD. Prognostic significance of the European leukemia net standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30(36):4515–4523. doi: 10.1200/JCO.2012.43.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcucci G, Haferlach T, Döhner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol. 2011;29(5):475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 5.Patel JP, Levine RL. How do novel molecular genetic markers influence treatment decisions in acute myeloid leukemia? Hematol Am Soc Hematol Educ Program. 2012;2012:28–34. doi: 10.1182/asheducation-2012.1.28. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94(6):776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trino S, Lamorte D, Caivano A, Laurenzana I, Tagliaferri D, Falco G, Del Vecchio L, Musto P, De Luca L. MicroRNAs as new biomarkers for diagnosis and prognosis, and as potential therapeutic targets in acute myeloid leukemia. Int J Mol Sci. 2018;19(2):460. doi: 10.3390/ijms19020460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma QL, Wang JH, Yang M, Wang HP, Jin J. MiR-362-5p as a novel prognostic predictor of cytogenetically normal acute myeloid leukemia. J Transl Med. 2018;16(1):68. doi: 10.1186/s12967-018-1445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan J, Wu G, Chen J, Xiong L, Chen G, Li P. Downregulated miR-217 expression predicts a poor outcome in acute myeloid leukemia. Cancer Biomark. 2018 doi: 10.3233/CBM-170936. [DOI] [PubMed] [Google Scholar]
- 11.Zhang TJ, Wu DH, Zhou JD, Li XX, Zhang W, Guo H, Ma JC, Deng ZQ, Lin J, Qian J. Overexpression of miR-216b: prognostic and predictive value in acute myeloid leukemia. J Cell Physiol. 2018;233(4):3274–3281. doi: 10.1002/jcp.26171. [DOI] [PubMed] [Google Scholar]
- 12.Bhayadia R, Krowiorz K, Haetscher N, Jammal R, Emmrich S, Obulkasim A, Fiedler J, Schwarzer A, Rouhi A, Heuser M, Wingert S, Bothur S, Döhner K, Mätzig T, Ng M, Reinhardt D, Döhner H, Zwaan CM, van den Heuvel Eibrink M, Heckl D, Fornerod M, Thum T, Humphries RK, Rieger MA, Kuchenbauer F, Klusmann JH. Endogenous tumor suppressor microRNA-193b: therapeutic and prognostic value in acute myeloid leukemia. J Clin Oncol. 2018;36(10):1007–1016. doi: 10.1200/JCO.2017.75.2204. [DOI] [PubMed] [Google Scholar]
- 13.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial–mesenchymal transition and cancer metastasis. RNA Biol. 2008;5(3):115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng X, Wang Z, Fillmore R, Xi Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014;344(2):166–173. doi: 10.1016/j.canlet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JS, Ahn YH, Won HS, Sun S, Kim YH, Ko YH. Prognostic Role of the MicroRNA-200 family in various carcinomas: a systematic review and meta-analysis. Biomed Res Int. 2017;2017:1928021. doi: 10.1155/2017/1928021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposed revised criteria for the classification of acute myeloid leukaemia. A report of the French–American–British Cooperative Group. Ann Intern Med. 1985;103(4):620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 17.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 18.Zhou JD, Zhang TJ, Li XX, Ma JC, Guo H, Wen XM, Zhang W, Yang L, Yan Y, Lin J, Qian J. Epigenetic dysregulation of ID4 predicts disease progression and treatment outcome in myeloid malignancies. J Cell Mol Med. 2017;21(8):1468–1481. doi: 10.1111/jcmm.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK, National Cancer Research Institute Adult Leukaemia Working Group Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 20.Cheng H, Huang C, Xu X, Hu X, Gong S, Tang G, Song X, Zhang W, Wang J, Chen L, Yang J. PIM-1 mRNA expression is a potential prognostic biomarker in acute myeloid leukemia. J Transl Med. 2017;15(1):179. doi: 10.1186/s12967-017-1287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS ONE. 2011;6(10):e26906. doi: 10.1371/journal.pone.0026906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 2012;91(4):519–525. doi: 10.1007/s00277-011-1352-7. [DOI] [PubMed] [Google Scholar]
- 23.Yang X, Qian J, Sun A, Lin J, Xiao G, Yin J, Xiao G, Yin J, Chen S, Wu D. RAS mutation analysis in a large cohort of Chinese patients with acute myeloid leukemia. Clin Biochem. 2013;46(7–8):579–583. doi: 10.1016/j.clinbiochem.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Qian J, Yao DM, Lin J, Qian W, Wang CZ, Chai HY, Yang J, Li Y, Deng ZQ, Ma JC, Chen XX. U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS ONE. 2012;7(9):e45760. doi: 10.1371/journal.pone.0045760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin J, Yang J, Wen XM, Yang L, Deng ZQ, Qian Z, Ma JC, Guo H, Zhang YY, Qian W, Qian J. Detection of SRSF2-P95 mutation by high-resolution melting curve analysis and its effect on prognosis in myelodysplastic syndrome. PLoS ONE. 2014;9(12):e115693. doi: 10.1371/journal.pone.0115693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen XM, Lin J, Yang J, Yao DM, Deng ZQ, Tang CY, Xiao GF, Yang L, Ma JC, Hu JB, Qian W, Qian J. Double CEBPA mutations are prognostically favorable in non-M3 acute myeloid leukemia patients with wild-type NPM1 and FLT3-ITD. Int J Clin Exp Pathol. 2014;7(10):6832–6840. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang TJ, Zhou JD, Zhang W, Lin J, Ma JC, Wen XM, Yuan Q, Li XX, Xu ZJ, Qian J. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47. doi: 10.1186/s13148-018-0486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang TJ, Lin J, Zhou JD, Li XX, Zhang W, Guo H, Xu ZJ, Yan Y, Ma JC, Qian J. High bone marrow miR-19b level predicts poor prognosis and disease recurrence in de novo acute myeloid leukemia. Gene. 2018;640:79–85. doi: 10.1016/j.gene.2017.10.034. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Mar BG, Zhang H, Puram RV, Vazquez F, Weir BA, Hahn WC, Ebert B, Pellman D. The EMT regulator ZEB2 is a novel dependency of human and murine acute myeloid leukemia. Blood. 2017;129(4):497–508. doi: 10.1182/blood-2016-05-714493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6(9):6472–6498. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi Y, Hur EH, Moon JH, Goo BK, Choi DR, Lee JH. Expression and prognostic significance of microRNAs in Korean patients with myelodysplastic syndrome. Korean J Intern Med. 2017 doi: 10.3904/kjim.2016.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.González-Gugel E, Villa-Morales M, Santos J, Bueno MJ, Malumbres M, Rodríguez-Pinilla SM, Piris MÁ, Fernández-Piqueras J. Down-regulation of specific miRNAs enhances the expression of the gene Smoothened and contributes to T-cell lymphoblastic lymphoma development. Carcinogenesis. 2013;34(4):902–908. doi: 10.1093/carcin/bgs404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The primer sequences for miR-200s clusters.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.


