ABSTRACT
In S. mutans, the expression of the surface glycoprotein Cnm mediates binding to extracellular matrix proteins, endothelial cell invasion and virulence in the Galleria mellonella invertebrate model. To further characterize Cnm as a virulence factor, the cnm gene from S. mutans strain OMZ175 was expressed in the non-pathogenic Lactococcus lactis NZ9800 using a nisin-inducible system. Despite the absence of the machinery necessary for Cnm glycosylation, Western blot and immunofluorescence microscopy analyses demonstrated that Cnm was effectively expressed and translocated to the cell wall of L. lactis. Similar to S. mutans, expression of Cnm in L. lactis enabled robust binding to collagen and laminin, invasion of human coronary artery endothelial cells and increased virulence in G. mellonella. Using an ex vivo human heart tissue colonization model, we showed that Cnm-positive strains of either S. mutans or L. lactis outcompete their Cnm-negative counterparts for tissue colonization. Finally, Cnm expression facilitated L. lactis adhesion and colonization in a rabbit model of infective endocarditis. Collectively, our results provide unequivocal evidence that binding to extracellular matrices mediated by Cnm is an important virulence attribute of S. mutans and confirm the usefulness of the L. lactis heterologous system for further characterization of bacterial virulence factors.
KEYWORDS: bacterial pathogenesis, collagen-binding protein, infective endocarditis, Lactococcus lactis, Streptococcus mutans
Introduction
The association of Streptococcus mutans with dental caries, the most prevalent and costly infectious disease worldwide, is well established.1,2 The capacity of S. mutans to form biofilms in the presence of sucrose, to produce organic acids upon fermentation of dietary carbohydrates, and to tolerate large fluctuations in pH are considered the major cariogenic traits of this bacterium.3,4 However, the medical implications of S. mutans are not limited to the oral cavity as this organism can cause extra-oral infections and may play an important and underestimated role in the onset of cardiovascular diseases.5-9 This facet of S. mutans pathogenesis is likely associated with its ability to interact with extracellular matrix (ECM) components such as collagen, which enables colonization of different host tissues.10,11
Infective endocarditis (IE) is a life-threatening bacterial infection of the endocardium with approximately 45,000 occurrences per year in the US12 and mortality rates around 15–20%.9,13,14 Along with staphylococci and enterococci, viridans streptococci (VS) are a major bacterial group associated with IE,12 and S. mutans is estimated to account for ∼20% of all VS-associated IE cases.9,15 In addition, S. mutans DNA has been detected at a high frequency in cardiovascular tissues from patients that underwent valve replacement surgery and atherosclerotic plaque removal.16,17 Yet, the mechanisms employed by S. mutans to colonize and persist in human heart tissues are still poorly understood.
The S. mutans surface protein Cnm is a collagen- and laminin- binding glycoprotein shown to mediate adhesion to collagen and laminin, invasion of endothelial cells, and virulence in an invertebrate model.18-20 Epidemiological studies have shown that cnm is present in approximately 15% of S. mutans isolates, being found at higher frequency among serotype e, f and k strains but rarely present in serotype c strains.18,21-23 Interestingly, isolates of S. mutans that belong to serotypes e, f and k are rare in the oral cavity but commonly detected in patients with IE, atherosclerosis and other systemic infections.1,22 Moreover, in vivo and cross-sectional studies suggest an association between S. mutans serotypes e, f and k expressing Cnm and extra-oral pathologies such as hemorrhagic stroke, cerebral microbleeds and non-alcoholic steatohepatitis,5,6,24 indicating that Cnm may contribute to the pathogenic potential of S. mutans during systemic infections.
The Gram-positive bacterium Lactococcus lactis is a non-pathogenic organism used in the dairy industry for the production of fermented milk products.25 In addition, L. lactis is used for the production of heterologous proteins,26 and in live mucosal vaccines.27 The presence of secretory and protein anchoring enzymes (e.g. SecA and SrtA) combined with the low number of genes encoding secreted and surface-anchored proteins makes L. lactis a desirable host for the characterization of surface virulence factors of pathogenic Gram-positive bacteria.26,28-30 For example, systemic or mucosal delivery of a recombinant L. lactis strain expressing the Streptococcus agalactiae pilus 1 genes was shown to protect mice from subsequent challenges with S. agalactiae.31 Other bacterial proteins successfully expressed in L. lactis include Staphylococcus aureus clumping factor A (ClfA) and fibronectin-binding protein A (FnBPA),30 Staphylococcus epidermidis SdrF,32 and Listeria monocytogenes LnlA.33 In each of these studies, heterologous expression of these foreign factors was sufficient to increase the pathogenic potential of this otherwise harmless microorganism. Here, the contribution of Cnm to bacterial virulence was further investigated using the L. lactis heterologous expression system. Our results provide unequivocal evidence that Cnm is a major virulence factor of S. mutans that contributes to systemic infections.
Results
Optimization of Cnm expression in L. lactis
After successful electroporation of pMSP3535::cnm into L. lactis NZ9800, the concentration of nisin that conferred optimal expression of Cnm without causing growth inhibitory effects was determined. The growth of L. lactis was only noticeably affected at 25 ng ml−1nisin or higher concentrations (data not shown). Western blot analysis showed a concentration-dependent induction of Cnm from pMSP3535::cnm by nisin with strong induction observed at 10 ng ml−1 and above (Fig. 1A). Non-induced cultures of L. lactis pMSP3535::cnm showed basal expression of Cnm, which is in line with previous reports demonstrating that the pMSP3535 system is leaky.34,39 Immunofluorescence labeling of intact cells confirmed expression and surface localization of Cnm (Fig. 1B). Based on these results, we chose 10 ng ml−1nisin for all subsequent experiments as it yielded robust protein production without interfering with cell growth.
Figure 1.
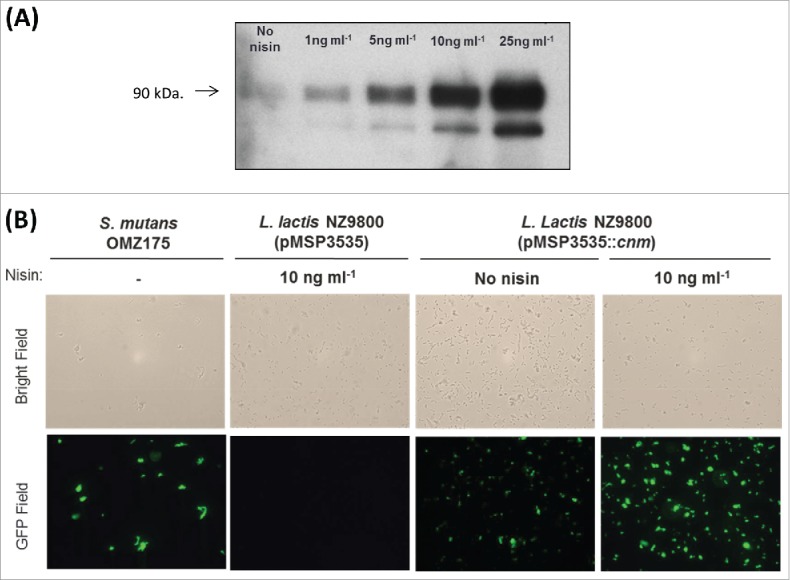
Expression of Cnm from pMSP3535::cnm in recombinant L. lactis. (A) Western blot analysis of protein lysates was performed with a polyclonal antibody specific to Cnm. A nisin concentration-dependent induction of Cnm was observed from pMSP3535::cnm, and the uninduced culture showed basal protein expression. (B) Immunofluorescence labeling analysis revealing Cnm expression on the surface of L. lactis.
As previously determined in S. mutans, L. lactis Cnm migrates as a higher molecular weight protein on SDS-PAGE (∼90 kDa) compared to its predicted size of 54 kDa (Fig. 2). This version of Cnm corresponds to the unglycosylated variant, which in S. mutans displays increased susceptibility to proteinase K degradation.19 Similar to unglycosylated Cnm from S. mutans, Cnm expressed by L. lactis was rapidly and completely degraded upon proteinase K treatment (Fig. 2). Nonetheless, unglycosylated Cnm in S. mutans retains its functional capacities,19 thus we proceeded with the characterization of Cnm-mediated phenotypes in L. lactis.
Figure 2.
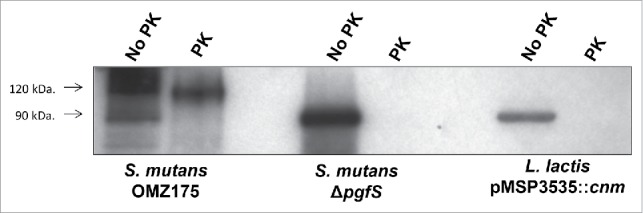
L. lactis expresses unglycosylated, proteinase K susceptible Cnm. Whole cells from S. mutans OMZ175, ΔpgfS and nisin-induced L. lactis pMSP3535::cnm were treated with 30 µg ml−1 of proteinase K for 30 min. After treatment, whole-cell lysates were prepared and Cnm degradation was monitored by protein gel blot using anti-rCnmA.
Cnm+ L. lactis binds to ECM and invades HCAEC
The ability of the recombinant L. lactis Cnm+ strain to bind to collagen and laminin in vitro was tested. When compared to the parent strain NZ9800 hosting the empty vector, expression of Cnm from pMSP3535::cnm led to robust binding to both collagen and laminin (Fig. 3A and B, respectively). In accordance with our expression analysis (Fig. 1), uninduced pMSP3535::cnm produced low levels of Cnm and therefore enabled the recombinant L. lactis strain to bind to both ECM proteins, albeit at lower levels than nisin-induced cells (P < 0.05). To further demonstrate that the ability to bind to collagen is mediated by Cnm, we performed the collagen-binding assay in the presence of anti-rCnmA antibody. As expected, co-incubation of L. lactis cells expressing Cnm with anti-rCnmA antibodies led to significant inhibition of collagen binding activity compared to L. lactis incubated with pre-immune serum (Fig. 3C). In agreement with increased collagen and laminin binding activities observed for nisin-induced cells, expression of Cnm enabled L. lactis to invade HCAEC (Fig. 3D). While ECM binding was observed in uninduced cultures, HCAEC invasion levels were negligible in the absence of nisin (P > 0.05).
Figure 3.
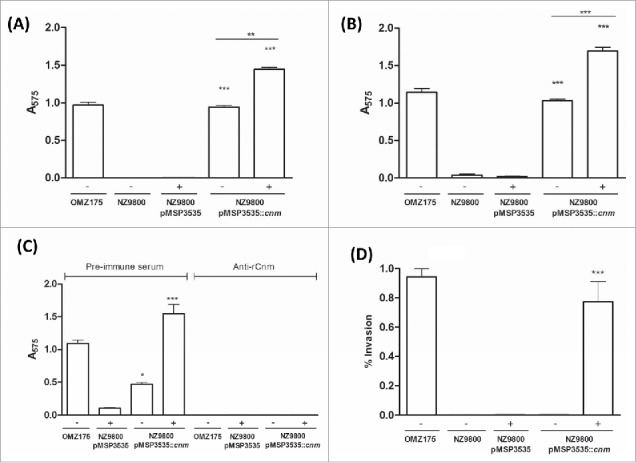
Cnm promotes binding to ECM proteins and invasion of endothelial cells. (A and B) Binding of bacterial cells to extracellular matrix proteins: (A) collagen type I and (B) laminin. (C) Inhibition of binding to collagen type I by anti-rCnmA antibody (1:20). (D) HCAEC invasion assay showing the normalized percentage of invasion for each strain after 3 h of co-incubation (*P < 0.05, **P < 0.01, ***P < 0.0001, ANOVA followed by Tukey's post-test). The S. mutans OMZ175 (Cnm+) strain was used as a positive control in all assays. The symbols “+” and “−“ in the graphs denote induction or no induction with nisin, respectively.
Expression of Cnm mediates adherence to aortic valve tissues ex vivo
To determine if Cnm bestows an enhanced heart tissue binding capacity to either S. mutans or L. lactis ex vivo, competition binding assays were performed using freshly extirpated human aortic valve tissues. In agreement with the ECM-binding in vitro data, the S. mutans OMZ175 and nisin-induced Cnm+ L. lactis strains outcompeted their Cnm− counterparts by approximately 10 and 100-fold, respectively (Fig. 4). In these experiments, the recovered CFUs were on average an order of magnitude higher for S. mutans and L. lactis Cnm+ strains (1.7 × 105 and 3.9 × 105 CFU ml−1, respectively) compared to Cnm− strains (1.7 × 104 and 2.7 × 104 CFU ml−1, respectively). Scanning electron microscopy (SEM) of tissues infected for 90 min with monocultures of L. lactis (Fig. 5) or S. mutans (data not shown) confirmed that only Cnm+ strains could effectively bind to the collagenous fibrils of damaged valve tissues (Fig. 5A). Moreover, while we cannot exclude the possibility that ECM components from heart valves became exposed during sample manipulation, SEM analysis revealed that Cnm+ strains could also adhere to tissue areas where no apparent damage can be observed (Fig. 5B).
Figure 4.
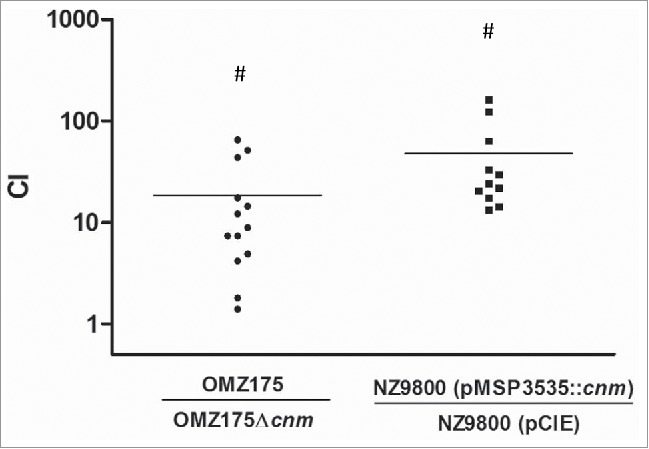
Expression of Cnm mediates bacterial adherence to aortic valve sections. A competition assay was developed to test the contribution of Cnm toward bacterial adherence to human aortic valves ex vivo. A 1:1 mixed infection of the valve sections was allowed for 90 min using Cnm+ and Cnm− strains. The competitive index (CI) was calculated based on the initial inocula, where a value of “=1” represents no difference and “>1” represents preferred binding by the numerator strain.
#P < 0.05
Figure 5.
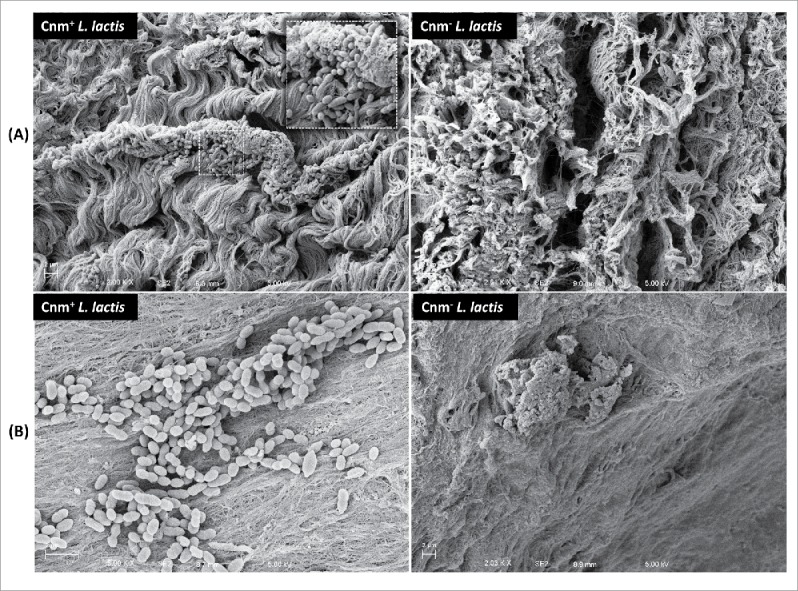
Cnm mediates binding to the collagenous fibrils of damaged and apparently undamaged infected human valve sections. SEM analysis of valve sections incubated with bacterial monocultures showed Cnm+ L. lactis binding to bundles of collagenous fibrils on the rough edges of the valves whereas no Cnm− L. lactis were found attached to this area (A). Cnm+ L. lactis could also adhere to tissue areas where no apparent damage was observed in contrast with their Cnm− counterparts (B).
Cnm contributes to virulence in galleria mellonella
The ability of the L. lactis strains to kill the larvae of G. mellonella, a model of systemic infection, was assessed. As shown in Figure 6, the mortality rates of G. mellonella were significantly higher in larvae infected with the Cnm+ strain compared to those infected with the wild-type strain harboring the empty vector control (P < 0.05). Remarkably when considering the extremely low virulence potential of L. lactis, the recombinant Cnm+ strain showed a similar pattern of killing when compared to the Cnm+ S. mutans OMZ175 strain.
Figure 6.
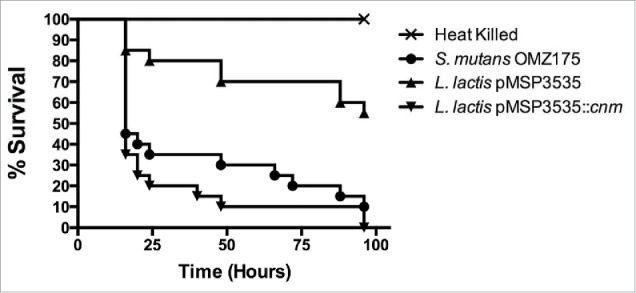
Cnm contributes to virulence in the G. mellonella systemic infection model. The Kaplan-Meier curves indicate the percent survival at selected intervals of larvae infected with S. mutans OMZ175 and L. lactis strains for 96 h. The mortality rates of G. mellonella were significantly higher in larvae infected with Cnm+ L. lactis compared to those infected with the wild-type NZ9800 harboring the empty vector control (pMSP3535) (P < 0.05).
Cnm contributes to endocardium adherence in vivo
A rabbit model of IE38 was employed to examine the effect of Cnm expression on L. lactis adherence to endocardium and vegetations. In one experiment, a mixed inoculum of NZ9800 (pCIE) and NZ9800 (pMSP3535::cnm) was injected into rabbits intravenously 1 h after surgery. Two hours later, the rabbits were euthanized and the hearts removed. This time was chosen so as to allow enough time for clearance of bacteria from the bloodstream,40 but not so long as to have excessive loss of Cnm due to growth without nisin. Catheters were clamped into place during necropsy to preserve their positions. At this early stage, vegetations were not visible, and the entire endocardial surface in apparent contact with the catheter was excised and homogenized. Following sonication, both the mixed inoculum and the homogenates were plated on selective media to determine strain ratios. In the second experiment, animals were inoculated 2 d after surgery. Vegetations, which were visible in all animals, were removed and homogenized. Because vegetations are composed primarily of platelets and fibrin rather than collagen,41 the endocardium beneath each vegetation was also collected, homogenized, and plated separately. As shown in Figure 7, the CI value of the cells harvested from the endocardium were similar in animals inoculated 1 hour or 2 d after catheterization (geometric mean = 1.68 and 1.66, respectively), indicating a ∼67% increase in infectivity of the Cnm-expressing strain relative to the control strain. These values were significantly greater from 1.0 — the value indicating no change in infectivity — whether the 2 experiments were analyzed separately or together. In contrast, there was no significant difference in the CI value for bacteria recovered from vegetations.
Figure 7.
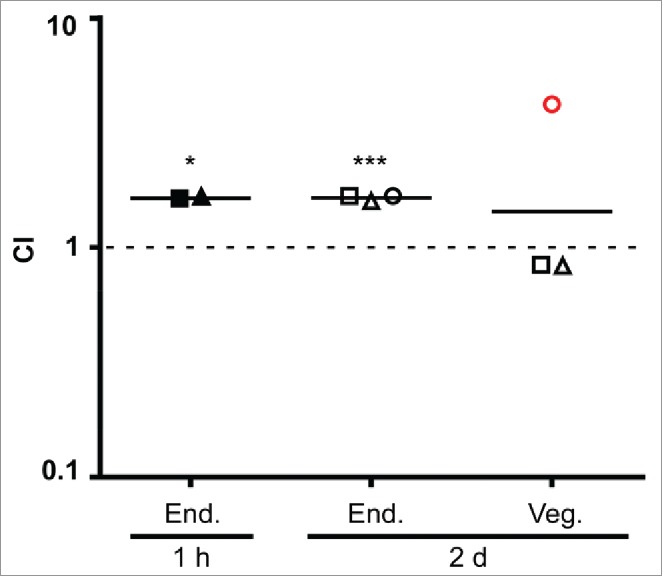
Cnm contributes to bacterial endocardium adherence in vivo. Rabbits (n=3) were co-inoculated with Cnm-expressing and control L. lactis strains 1 hour or 2 days after insertion of an intracardiac catheter as indicated and then sacrificed 2 hours later. Bacteria were recovered from endocardial surfaces (End.), and in the 2-day samples, from adjacent vegetations (Veg.). CI values (for L. lactis Cnm+ over L. lactis Cnm−) are indicated for each sample, with geometrical mean CIs indicated by a horizontal line. Means significantly different from 1.0 (dashed line) are indicated: *P < 0.05; ***P < 0.001. Like symbols in the 2-day experiment indicate samples recovered from the same animal. Red circle, sample from which one of the 2 strains was not recovered (L. lactis Cnm−); the limit of detection was used for the calculation.
Discussion
The presence of Cnm has been correlated with an increased ability of S. mutans to invade and persist intracellularly, and to cause disease.6,18,20,21,42,43 In the present study, by expressing Cnm in the non-pathogenic L. lactis host we provided unequivocal evidence that Cnm contributes to bacterial virulence.
Various properties of L. lactis have justified its attractiveness for heterologous expression of surface proteins. For instance, the fermentative metabolism of L. lactis is relatively simple and well known, the genomes of various strains are available and known to encode a very low number of genes encoding secreted and surface-anchored proteins.25,44,45 Here, we showed that Cnm expression increased the ability of L. lactis to bind to type I collagen and laminin in vitro, adhere to aortic valve tissues ex vivo, and colonize endocardial surfaces in vivo. Of note, type I collagen is the most abundant ECM component in human46,47 and rabbit48 aortic valves. Although ex vivo experiments cannot reproduce the immunologic responses and constant shear force of blood flow, our findings highlighted the contribution of Cnm in binding of organisms to collagen-rich tissues in both humans and rabbits. Also, while we observed higher levels of ECM binding and virulence G. mellonella by Cnm+L. lactis compared to S. mutans OMZ175, during this study we restrained ourselves from making direct comparisons as these species possess fundamental differences that may affect Cnm-dependent phenotypes (e.g., Cnm expression levels, additional surface proteins, growth conditions).
Recently, our group reported that Cnm is a glycoprotein and identified the glycosylation machinery (Pgf) responsible for its post-translational modification. In SDS-PAGE, the native Cnm migrates at approximately 120 kDa whereas the variant from the deletion of the pgfS gene was found to migrate at ∼90 kDa.19 When analyzed by Western blot, the Cnm variant produced by L. lactis was found to migrate at ∼90 kDa, the same size as the unglycosylated version of Cnm produced in S. mutans.19 Moreover, like the unglycosylated Cnm produced by the ΔpgfS strain in S. mutans, the Cnm expressed by L. lactis was highly susceptible to proteinase K degradation.19 These findings are supported by in silico analysis that indicated the absence of Pgf homologs in the available genomes of L. lactis. Most importantly, the lack of Cnm glycosylation may explain the difference in adhesion between endocardial tissue in vivo and ECM-binding in vitro or ex vivo. Whether this difference is due to the considerable shear forces and host proteases present in the in vivo model, or to some other factor(s) remains to be determined. However, we hypothesize that L. lactis expressing glycosylated Cnm will show further increased levels of binding in vitro, ex vivo and, possibly, in vivo. Expression of Cnm had no effect on colonization of preexisting vegetations, which is not surprising considering these vegetations are composed primarily of platelets and fibrin.41 Nevertheless, adhesins for collagen and other matrix proteins have been identified previously as virulence factors for IE.29,49,50 As with these other adhesins, Cnm may allow adherence to damaged endocardium prior to vegetation formation, thereby providing another route for infection in IE.51 The increased adherence to endocardium observed is also consistent with previous reports of association of Cbm-expressing strains, with both IE and non-IE cardiovascular diseases.7,52
Collectively, our results clearly show that expression of Cnm alone confers virulence to an otherwise non-pathogenic bacterium, and promotes bacterial adherence to cardiac tissue in vivo and ex vivo. Thus, Cnm could be considered a target for the development of antimicrobial strategies to mitigate virulence of invasive S. mutans strains.
Material and methods
Bacterial strains and growth conditions
All strains used in this study are listed in Table 1. The L. lactis strains were routinely cultured aerobically at 30°C in M17 broth (Difco) supplemented with 0.5% (w/v) glucose (GM17). When necessary, 10 µg ml−1 erythromycin or 15 µg ml−1 chloramphenicol (Sigma-Aldrich) was added to the growth media (GM17-Erm or GM17-Chlor, respectively). The S. mutans strains were routinely cultured in brain heart infusion (BHI) medium at 37°C in a 5% CO2 atmosphere.
Table 1.
Strains used in this study.
| Strain | Relevant phenotype | Reference |
|---|---|---|
| Streptococcus mutans | ||
| OMZ175 | Cnm+ | Lab collection |
| OMZ175 (pBGE) | Cnm+Ermr | (21) |
| Δcnm ΔpgfS | Cnm− KanrCnm+ Kanr | (21) (19) |
| Lactococcus lactis subsp. lactis | ||
| NZ9800 | WT, Cnm− | Lab collection |
| NZ9800 (pMSP3535) | Cnm−Ermr | This study |
| NZ9800 (pMSP3535::cnm) | Cnm+Ermr | This study |
| NZ9800 (pCIE) | Cnm−, Chlorr | This study |
Note. WT (wild type); Kan (kanamycin); Erm (erythromycin); Chlor (chloramphenicol).
Expression of Cnm in L. lactis
The nisin-inducible plasmid pMSP353534 harboring the cnm gene (pMSP3535::cnm) from S. mutans OMZ175,21 was used to express Cnm in L. lactis. The pMSP3535::cnm plasmid as well as the empty pMSP3535 vector 34 and the pCIE plasmids (Dunny et al., unpublished) were electroporated into L. lactis NZ9800 as detailed elsewhere 35 with some modifications. The pCIE plasmid, conferring resistance to chloramphenicol, was generated from shuttle plasmid pCI13340 36 and introduced into L. lactis for selection purposes in competition assays. Briefly, cells were grown overnight at 30°C in M17 broth supplemented with 0.5% glucose (GM-17) and sub-cultured (1:100) in fresh GM-17 containing 1% glycine and 0.5 M sucrose (GM17S). The culture was grown to OD600 of 0.2, centrifuged at 5,000 rpm for 10 min at 4°C and washed 3 times with cold electroporation buffer (EB) (0.5 M sucrose, 1 mM MgCl2, 7 mM K2HPO4-KH2PO4 [pH 7.4]). Electrocompetent cells were resuspended in EB, quickly frozen on dry ice/ethanol bath and stored at −80°C until use. A micro-pulser electroporation apparatus (Bio-Rad Laboratories) (0.1cm cuvette, 1.4 kV, 600 Ω, 10 µF) was used to electroporate the desired plasmids into L. lactis. After electroporation, cells were incubated in GM17S for 3 h at 30°C and spread on GM17 plates containing the desired antibiotic. Resulting colonies were screened for the presence of the desired plasmid by PCR. For induction of Cnm, cells were grown overnight in GM17-Erm in the presence of increasing concentrations of nisin (Sigma-Aldrich). The production and cell surface localization of Cnm was confirmed by Western blotting and by immunofluorescence labeling as detailed below.
Western blot (WB) analysis
Overnight cultures of S. mutans OMZ175 and recombinant L. lactis strains were used to prepare protein lysates by homogenization with 0.1 mm glass beads using a bead beater (Biospec). Protein lysates were separated on a 10% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore) using standard protocols. The presence of Cnm was detected using rabbit anti-rCnmA polyclonal antibody 19 diluted 1:2000 in 1 x phosphate buffered saline (PBS) with 0.1% Tween 20 pH 7.2 and anti-IgG (goat anti-rabbit) horseradish peroxidase-coupled antibody diluted 1:10000 (Sigma-Aldrich). Membranes were developed using an enhanced chemiluminescent detection kit (GE Life Sciences).
Proteinase K susceptibility assay
The susceptibility of Cnm to protease degradation was assessed as described previously.19 Briefly, overnight cultures of S. mutans OMZ175, ΔpgfS and nisin-induced L. lactis pMSP3535::cnm were pelleted and resuspended in 1x PBS pH 7.4 containing 30 µg ml−1 of proteinase K (Sigma-Aldrich). After 30 min incubation on ice, protease activity was neutralized with the addition of 1x protease inhibitor cocktail (Thermo Scientific). Bacterial cells were then washed once with PBS, and Cnm stability was analyzed by western blotting.
Immunofluorescence labeling of Cnm
Cultures of recombinant L. lactis and S. mutans OMZ175 were used for cellular localization of Cnm. Cells grown to mid-exponential phase were washed in PBS and incubated in the presence of anti-rCnmA antibody (1:100) followed by anti-rabbit Alexa-488 conjugate antibody (1:100) with 3 washes between each antibody exposure. Cell-antibody complexes were visualized using an Olympus BX41 fluorescent microscope and images were captured with the QCapture Software.
Binding to extracellular matrix (ECM) proteins
Collagen and laminin binding assays were performed as previously described.20,21 Briefly, 96-well plates were coated with 40 µg ml−1 type I collagen from rat tail (Sigma-Aldrich), or 50 µg ml−1 mouse laminin (Becton-Dickinson) for 18 h at 4°C. Individual wells were washed and blocked with 5% bovine serum albumin for 1 h at 37°C. Overnight bacterial cultures were washed twice in PBS and 100 μl aliquots of each bacterial suspension added to the wells, followed by incubation for 3 h at 37°C. Loosely bound bacteria were removed by washing with PBS and adhered cells were stained with 0.05% crystal violet solution and then quantified at 575 nm. Experiments were also performed with bacteria pre-incubated with anti-rCnmA antibody (1:20 dilution in PBS) for 30 min before incubation with collagen. To confirm the requirement of Cnm in collagen binding, experiments were performed as described above but bacteria were pre-incubated with anti-rCnmA or pre-immune serum (1:100) for 30 min before exposure to collagen.19
Invasion of human coronary artery endothelial cells (HCAEC)
Cultivation of primary HCAECs and invasion assays were performed as previously described.18,21 The HCAEC monolayer was incubated with 1 ml of 2% fetal bovine serum (FBS)-endothelial basal medium (EBM-2) containing 1-2 × 107 CFU ml−1 of S. mutans OMZ175 or Δcnm, or recombinant L. lactis strains for 2 h at a multiplicity of infection of 100:1, followed by 3 h of incubation in 2% FBS-EBM-2 medium containing 300 μg ml−1gentamicin and 50 μg ml−1 penicillin G to kill extracellular bacteria. After incubation with antibiotics, HCAEC were lysed with 1 ml of sterile water and the mixture of lysed HCAEC and bacteria serially diluted and plated onto tryptic soy agar (TSA) plates to determine the number of intracellular bacteria. The percentage of invasion for each strain was calculated based on the total colony forming units (CFU) number present in the initial inoculum and the number of intracellular bacteria recovered from HCAEC lysates.
Adherence to human cardiac valve sections ex vivo
The ability of S. mutans and L. lactis strains to bind to human aortic valve tissues ex vivo was assessed by adapting a protocol originally described for porcine heart tissues.37 Human aortic valve tissues were obtained from patients undergoing valve replacement surgery due to calcific stenosis (Institutional Review Board protocol no. RSRB00053016, University of Rochester). Valvular tissues with no signs of calcification, bleeding or inflammation were selected and 3-mm diameter valve sections were obtained using a biopsy punch tool (Acuderm). Valve sections were incubated overnight in 12-well plates with endothelial cell medium (ECM) supplemented with 5% FBS, 1000 mg L−1 hydrocortisone, 12 mg L−1 bovine brain extract, 0.1% human recombinant epidermal growth factor (Lonza) plus 300 µg ml−1 gentamicin. On the following day, sections were washed twice with Hank's Balanced Salt Solution (HBSS, Corning Inc.) to remove residual antibiotic and then transferred to a new plate. A competition assay was then performed to determine the capacity of Cnm+ strains to compete with Cnm− strains. For this, valve sections were infected with approximately 5 × 107 CFU ml−1 of a 1:1 suspension of Cnm+ and Cnm−strains bearing distinct antibiotic markers for CFU determination of each strain. Plates were incubated for 90 min at 37°C (for S. mutans) or 30°C (for L. lactis) with gentle rocking. Valve sections were transferred to sterile centrifuge tubes containing 1 ml of HBSS and vigorously agitated 3 times for 5 seconds using a vortex to remove loosely bound bacteria from the tissues. Washed valve sections were homogenized in 500 µl PBS for 2 min using a motorized pestle (Fisher Scientific), serially diluted and plated onto TSA plates with the corresponding antibiotics. The competitive index (CI) was calculated based on the CFUs present in the initial inoculum and the final enumeration of bacteria recovered from the tissue.
Scanning electron microscopy (SEM) analysis of valve sections
To visualize bacterial attachment to heart valve sections, tissue samples were prepared and incubated in the presence of 5 × 107 CFU ml−1 S. mutans or L. lactis monocultures as described in the previous section. Valve samples were washed and fixed with 4% paraformaldehyde, 2.5% glutaraldehyde and 0.1 M sodium cacodylate for 2 d at 4°C, with slow rocking. The samples were analyzed using a Zeiss-Auriga focused ion beam field emission scanning electron microscope (FIB-FE-SEM) and the images captured using the attached Gatan Erlangshen digital camera at the University of Rochester Medical Center Electron Microscopy Core Facility.
Galleria mellonella infection
The systemic infection of G. mellonella has been previously detailed elsewhere.19,21 Briefly, 5 µl aliquots containing 1-1.5x106 CFU of S. mutans OMZ175 (positive control), or recombinant L. lactis were injected in the larvae hemocoel via the last left proleg. Larvae injected with heat-killed bacteria (45 min, 80°C) were used as negative controls. After injection, larvae were kept in the dark at 37°C, and survival was periodically monitored for up to 4 d.
In vivo model of infective endocarditis in rabbits
The rabbit model of IE employed for this study was modified from a previously used protocol.38 Specific-pathogen-free, male New Zealand White Rabbits 2 to 3 kg in size were purchased from Robinson Services Inc. After acclimation, rabbits were injected with acepromazine as a sedative, a cocktail of ketamine, xylazine, and buprenorphine for general anesthesia, glycopyrrolate for control of respiratory secretions, and bupivacaine at the incision site. An incision was then made in the neck region, and a 19-gauge catheter (Intracath, BD Bioscience) was inserted through the right internal carotid artery, contacting or passing through the aortic valve to cause minor damage. The catheter was tied off, trimmed, and sutured into place, and the incision closed. Buprenorphine SR Lab was used for post-surgical analgesia. The day prior to inoculation, L. lactis strains were cultured overnight in GM17 and antibiotics as described above with 10 ng ml−1 nisin. Twenty hours later, the cultures were diluted 10-fold into pre-warmed media of the same composition and cultured another 3 h. Cells were harvested by centrifugation, washed twice with cold PBS, diluted in PBS, and combined to achieve a concentration of 3 – 5 × 108 CFU ml−1 for each strain. One hour or 2 d after the surgeries, rabbits were inoculated with 0.5 ml of bacterial cells via a peripheral ear vein. Remaining cells were sonicated for 1.5 min at 50% power in a Biologics Model 150 sonicator (Biologic Inc.) to break up clumps and chains and plated with an Eddy Jet spiral plater (Neutec Group Inc.) on GM17 plates containing erythromycin or chloramphenicol to establish the ratio of the 2 strains in the inoculum. Two hours after inoculation, each rabbit was euthanized with intravenous Euthasol (Virbac) and was then checked for proper placement of the catheter tip in the valve region or beneath it in the left ventricle. Any rabbit in which the catheter was not correctly situated was excluded from the study. For rabbits inoculated on the day of surgery, the aortic valves and surrounding endocardium were removed and homogenized in 5 ml PBS using an electric homogenizer (Omni International). For rabbits inoculated 2 d post-surgery, visible vegetations and the endocardium underlying each vegetation were harvested separately and homogenized in 2 ml PBS each using a disposable 15-ml tissue grinder. In all cases, homogenates were sonicated and plated as above for enumeration.
Statistical analysis
All results represent a minimum of 3 independent experiments. Descriptive and inferential statistics were used to analyze the data on Graphpad Prism version 5.0. One-way ANOVA followed by Tukey's post-test was used to analyze the significance of binding and invasion, with type I error set as 0.05. Analysis of log10-transformed competitive indices was performed using one-way ANOVA with Bonferroni post hoc comparisons to determine substrate preferences among strain pairs. For the G. mellonella studies, Kaplan-Meier killing curves were plotted and estimation of differences in survival was compared using the log-rank test. Log10-transformed competitive indices were compared to 0 using a one-sample T test for rabbit studies.
Abbreviations
- BHI
brain heart infusion
- CFU
colony forming unit
- CI
competition/competitive index
- EB
electroporation buffer
- EBM
endothelial basal medium
- ECM
endothelial cell medium
- ECM
extracellular matrix
- FBS
fetal bovine serum
- FIB-FE-SEM
focused ion beam field emission scanning electron microscope
- GM17
M17 broth supplemented with 0.5% glucose
- GM17-Erm or GM17-Chlor
M17 broth supplemented with 0.5% glucose plus erythromycin or chloramphenicol (Chlor)
- GM17S
GM-17 containing 1% glycine and 0.5 M sucrose
- HBSS
Hank's balanced salt solution
- HCAEC
human coronary artery endothelial cells
- IE
infective endocarditis
- PVDF
polyvinylidene fluoride
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SEM
scanning electron microscopy
- TSA
tryptic soy agar
- VS
viridans streptococci
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Dr. Gary Dunny (University of Minnesota) for kindly providing L. lactis NZ9800 and the pCIE plasmid.
Funding
This work was supported by the National Institute of Dental and Craniofacial Research R01 DE022559 to JL and JA, National Heart, Lung, and Blood Institute F31 HL124952 to AAR, São Paulo Research Foundation (FAPESP, Brazil, grants no. 2014/07231-0 and no. 2013/25080-7) to IAF, and the National Council for Scientific and Technological Development (CNPq, Brazil, grant no. 308644/2011-5) to PLR.
References
- [1].National Institutes of Health Diagnosis and management of dental caries throughout life. NIH Consens Statement 2001; 18:1-23; PMID:11699634 [PubMed] [Google Scholar]
- [2].Bagramian RA, Garcia-Godoy F, Volpe AR. The global increase in dental caries. A pending public health crisis. Am J Dent 2009; 22:3-8; PMID:19281105 [PubMed] [Google Scholar]
- [3].Bowen WH. The Stephan Curve revisited. Odontology 2013; 101:2-8; PMID:23224410; http://dx.doi.org/ 10.1007/s10266-012-0092-z [DOI] [PubMed] [Google Scholar]
- [4].Lemos JA, Burne RA. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 2008; 154:3247-55; PMID:18957579; http://dx.doi.org/ 10.1099/mic.0.2008/023770-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Miyatani F, Kuriyama N, Watanabe I, Nomura R, Nakano K, Matsui D, Ozaki E, Koyama T, Nishigaki M, Yamamoto T, et al.. Relationship between Cnm-positive Streptococcus mutans and cerebral microbleeds in humans. Oral Dis 2015; 21:886-93; PMID:26205098; http://dx.doi.org/ 10.1111/odi.12360 [DOI] [PubMed] [Google Scholar]
- [6].Nakano K, Hokamura K, Taniguchi N, Wada K, Kudo C, Nomura R, Kojima A, Naka S, Muranaka Y, Thura M, et al.. The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat Commun 2011; 2:485; PMID:21952219; http://dx.doi.org/ 10.1038/ncomms1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nomura R, Naka S, Nemoto H, Inagaki S, Taniguchi K, Ooshima T, Nakano K. Potential involvement of collagen-binding proteins of Streptococcus mutans in infective endocarditis. Oral Dis 2013; 19:387-93; PMID:22998492; http://dx.doi.org/ 10.1111/odi.12016 [DOI] [PubMed] [Google Scholar]
- [8].Nomura R, Nakano K, Nemoto H, Fujita K, Inagaki S, Takahashi T, Taniguchi K, Takeda M, Yoshioka H, Amano A, et al.. Isolation and characterization of Streptococcus mutans in heart valve and dental plaque specimens from a patient with infective endocarditis. J Med Microbiol 2006; 55:1135-40; PMID:16849735; http://dx.doi.org/ 10.1099/jmm.0.46609-0 [DOI] [PubMed] [Google Scholar]
- [9].Que YA, Moreillon P. Infective endocarditis. Nat Rev Cardiol 2011; 8:322-36; PMID:21487430; http://dx.doi.org/ 10.1038/nrcardio.2011.43 [DOI] [PubMed] [Google Scholar]
- [10].Nakano K, Nomura R, Nakagawa I, Hamada S, Ooshima T. Demonstration of Streptococcus mutans with a cell wall polysaccharide specific to a new serotype, k, in the human oral cavity. J Clin Microbiol 2004; 42:198-202; PMID:14715753; http://dx.doi.org/ 10.1128/JCM.42.1.198-202.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shun CT, Lu SY, Yeh CY, Chiang CP, Chia JS, Chen JY. Glucosyltransferases of viridans streptococci are modulins of interleukin-6 induction in infective endocarditis. Infect Immun 2005; 73:3261-70; PMID:15908350; http://dx.doi.org/ 10.1128/IAI.73.6.3261-3270.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pant S, Patel NJ, Deshmukh A, Golwala H, Patel N, Badheka A, Hirsch GA, Mehta JL. Trends in infective endocarditis incidence, microbiology, and valve replacement in the United States from 2000 to 2011. J Am Coll Cardiol 2015; 65:2070-6; PMID:25975469; http://dx.doi.org/ 10.1016/j.jacc.2015.03.518 [DOI] [PubMed] [Google Scholar]
- [13].Murdoch DR, Corey GR, Hoen B, Miro JM, Fowler VG Jr., Bayer AS, Karchmer AW, Olaison L, Pappas PA, Moreillon P, et al.. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: the International Collaboration on Endocarditis-Prospective Cohort Study. Arch Intern Med 2009; 169:463-73; PMID:19273776; http://dx.doi.org/ 10.1001/archinternmed.2008.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yew HS, Murdoch DR. Global trends in infective endocarditis epidemiology. Curr Infect Dis Rep 2012; 14:367-72; PMID:22592632; http://dx.doi.org/ 10.1007/s11908-012-0265-5 [DOI] [PubMed] [Google Scholar]
- [15].Banas JA. Virulence properties of Streptococcus mutans. Front Biosci 2004; 9:1267-77; PMID:14977543; http://dx.doi.org/ 10.2741/1305 [DOI] [PubMed] [Google Scholar]
- [16].Nakano K, Ooshima T. Serotype classification of Streptococcus mutans and its detection outside the oral cavity. Future Microbiol 2009; 4:891-902; PMID:19722842; http://dx.doi.org/ 10.2217/fmb.09.64 [DOI] [PubMed] [Google Scholar]
- [17].Oliveira FA, Forte CP, Silva PG, Lopes CB, Montenegro RC, Santos AK, Sobrinho CR, Mota MR, Sousa FB, Alves AP. Molecular analysis of oral bacteria in heart valve of patients with cardiovascular disease by real-time polymerase chain reaction. Medicine (Baltimore) 2015; 94:e2067; PMID:26632711; http://dx.doi.org/ 10.1097/MD.0000000000002067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Abranches J, Zeng L, Belanger M, Rodrigues PH, Simpson-Haidaris PJ, Akin D, Dunn WA Jr., Progulske-Fox A, Burne RA. Invasion of human coronary artery endothelial cells by Streptococcus mutans OMZ175. Oral Microbiol Immunol 2009; 24:141-5; PMID:19239641; http://dx.doi.org/ 10.1111/j.1399-302X.2008.00487.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Aviles-Reyes A, Miller JH, Simpson-Haidaris PJ, Hagen FK, Abranches J, Lemos JA. Modification of Streptococcus mutans Cnm by PgfS contributes to adhesion, endothelial cell invasion, and virulence. J Bacteriol 2014; 196:2789-97; PMID:24837294; http://dx.doi.org/ 10.1128/JB.01783-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Aviles-Reyes A, Miller JH, Simpson-Haidaris PJ, Lemos JA, Abranches J. Cnm is a major virulence factor of invasive Streptococcus mutans and part of a conserved three-gene locus. Mol Oral Microbiol 2014; 29:11-23; PMID:24103776; http://dx.doi.org/ 10.1111/omi.12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Abranches J, Miller JH, Martinez AR, Simpson-Haidaris PJ, Burne RA, Lemos JA. The collagen-binding protein Cnm is required for Streptococcus mutans adherence to and intracellular invasion of human coronary artery endothelial cells. Infect Immun 2011; 79:2277-84; PMID:21422186; http://dx.doi.org/ 10.1128/IAI.00767-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nakano K, Nomura R, Taniguchi N, Lapirattanakul J, Kojima A, Naka S, Senawongse P, Srisatjaluk R, Gronroos L, Alaluusua S, et al.. Molecular characterization of Streptococcus mutans strains containing the cnm gene encoding a collagen-binding adhesin. Arch Oral Biol 2010; 55:34-9; PMID:20005510; http://dx.doi.org/ 10.1016/j.archoralbio.2009.11.008 [DOI] [PubMed] [Google Scholar]
- [23].Nomura R, Nakano K, Taniguchi N, Lapirattanakul J, Nemoto H, Gronroos L, Alaluusua S, Ooshima T. Molecular and clinical analyses of the gene encoding the collagen-binding adhesin of Streptococcus mutans. J Med Microbiol 2009; 58:469-75; PMID:19273643; http://dx.doi.org/ 10.1099/jmm.0.007559-0 [DOI] [PubMed] [Google Scholar]
- [24].Naka S, Nomura R, Takashima Y, Okawa R, Ooshima T, Nakano K. A specific Streptococcus mutans strain aggravates non-alcoholic fatty liver disease. Oral Dis 2014; 20:700-6; PMID:25360469; http://dx.doi.org/ 10.1111/odi.12191 [DOI] [PubMed] [Google Scholar]
- [25].Mierau I, Kleerebezem M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol 2005; 68:705-17; PMID:16088349; http://dx.doi.org/ 10.1007/s00253-005-0107-6 [DOI] [PubMed] [Google Scholar]
- [26].Morello E, Bermudez-Humaran LG, Llull D, Sole V, Miraglio N, Langella P, Poquet I. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J Mol Microbiol Biotechnol 2008; 14:48-58; PMID:17957110; http://dx.doi.org/ 10.1159/000106082 [DOI] [PubMed] [Google Scholar]
- [27].Wyszynska A, Kobierecka P, Bardowski J, Jagusztyn-Krynicka EK. Lactic acid bacteria–20 years exploring their potential as live vectors for mucosal vaccination. Appl Microbiol Biotechnol 2015; 99:2967-77; PMID:25750046; http://dx.doi.org/ 10.1007/s00253-015-6498-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Danne C, Entenza JM, Mallet A, Briandet R, Debarbouille M, Nato F, Glaser P, Jouvion G, Moreillon P, Trieu-Cuot P, et al.. Molecular characterization of a Streptococcus gallolyticus genomic island encoding a pilus involved in endocarditis. J Infect Dis 2011; 204:1960-70; PMID:22043018; http://dx.doi.org/ 10.1093/infdis/jir666 [DOI] [PubMed] [Google Scholar]
- [29].Piroth L, Que YA, Widmer E, Panchaud A, Piu S, Entenza JM, Moreillon P. The fibrinogen- and fibronectin-binding domains of Staphylococcus aureus fibronectin-binding protein A synergistically promote endothelial invasion and experimental endocarditis. Infect Immun 2008; 76:3824-31; PMID:18541660; http://dx.doi.org/ 10.1128/IAI.00405-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Que YA, Haefliger JA, Piroth L, Francois P, Widmer E, Entenza JM, Sinha B, Herrmann M, Francioli P, Vaudaux P, et al.. Fibrinogen and fibronectin binding cooperate for valve infection and invasion in Staphylococcus aureus experimental endocarditis. J Exp Med 2005; 201:1627-35; PMID:15897276; http://dx.doi.org/ 10.1084/jem.20050125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Buccato S, Maione D, Rinaudo CD, Volpini G, Taddei AR, Rosini R, Telford JL, Grandi G, Margarit I. Use of Lactococcus lactis expressing pili from group B Streptococcus as a broad-coverage vaccine against streptococcal disease. J Infect Dis 2006; 194:331-40; PMID:16826481; http://dx.doi.org/ 10.1086/505433 [DOI] [PubMed] [Google Scholar]
- [32].Arrecubieta C, Toba FA, von Bayern M, Akashi H, Deng MC, Naka Y, Lowy FD. SdrF, a Staphylococcus epidermidis surface protein, contributes to the initiation of ventricular assist device driveline-related infections. PLoS Pathog 2009; 5:e1000411; PMID:19412528; http://dx.doi.org/ 10.1371/journal.ppat.1000411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Innocentin S, Guimaraes V, Miyoshi A, Azevedo V, Langella P, Chatel JM, Lefevre F. Lactococcus lactis expressing either Staphylococcus aureus fibronectin-binding protein A or Listeria monocytogenes internalin A can efficiently internalize and deliver DNA in human epithelial cells. Appl Environ Microbiol 2009; 75:4870-8; PMID:19482952; http://dx.doi.org/ 10.1128/AEM.00825-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bryan EM, Bae T, Kleerebezem M, Dunny GM. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 2000; 44:183-90; PMID:10964628; http://dx.doi.org/ 10.1006/plas.2000.1484 [DOI] [PubMed] [Google Scholar]
- [35].Holo H, Nes IF. Transformation of Lactococcus by electroporation. Methods Mol Biol 1995; 47:195-9; PMID:7550735 [DOI] [PubMed] [Google Scholar]
- [36].Hayes F, Daly C, Fitzgerald GF. Identification of the minimal replicon of lactococcus lactis subsp. lactis UC317 Plasmid pCI305. Appl Environ Microbiol 1990; 56:202-9; PMID:16348092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chuang-Smith ON, Wells CL, Henry-Stanley MJ, Dunny GM. Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PLoS One 2010; 5:e15798; PMID:21209892; http://dx.doi.org/ 10.1371/journal.pone.0015798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Crump KE, Bainbridge B, Brusko S, Turner LS, Ge X, Stone V, Xu P, Kitten T. The relationship of the lipoprotein SsaB, manganese and superoxide dismutase in Streptococcus sanguinis virulence for endocarditis. Mol Microbiol 2014; 92:1243-59; PMID:24750294; http://dx.doi.org/ 10.1111/mmi.12625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fu RY, Chen J, Li Y. Heterologous leaky production of transglutaminase in Lactococcus lactis significantly enhances the growth performance of the host. Appl Environ Microbiol 2005; 71:8911-9; PMID:16332889; http://dx.doi.org/ 10.1128/AEM.71.12.8911-8919.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Durack DT, Beeson PB. Experimental bacterial endocarditis. I. Colonization of a sterile vegetation. Br J Exp Pathol 1972; 53:44-9; PMID:5014243 [PMC free article] [PubMed] [Google Scholar]
- [41].Durack DT. Experimental bacterial endocarditis. IV. Structure and evolution of very early lesions. J Pathol 1975; 115:81-9; PMID:1151519; http://dx.doi.org/ 10.1002/path.1711150204 [DOI] [PubMed] [Google Scholar]
- [42].Miller JH, Aviles-Reyes A, Scott-Anne K, Gregoire S, Watson GE, Sampson E, Progulske-Fox A, Koo H, Bowen WH, Lemos JA, et al.. The collagen binding protein Cnm contributes to oral colonization and cariogenicity of Streptococcus mutans OMZ175. Infect Immun 2015; 83:2001-10; PMID:25733523; http://dx.doi.org/ 10.1128/IAI.03022-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kojima A, Nomura R, Naka S, Okawa R, Ooshima T, Nakano K. Aggravation of inflammatory bowel diseases by oral streptococci. Oral Dis 2014; 20:359-66; PMID:23679203; http://dx.doi.org/ 10.1111/odi.12125 [DOI] [PubMed] [Google Scholar]
- [44].Kunji ER, Slotboom DJ, Poolman B. Lactococcus lactis as host for overproduction of functional membrane proteins. Biochim Biophys Acta 2003; 1610:97-108; PMID:12586384; http://dx.doi.org/ 10.1016/S0005-2736(02)00712-5 [DOI] [PubMed] [Google Scholar]
- [45].Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, et al.. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 2007; 189:3256-70; PMID:17307855; http://dx.doi.org/ 10.1128/JB.01768-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Latif N, Sarathchandra P, Taylor PM, Antoniw J, Yacoub MH. Localization and pattern of expression of extracellular matrix components in human heart valves. J Heart Valve Dis 2005; 14:218-27; PMID:15792183 [PubMed] [Google Scholar]
- [47].Rodriguez KJ, Piechura LM, Porras AM, Masters KS. Manipulation of valve composition to elucidate the role of collagen in aortic valve calcification. BMC Cardiovasc Disord 2014; 14:29; PMID:24581344; http://dx.doi.org/ 10.1186/1471-2261-14-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hinton RB Jr., Lincoln J, Deutsch GH, Osinska H, Manning PB, Benson DW, Yutzey KE. Extracellular matrix remodeling and organization in developing and diseased aortic valves. Circ Res 2006; 98:1431-8; PMID:16645142; http://dx.doi.org/ 10.1161/01.RES.0000224114.65109.4e [DOI] [PubMed] [Google Scholar]
- [49].Singh B, Fleury C, Jalalvand F, Riesbeck K. Human pathogens utilize host extracellular matrix proteins laminin and collagen for adhesion and invasion of the host. FEMS Microbiol Rev 2012; 36:1122-80; PMID:22537156; http://dx.doi.org/ 10.1111/j.1574-6976.2012.00340.x [DOI] [PubMed] [Google Scholar]
- [50].Singh KV, La Rosa SL, Somarajan SR, Roh JH, Murray BE. The fibronectin-binding protein EfbA contributes to pathogenesis and protects against infective endocarditis caused by Enterococcus faecalis. Infect Immun 2015; 83:4487-94; PMID:26351286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bashore TM, Cabell C, Fowler V Jr. Update on infective endocarditis. Curr Probl Cardiol 2006; 31:274-352; PMID:16546554; http://dx.doi.org/ 10.1016/j.cpcardiol.2005.12.001 [DOI] [PubMed] [Google Scholar]
- [52].Nakano K, Nomura R, Nemoto H, Mukai T, Yoshioka H, Shudo Y, Hata H, Toda K, Taniguchi K, Amano A, et al.. Detection of novel serotype k Streptococcus mutans in infective endocarditis patients. J Med Microbiol 2007; 56:1413-5; PMID:17893184; http://dx.doi.org/ 10.1099/jmm.0.47335-0 [DOI] [PubMed] [Google Scholar]


