SUMMARY
Imprinted genes are differentially expressed by adult stem cells, but their functions in regulating adult stem cell fate are incompletely understood. Here, we show that growth factor receptor bound protein 10 (Grb10), an imprinted gene, regulates hematopoietic stem cell (HSC) self-renewal and regeneration. Deletion of the maternal allele of Grb10 in mice (Grb10 m/+ mice) substantially increased HSC long-term repopulating capacity compared to Grb10 +/+ mice. Following total body irradiation (TBI), Grb10 m/+ mice demonstrated accelerated HSC regeneration and hematopoietic reconstitution compared to Grb10 +/+ mice. Grb10-deficient HSCs displayed increased proliferation following competitive transplantation or TBI, commensurate with upregulation of CDK4 and Cyclin E. Furthermore, the enhanced HSC regeneration observed in Grb10 – deficient mice was dependent on activation of the Akt/mTORC1 pathway. This study reveals a function for the imprinted gene, Grb10, in regulating HSC self-renewal and regeneration and suggests that inhibition of Grb10 can promote hematopoietic regeneration in vivo.
Graphical abstract
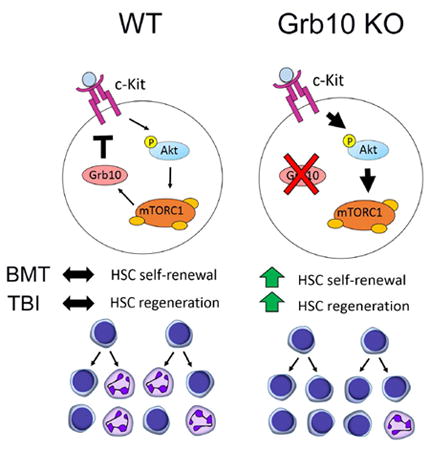
INTRODUCTION
HSC self-renewal is regulated by intrinsic mechanisms and extrinsic signals emanating from the bone marrow (BM) microenvironment or niche (Zon, 2008; Morrison and Scadden, 2014; Mendelson and Frenette, 2014). A fundamental property of HSCs is the capacity to regenerate following various genotoxic stresses, including ionizing radiation, chemotherapy, and inflammatory processes. However, the mechanisms which govern HSC regeneration are not well understood (Mendelson and Frenette, 2014; Zhao et al., 2014; Himburg et al., 2010; Himburg et al., 2012; Doan et al., 2013; Hooper et al., 2009; Poulos et al., 2013). Several imprinted genes have been shown to be essential for normal development and imprinted genes have been postulated to have an important role in regulating adult stem cell fate (Berg et al., 2011; Wood and Oakey, 2006; Tycko and Morison, 2002). However, few imprinted genes have been shown to have a role in regulating adult stem cell function (Ferron et al., 2011; Zacharek et al., 2011; Venkatraman et al., 2013; Qian et al., 2015).
In order to identify candidate genes that regulate HSC regeneration, we performed gene expression analysis of purified murine BM HSCs in steady state and following myeloablative TBI. Via comprehensive gene expression analysis, we discovered that the expression of an imprinted gene, Grb10, was substantially increased in regenerating BM HSCs compared to steady state HSCs. Expression of the paternal allele of Grb10 is restricted to the nervous system, whereas the maternal allele of Grb10 is expressed in most non-neuronal tissues (Plasschaert et al., 2015). Grb10 encodes for an adaptor protein which regulates signaling through multiple tyrosine kinases, and is most well characterized as an inhibitor of insulin receptor (IR) and insulin-like growth factor 1 receptor (IGF1R) signaling in adipose tissue and muscle (Smith et al., 2007; Charalambous et al., 2003; Vecchione et al., 2003; Liu et al., 2014). Of note, biochemical analyses suggest that Grb10 may also bind tyrosine kinases that are expressed by HSCs, including c-kit and VEGFR (Holt et al., 2005), but the functional role of Grb10 in regulating HSC fate has not been established. Here, we show that Grb10 regulates HSC self-renewal and regeneration following injury. Maternal deletion of Grb10 markedly increased HSC self-renewal in vivo following competitive transplantation and also promoted HSC regeneration following TBI. Furthermore, the augmentation in HSC regeneration that we observed in Grb10 – deficient mice was dependent on activation of the Akt/mTORC1 pathway. These studies reveal an inhibitory role for the imprinted gene, Grb10, in regulating HSC self-renewal and regeneration.
RESULTS
Grb10 is differentially expressed by regenerating HSCs
In order to identify candidate genes that regulate HSC regeneration, we compared the gene expression of BM ckit+sca-1+lineage- (KSL) cells in non-irradiated mice with that of BM KSL cells isolated from mice at day +14 following 550 cGy TBI. We selected the day +14 time point because this was the earliest time point at which BM KSL cells were readily detectable by flow cytometry following myeloablative TBI (Figure 1A). Gene expression analysis of KSL cells at this time point revealed several genes that were upregulated and down-regulated in expression compared to KSL cells in steady state (Figure 1B, Table S1). The expression of Grb10 was 5.5-fold higher in irradiated BM KSL cells compared to non-irradiated KSL cells (Figure 1C). Conversely, Grb10 expression was not altered in lineage committed ckit+sca-1-lin- myeloid progenitor cells, suggesting an HSC-specific alteration of Grb10 expression in response to irradiation. Interestingly, Grb10 expression was highest in BM CD34-KSL cells, which are enriched for long-term HSCs, compared to whole BM or committed progenitor cells (Figure 1D).
Figure 1. Grb10 expression is increased in regenerating BM HSCs.
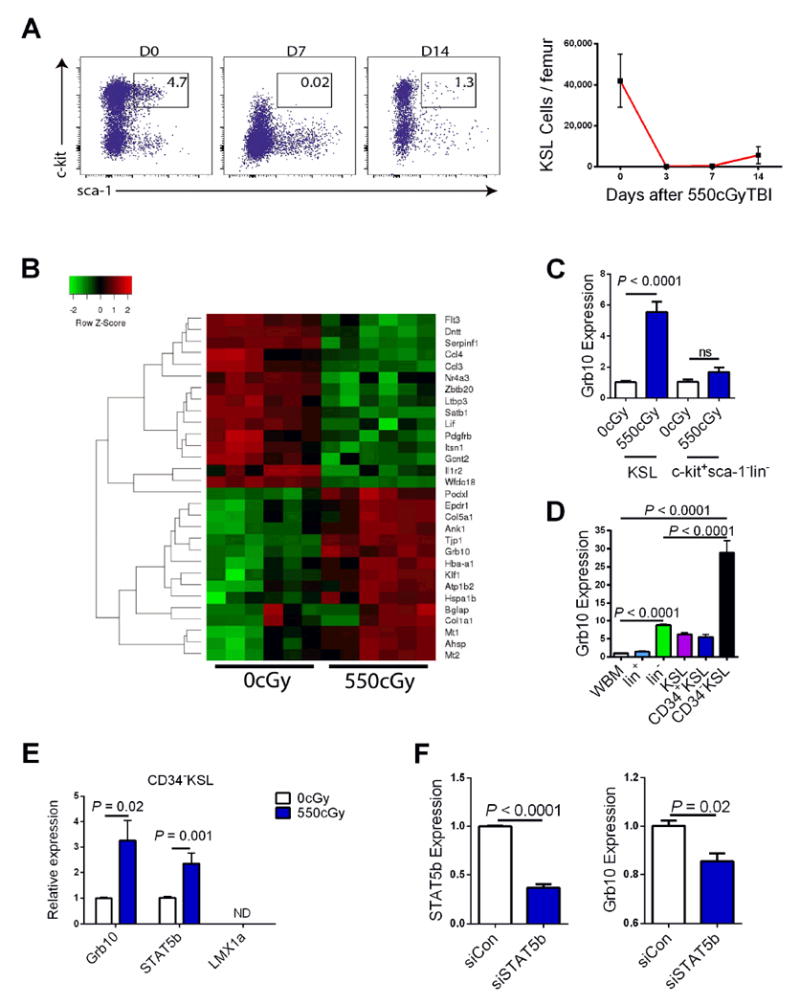
(A) At left, representative flow cytometric analysis of BM KSL cells in non-irradiated, adult C57Bl6 mice and at day +7 and day +14 following 550 cGy TBI. At right, mean numbers of BM KSL cells/femur are shown over time following TBI (n=8/group, means ± SEM). (B) The heat map shows the genes whose expression was most highly up- or down-regulated following 550 cGy TBI (n = 6 mice/sample, 6 samples/group. Red=increased expression; green=decreased expression). (C) Mean expression of Grb10 by qRT-PCR analysis of BM KSL cells or c-kit+sca-1-lin- progenitor cells in non-irradiated mice and at day +14 following 550cGy TBI (n = 6/group, ns=not significant). (D) Mean expression of Grb10 in BM CD34-KSL HSCs, KSL stem/progenitors and other committed hematopoietic populations by qRT-PCR. WBM=whole bone marrow cells (n=6-10 mice/group). (E) Expression of Grb10, STAT5b and LMX1a in BM CD34-KSL cells in steady state and at day +10 following 550cGy TBI (n = 6/group). (F) Expression of STAT5b (left) and Grb10 (right) in BM CD34-KSL cells at day +3 following treatment with siRNA-STAT5b or scramble siRNA (n = 6/group)(all panels, means ± SEM). See also Table S1.
We next sought to determine whether specific transcription factors were involved in regulating the expression of Grb10 in HSCs. STAT5b and LMX1a are transcription factors that have been suggested to bind to or regulate the expression of Grb10 (Hoekstra et al., 2013; Cowley et al., 2014). We found that LMX1a was not expressed by BM CD34-KSL cells in steady state or following 550 cGy irradiation, but STAT5b was expressed by BM KSL cells and increased in expression following 550 cGy (Figure 1E). Further, when we suppressed STAT5b expression in BM CD34-KSL cells via STAT5b-siRNA, we observed significant reduction in Grb10 expression (Figure 1F). Taken together, these data suggested that STAT5b regulates the expression of Grb10 in BM CD34-KSL cells and likely contributes to Grb10 upregulation in response to irradiation.
Maternal deletion of Grb10 increases HSC repopulating capacity
In order to test whether Grb10 regulates hematopoiesis, we obtained Grb10 gene trap mutant mice (Grb10Δ2-4 mice)(Charalambous et al., 2003) and extensively back-crossed this strain into the C57Bl6 strain. Paternal inheritance of Grb10Δ2-4 (Grb10 +/p mice) caused no significant alteration in Grb10 expression in BM cells, but caused significantly decreased expression in the brain (Figure S1A). In contrast, maternal inheritance of Grb10Δ2-4 (Grb10m/+ mice) caused significantly decreased expression of Grb10 in BM hematopoietic lineage- cells, with no effect on expression in the brain (Figure S1A). We therefore focused on evaluating the effect of maternal inheritance of Grb10Δ2-4 on the hematopoietic system. Adult Grb10 m/+ mice displayed moderately increased peripheral blood (PB) WBCs, hemoglobin, platelet counts and Mac-1+ myeloid cells compared to Grb10 +/+ mice (Figure 2A, Figure S1B). Interestingly, Grb10 m/+ mice also displayed increased percentages of eythroid progenitors (EPs), red blood cell counts, and megakaryotic progenitors (MkPs) in steady state compared to Grb10 +/+ mice (Figure 2B, Figure S1C). However, no differences were observed in spleen sizes compared to Grb10 +/+ littermates. No significant differences were observed in BM cell counts, BM KSL cells or SLAM+KSL HSCs between Grb10 m/+ mice and Grb10 +/+ littermates (Figure 2C, Figure S1D). However, Grb10m/+ mice contained significantly increased BM colony forming cells (CFCs, Figure 2C). Furthermore, mice transplanted competitively with 5 × 104 BM cells from Grb10 m/+ mice, along with 2 × 105 host BM cells, displayed approximately 6-fold increased donor-derived, multilineage hematopoietic cell repopulation in the PB through 20 weeks post-transplant (Figure 2D-F, Figure S1E). Mice competitively transplanted with BM cells from Grb10 m/+ mice had increased donor cell contribution to BM SLAM+KSL HSCs compared to mice transplanted with BM from Grb10 +/+ mice (Figure 2E). Recipients of BM cells from Grb10 m/+ mice also displayed increased donor-derived multipotent progenitor cells (MPPs) and megakaryocytic-erythroid progenitors (MEPs) compared to recipients of Grb10 +/+ BM cells (Figure 2F). Secondary competitive repopulation assays in which recipient mice were transplanted with 3 × 106 BM cells from primary recipient mice showed that Grb10 m/+ BM cells provided significantly increased, long-term multilineage hematopoietic repopulation compared to Grb10 +/+ BM cells (Figure 2G). These data suggested that maternal deletion of Grb10 substantially increased the long-term repopulating capacity of HSCs. Of note, primary recipients of competitively transplanted BM cells from Grb10 m/+ mice displayed moderately increased B cell differentiation relative to myeloid cell differentiation at 20 weeks after transplant, but this lineage skewing was not evident in secondary transplanted mice (Figure S1F, G).
Figure 2. Maternal deletion of Grb10 increases HSC repopulating capacity.
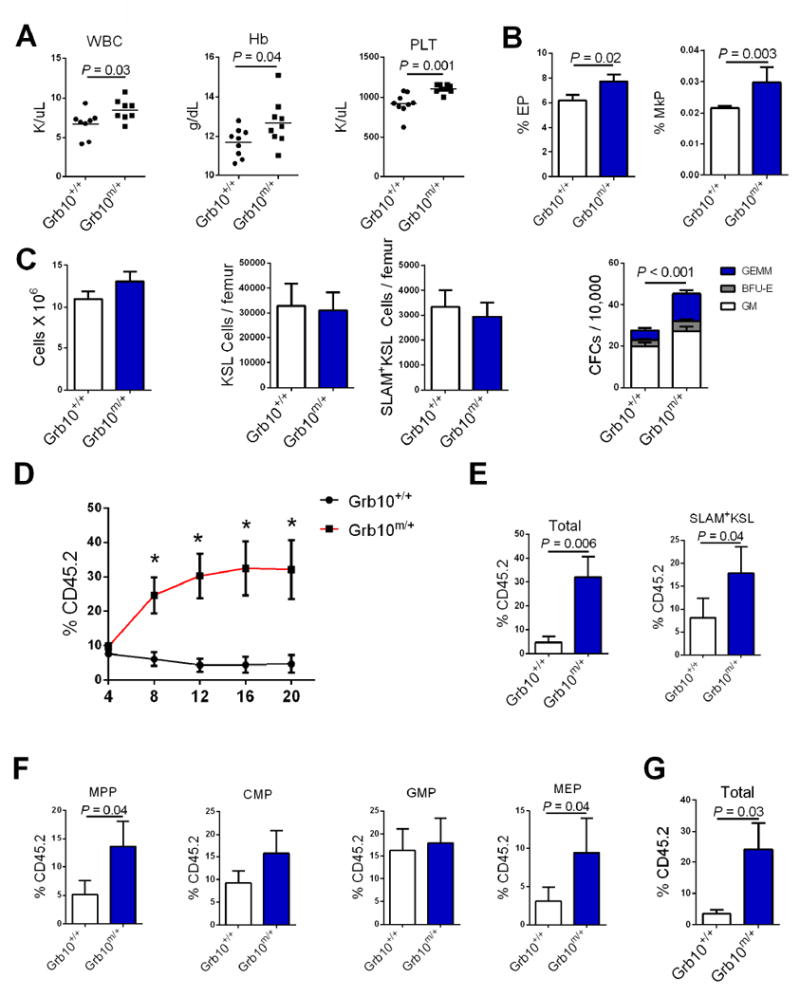
(A) Scatter plots of PB white blood counts (WBC), hemoglobin (Hb) and platelet counts (PLT) in 8 week old Grb10 m/+ mice and Grb10 +/+ mice (n=8–10 mice/group). (B) Mean percentages of erythroid progenitors (EPs) and megakaryocyte progenitors (MkPs) in Grb10 m/+ mice and Grb10 +/+ mice (n=8/group). (C) Mean BM cell counts, KSL cells, SLAM+KSL cells and CFCs in Grb10 m/+ mice and Grb10 +/+ mice (n=6-12/group; GEMM=colony forming unit-granulocyte erythroid monocyte megakaryocyte; BFU-E=burst forming unit-erythroid; GM=colony forming unit-granulocyte macrophage. (D) Donor (CD45.2+) cell engraftment over time in recipient CD45.1+ mice transplanted with 5 × 104 BM cells from Grb10 m/+ mice or Grb10 +/+ mice, together with 2 × 105 CD45.1+ competitor BM cells (n=8/ group. P=0.007, P=0.001, P=0.003, P=0.006 for 8, 12, 16, and 20 weeks, respectively, Mann-Whitney test). (E) Left, mean total donor CD45.2+ cells in the PB of recipient CD45.1+ mice at 20 weeks following competitive transplantation in each group; at right, mean donor-derived HSCs (SLAM+KSL cells) in the BM of recipient mice following competitive transplantation (n=8/group, Mann-Whitney test). (F) Mean donor-derived MPPs, CMPs, GMPs and MEPs in the BM of recipient mice following competitive transplantation (n=8/group, Mann-Whitney test). (G) Mean total donor CD45.2+ cells in secondary recipient mice at 12 weeks following competitive transplantation (3 × 106 donor BM cells, 2 × 105 competitor cells, n = 10-12/group; Mann-Whitney test)(all panels, means ± SEM). See also Figure S1.
Grb10 deletion promotes HSC regeneration after irradiation
Since the expression of Grb10 increased in HSCs during recovery from TBI, we sought to determine if Grb10 regulated HSC regeneration in mice. At day +10 following 550 cGy TBI, a myelosuppressive but sublethal radiation dose, Grb10 m/+ mice displayed increased recovery of BM cell counts, KSL cells, SLAM+KSL HSCs, CFCs and erythroid progenitors compared to Grb10 +/+ mice (Figure 3A-D). Furthermore, mice that were competitively transplanted with BM cells collected at day +10 from irradiated, Grb10 m/+ mice displayed approximately 8-fold higher multilineage donor hematopoietic cell repopulation through 20 weeks post-transplant compared to mice transplanted with BM cells from identically irradiated, Grb10 +/+ mice (Figure 3E, F). These results suggested that deletion of Grb10 accelerated the regeneration of long-term repopulating HSCs and progenitor cells following TBI.
Figure 3. Maternal deletion of Grb10 promotes HSC regeneration following irradiation.
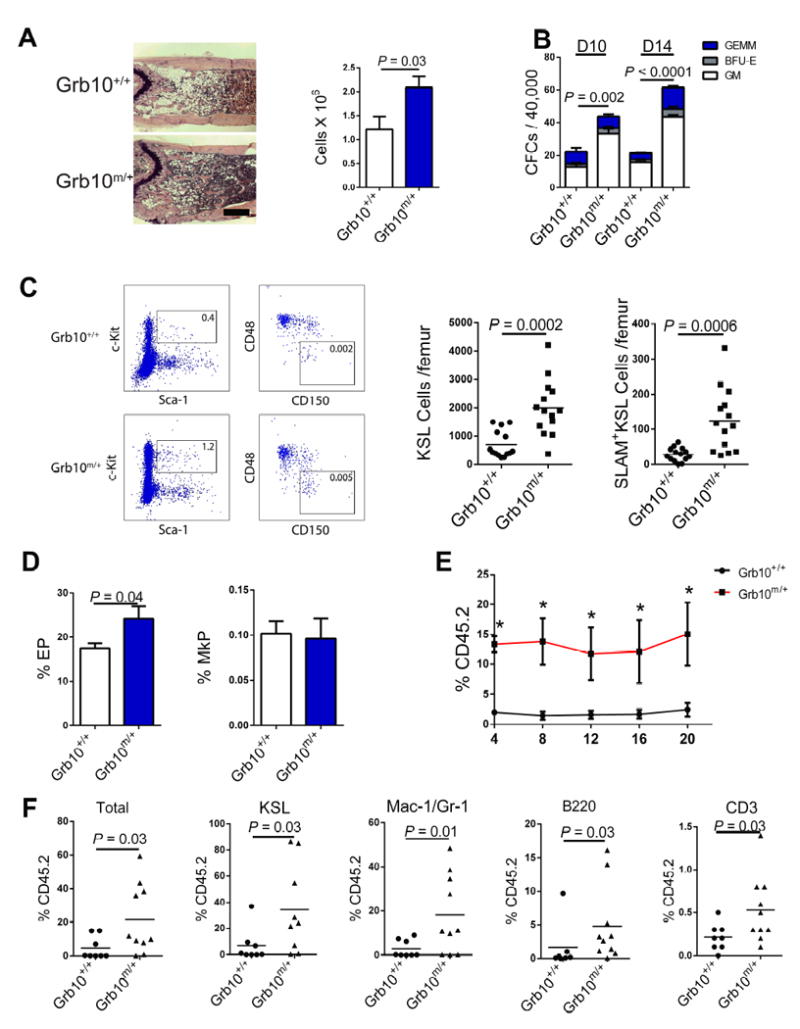
(A) Representative H & E stained femurs from Grb10 m/+ mice and Grb10 +/+ mice at day +10 following 550 cGy TBI and mean BM cell counts in each group (n=8 mice/group; 5x magnification, scale bar=500 μm). (B) Mean BM CFCs in Grb10 m/+ mice and Grb10 +/+ mice at day +10 and day +14 following 550 cGy TBI (n=6/group). (C) A representative flow cytometric plot of BM KSL cells and SLAM+KSL cells and numbers of BM KSL cells and SLAM+KSL cells are shown in Grb10 +/+ mice and Grb10 m/+ mice at day +10 following 550 cGy TBI (n=12 – 14 mice/group; percentages of KSL and SLAM+KSL cells are shown in the gates. Mean values represented by horizontal lines). (D) Mean percentages of erythroid progenitors (EPs) and megakaryocyte progenitors (MkPs) in Grb10 m/+ mice and Grb10 +/+ mice at day +10 following 550 cGy TBI (n=5/group). (E) Donor (CD45.2+) cell engraftment over time in the PB of recipient CD45.1+ mice transplanted with 5×105 BM cells from Grb10 m/+ or Grb10 +/+ mice at day +10 following 550 cGy TBI, together with 1×105 CD45.1+ competitor BM cells (n = 8 - 10/group, P=0.002, P=0.01, P=0.001, P=0.008, and P=0.03, for 4, 8, 12, 16 and 20 weeks, respectively; Mann-Whitney test). (F) Scatter plots show the percentage donor CD45.2+ cell, KSL cell, Mac-1/Gr-1+, B220+ and CD3+ engraftment in the BM at 20 weeks following competitive transplantation of irradiated donor BM cells from each group into recipient CD45.1+ mice (n = 8 - 10/group, Mann-Whitney test)(all panels, means ± SEM).
Grb10 deletion increases hematopoietic stem/progenitor cell proliferative potential
Since Grb10 – deficient HSCs displayed increased self-renewal capacity following competitive transplantation and following radiation injury, we sought to determine the cellular mechanism responsible for these effects. We observed no differences in BM SLAM+KSL cell proliferation or survival of BM KSL cells between Grb10 m/+ mice and Grb10 +/+ mice in steady state (Figure S2A). Further, we observed no difference in the homing capacity of transplanted Grb10m/+ BM sca-1+lin- cells compared to Grb10+/+ BM sca-1+lin- cells (Figure S2A). However, at 48 hours following transplantation, we observed significantly increased BrdU incorporation in transplanted Grb10 m/+ cells in the BM of recipient mice compared to Grb10 +/+ BM cells (Figure 4A). These data suggested that Grb10 deletion primed transplanted BM cells to proliferative more rapidly following homing to the BM niche.
Figure 4. Grb10 deletion promotes HSC regeneration via induction of Akt/mTORC1 signaling.
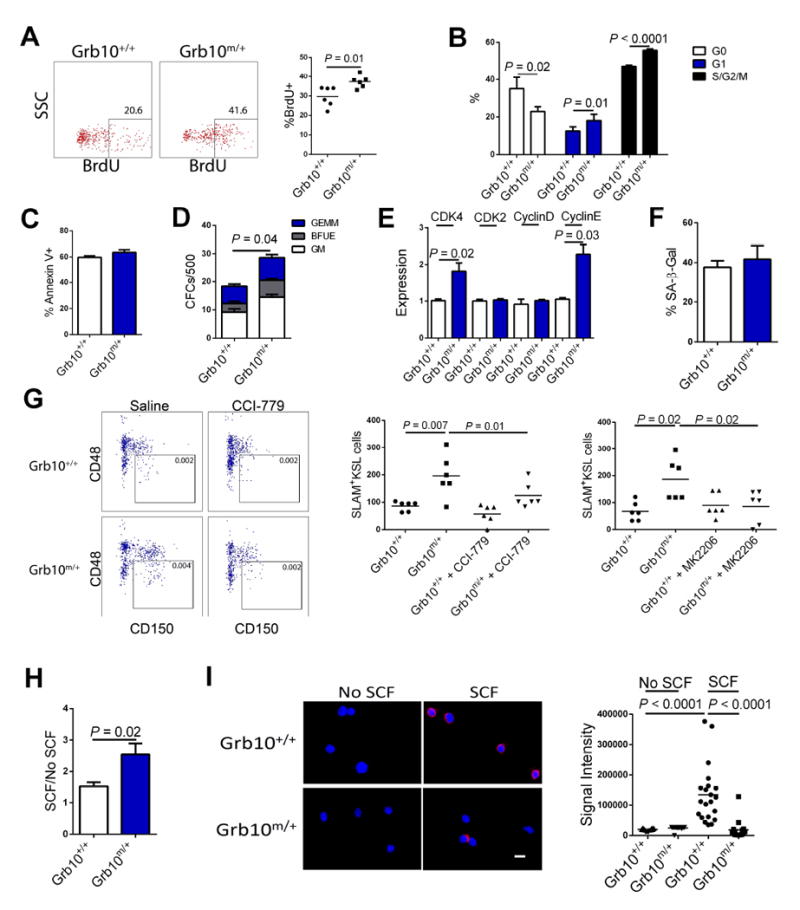
(A) At left, representative flow cytometric plots are shown of BrdU+ donor CD45.2+ BM cells at 48 hours following transplantation of BM cells from Grb10 +/+ mice or Grb10 m/+ mice into CD45.1+ recipients. At right, the scatter plot shows the mean percentage of BrdU+ BM cells in each group (n=6/group). (B) Mean percentages of Grb10 +/+ and Grb10 m/+ KSL cells in G0, G1 and G2/S/M phase at 3 hours after 300 cGy in vitro irradiation (n = 6/group). (C) Mean percentages of Annexin V+ cells in BM KSL cells from Grb10 +/+ and Grb10 m/+ mice, at 3 hours after 300 cGy in vitro irradiation (n = 8/group). (D) Mean numbers of CFCs from 500 BM KSL cells isolated from Grb10 +/+ and Grb10 m/+ mice at day +3 following 300 cGy in vitro (n = 6/group). (E) Expression of cyclin dependent kinases and cyclin proteins in BM KSL cells from Grb10 +/+ mice and Grb10 m/+ mice at 3 hours after 300 cGy (n = 6/group). (F) Mean percentages of senescence associated-β-galactosidase (SA-β-Gal) positive CD34-KSL cells in Grb10 +/+ and Grb10 m/+ mice at 24h following 700cGy TBI (n = 8/group). (G) At left, representative flow cytometric analysis of SLAM+KSL cells in Grb10 +/+ mice and Grb10 m/+ mice at day +10 following 550 cGy TBI and treatment with and without the mTORC1 inhibitor, CCI-779. At right, scatter plot shows the numbers of BM SLAM+KSL cells in Grb10 +/+ and Grb10 m/+ mice at day + 10 following 550 cGy TBI and treatment with and without CCI-779 or MK2206, an Akt inhibitor (n = 6/group). (H) Mean ratios of CFC numbers from BM KSL cells in response to SCF treatment or no SCF treatment for 3 days, showing enhanced response in BM KSL cells from Grb10 m/+ mice (n = 6/group). (I) At left, representative fluorescence microscopic images (63x, scale bar=10μm) show the binding of Grb10 and c-kit proteins via proximity ligation assay in BM KSL cells from Grb10 +/+ mice and Grb10 m/+ mice following 15 min of SCF treatment. Blue = DAPI; red = Grb10 - c-kit complex; at right, scatter plot shows the mean fluorescence signal intensity of BM KSL cells in each group (n = 21-30, t test)(all panels, means ± SEM). See also Figure S2 and Figure S3.
Following in vitro irradiation with 300 cGy, increased percentages of KSL cells from Grb10 m/+ mice were in G1 and G2/S/M phase compared to KSL cells from Grb10 +/+ mice (Figure 4B). In vivo, at 24 hours following 700 cGy irradiation, Grb10 m/+ mice also displayed increased percentages of BM KSL cells in G1 and G2/S/M phase compared to Grb10 +/+ mice (Figure S2B). Conversely, no differences were observed in apoptosis of BM KSL cells from Grb10 m/+ mice or Grb10 +/+ following 300 cGy irradiation in vitro or 700 cGy in vivo (Figure 4C, Figure S2C). Nonetheless, CFC recovery from BM KSL cells from Grb10 m/+ mice following 300 cGy was significantly increased compared to irradiated KSL cells from Grb10 +/+ mice (Figure 4D). Taken together, these results suggested that BM KSL cells from Grb10 m/+ mice possessed increased proliferative potential following injury. Consistent with this observation, we detected increased expression of the cyclin-dependent kinase, CDK4, and Cyclin E, in BM KSL cells in Grb10 m/+ mice at 3 hours following 300 cGy irradiation (Figure 4E). We observed no differences in the expression of cyclin dependent kinase (CDK) inhibitors between Grb10 m/+ BM KSL cells compared to Grb10 +/+ BM KSL cells (Figure S2D) and also no difference in HSC senescence (Figure 4F) between these populations after irradiation.
Grb10 deletion potentiates HSC regeneration via Akt/mTORC1 activation
Grb10 is an adaptor protein that lacks catalytic function but contains several protein binding motifs which implicate Grb10 in mediating interactions between other proteins (Smith et al., 2007). Grb10 can bind several receptor tyrosine kinases (RTKs), including IR, IGF1-R, and c-kit, and genetic studies have demonstrated that Grb10 is a negative regulator of insulin and IGF1 signaling in adipose tissue (Charalambous et al., 2003; Smith et al., 2007; Cao, et al., 2008). Grb10 is also a substrate for the mammalian target of rapamycin (mTOR) and Grb10 negatively regulates mTORC1 signaling in adipocytes (Yu et al., 2011; Liu et al., 2014). Interestingly, BM KSL cells from Grb10 m/+ mice showed no increase in Akt or mTORC1 activation in steady state (Figure S3A). However, following 700 cGy TBI, BM KSL cells from Grb10 m/+ mice displayed significantly increased mTORC1 activation compared to Grb10 +/+ cells (Figure S3B). Similarly, treatment with c-kit ligand (SCF) significantly increased Akt and mTORC1 activation in Grb10 m/+ KSL cells compared to Grb10+/+ KSL cells (Figure S3B). Taken together, these data suggested that maternal deletion of Grb10 did not alter baseline Akt or mTORC1 activation in KSL cells, but rather potentiated the responsiveness of the Akt/mTORC1 pathway in BM KSL cells in the setting of stress or cytokine stimulation. Importantly, irradiated Grb10 m/+ mice displayed accelerated recovery of BM SLAM+KSL HSCs at day +10 following 550 cGy TBI, but systemic administration of a specific inhibitor of Akt, MK2206, or mTORC1, CCI-779, completely abrogated this accelerated recovery of HSCs in Grb10 m/+ mice (Figure 4G). These results suggested that the early HSC regeneration observed in irradiated Grb10 m/+ mice was dependent on Akt/mTORC1 pathway activation.
Since Grb10 has been shown to inhibit IR signaling in adipocytes via binding and inhibition of the catalytic domains of IR (Liu et al., 2014), we postulated that Grb10 might mediate a similar inhibitory action on the c-kit receptor in HSCs, since c-kit activation induces Akt/mTORC1 signaling in HSCs (Lennartson et al., 2012; Ding et al., 2012). Interestingly, treatment of BM KSL cells from Grb10 m/+ mice with SCF produced increased numbers of CFCs compared to the yield from SCF treatment of KSL cells from Grb10 +/+ mice (Figure 4H). Further, proximity ligation assay (Lu et al., 2012) demonstrated increased binding of Grb10 with c-kit in wild type BM KSL cells following SCF treatment (Figure 4I). Conversely, when BM KSL cells from Grb10 m/+ mice were treated with SCF, Grb10 binding to c-kit was markedly decreased. Taken together, these results suggest that Grb10 binds c-kit in BM hematopoietic stem/progenitor cells in response to SCF treatment and that the enhanced Akt/mTORC1 activation in Grb10 deficient KSL cells may be related to the absence of inhibitory Grb10 binding to c-kit.
DISCUSSION
These studies reveal a previously unrecognized role for the imprinted gene, Grb10, in regulating adult HSC self-renewal. Maternal deletion of Grb10 substantially increased HSC repopulating capacity as measured in primary and secondary transplanted mice and also increased the frequencies of BM erythroid progenitors and megakaryocytic progenitor cells. These functional results are consistent with the prior observation by Berg et al. (Berg et al., 2011) that Grb10 is differentially expressed in LT-HSCs and myelo-erythroid progenitor cells. Further, primary mice transplanted with Grb10 m/+ BM cells displayed moderately increased B lymphoid differentiation and decreased myeloid differentiation compared to mice transplanted with Grb10 +/+ BM cells. Taken together, these results suggest that in addition to effects on HSC repopulating capacity, Grb10 likely regulates myeloid differentiation potential.
Our findings regarding Grb10 add significantly to the general understanding of the function of imprinted genes in adult stem cells. Recently, two imprinted genes, Cdkn1c (p57) and delta-like 1 homolog (Dlk1), were found to inhibit the regeneration of adult lung and muscle stem cells, respectively (Zacharek et al., 2011; Anderson et al., 2013). Epigenetic silencing of the imprinted gene, Cdkn1c, by BMI1 was reported to be necessary for bronchioalveolar stem cell (BASC) regeneration and silencing of Cdkn1c rescued BASC regenerative function (Zacharek et al., 2011). Similarly, deletion of Dlk1 was shown to promote adult skeletal muscle regeneration in vivo (Anderson et al., 2013). Conversely, expression of long non-coding RNAs from the Dlk1-Gtl2 imprinted gene locus was shown to promote HSC maintenance via suppression of reactive oxygen species (Qian et al., 2015). Another imprinted gene locus, H19-Igf2, was found to be important for HSC repopulating capacity and long-term maintenance of quiescent HSCs (Venkatraman et al., 2013). Here, we demonstrate that Grb10 deletion promotes HSC repopulating capacity and regeneration in vivo, suggesting that Grb10 negatively regulates these processes.
We have further demonstrated that Grb10-deficient hematopoietic stem/progenitor cells (HSPCs) possess enhanced proliferative potential and regenerative capacity in vivo following transplantation or radiation injury. Grb10 expression increased in wild type BM 34-KSL cells and KSL cells following high dose irradiation and siRNA studies suggested that Grb10 expression in HSCs is under the control of STAT5b. Interestingly, following radiation injury, BM KSL cells from Grb10 m/+ mice manifested no alterations in apoptosis or senescence compared to Grb10 +/+ mice, but rather displayed increased percentages of cells in G2/S/M phase, concordant with increased expression of CDK4 and Cyclin E. These results are consistent with the established functions of CDK4 and Cyclin E in promoting cell cycle entry and G1 – S phase transition (Mende et al., 2015).
Our results further suggest that enhancement of HSC regeneration in Grb10 – deficient mice is dependent upon activation of Akt and mTORC1 signaling. mTORC1 has been shown to promote cell proliferation via induction of CDK4 and Cyclin E expression (Lee et al., 2015). Miao et al. reported that mTORC1 activation was essential for Akt-mediated axon regeneration in the central nervous system (Miao et al., 2016) and Akt/mTORC1 pathway activation has been shown to promote hepatic regeneration in aged mice (Gielchinsky et al., 2010). We found that Grb10 deletion in HSCs did not alter mTORC1 activation in steady state, but did augment mTORC1 signaling in HSCs in response following irradiation or SCF treatment. Importantly, we have shown that the accelerated regeneration of HSCs in Grb10 – deficient mice was dependent upon activation of Akt and mTORC1. In this regard, our findings are consistent with the recently described role for mTORC1 in the adaptive response of muscle stem cells to injury (Rodgers et al., 2014). Since Grb10 deletion did not cause constitutive activation of the mTOR pathway in HSCs, we did not observe exhaustion of the HSC pool as has been described following deletion of the mTOR inhibitors, phosphatase and tensin homologue (PTEN) and the Tuberous Sclerosis Complex 1 (TSC1)(Yilmaz et al., 2006; Lee et al., 2010; Gan et al., 2009). Rather, our results support a functional role for mTORC1 signaling in HSC self-renewal, as was recently suggested by Ghosh et al. (Ghosh et al., 2016).
In summary, we describe a function for the adaptor protein, Grb10, in regulating HSC self-renewal following transplantation and HSC regeneration in response to injury. Targeted inhibition of Grb10 has therapeutic potential as a means to promote hematopoietic regeneration in the setting of HSC transplantation or following myelosuppressive injury. For this reason, the long-term consequences of Grb10 inhibition on the hematopoietic system of irradiated or transplanted animals will be important questions to address. Clarification of the effects of Grb10 inhibition on myeloid differentiation will be particularly critical to address in pre-clinical studies. In this regard, it is noteworthy that we have observed no signs of myelodysplasia or leukemia in Grb10-deficient mice through 1 year of age. Furthermore, the potency of Grb10 deletion in augmenting HSC self-renewal and regeneration highlights the importance of developing pharmacologic agents to inhibit Grb10 as a potential means to accelerate hematopoietic recovery in patients receiving myelosuppressive therapies or undergoing hematopoietic cell transplantation.
EXPERIMENTAL PROCEDURES
Statistics
Unless stated otherwise in the Figure Legends, all graphical data are presented as means ± SEM, except histograms, and significance was calculated using two-tailed Student’s t test. P < 0.05 was considered significant. Competitive repopulating assays were compared with a two-tailed Mann-Whitney test. For all animal studies, we used a power test to determine the sample size needed for a 2-fold difference in mean with 0.8 power using a two-tailed Student’s t-test. Animals in our studies were sex and age-matched and littermates were used as controls.
Mice
All mouse procedures were performed in accordance with UCLA animal research and oversight committee-approved protocols. Grb10Δ2-4 mice were a kind gift from Dr. Andrew Ward (University of Bath, UK). Grb10Δ2-4 mice were extensively backcrossed into a C57BL/6 background for these studies. Grb10 m/+ mice, Grb10 +/+ mice and Grb10+/p mice were evaluated at 8-12 weeks of age. C57BL/6 mice and B6.SJL mice between 8-12 weeks old were obtained from the Jackson Laboratory (Bar Harbor, ME).
Microarray analysis
C57BL6 mice were irradiated with 550 cGy TBI. At day +14, BM KSL cells were isolated by FACS, as previously described (Himburg et al., 2014). Genomic RNAs from irradiated BM KSL cells and non-irradiated KSL cells were isolated and amplified using an RNA amplification kit (Ovation RNA-Seq System V2, Nugen). Amplified RNA samples then were loaded onto GeneChip Mouse Genome 430A 2.0 array (Affymetrix). We analyzed the microarray data with Partek Genomics Suite and generated lists of differentially expressed genes triggered by irradiation, as previously described (Zheng et al., 2014).
STAT5b siRNA studies
BM CD34-KSL cells from Grb10m/+ and Grb10+/+ mice were sorted with a BD ARIA sorter and resuspended in Accell Delivery Medium (Dharmacon) supplemented with 100 ng/ml stem cell factor (SCF, R&D Systems) and 20ng/ml thrombopoietin (TPO, R&D Systems). 1uM Accell siRNA (Dharmacon) targeting STAT5b was added to the medium. At 72 hours after siRNA treatment, cells were then collected and qRT-PCR analysis for STAT5b and Grb10 was performed.
Mouse competitive repopulation assays
For primary competitive repopulation assays, BM cells were isolated from 10-12 week old donor Grb10m/+ mice and Grb10+/+ mice which express CD45.2. Ten week old recipient CD45.1+ B6.SJL mice were irradiated with 950 cGy TBI using a Cs 137 irradiator 12 hours prior to transplantation. BM cells (5 × 104) from Grb10m/+ mice or Grb10+/+ mice were injected via tail vein into recipient mice along with 2 × 105 host competitor BM cells. Multilineage hematopoietic cell engraftment was measured by flow cytometric analysis of the PB every 4 weeks through week 20 post-transplant, as previous described (Himburg et al., 2014; Quarmyne et al., 2015). For secondary competitive repopulation assays, 3 × 106 total BM cells from primary recipient mice were injected via tail vein into irradiated B6.SJL mice with 2 × 105 host competitor BM cells from the B6.SJL mice. Donor hematopoietic cell engraftment was measured by flow cytometry at 4 through 20 weeks post-transplant.
Flow cytometric analysis
BM cells were collected from tibias and femurs and stained with anti-c-kit PE-Cy7 (BD Biosciences), anti-Sca-1 APC-Cy7 (BD Biosciences), anti-lineage V450 (BD Biosciences), anti-CD48 FITC (Biolegend), anti-CD127 FITC (BD Biosciences), anti-CD41 Alexa Fluor 488 (Biolegend), anti-CD34 APC (BD Biosciences), anti-CD135 PE (BD Biosciences), anti-CD16/32 PerCP (BD Biosciences) and anti-CD150 Alexa Fluor 647 (Biolegend). BM HSCs (CD150+CD48-Lineage-Sca-1+c-Kit+), MPPs (CD150-CD48-Lineage-Sca-1+c-Kit+), CMPs (CD34+CD16/32lowCD127-Lineage-Sca-1-c-Kit+), GMPs (CD34+CD16/32highCD127-Lineage-Sca-1-c-Kit+), MEPs (CD34-CD16/32-/lowCD127-Lineage-Sca-1-c-Kit+), MkPs (CD150+CD41+Lineage-Sca-1-c-Kit+) and EPs (CD71+Ter119+) were stained with the indicated cell surface marker antibodies. For intracellular phosphoprotein analysis, BM cells were flushed into serum-free PBS. BM cells were stained with surface marker antibodies, then washed with PBS and fixed for 15 minutes at 4°C, followed by incubation with BD c ytoperm buffer for 30 minutes at room temperature. Rabbit anti-pS6K (Thr389) (Cell Signaling Technology) or anti-pAkt PE (Cell Signaling Technology) were added at 1:50 dilution for 30 minutes. For pS6K staining, cells were incubated with anti-rabbit IgG PE (Cell Signaling Technology) at 1:500 for 30 minutes. Flow cytometric analysis was performed on a FACS CANTO II (BD Biosciences).
Immunohistochemistry
Hematoxylin and eosin (H&E) staining of femurs from mice was performed as previously described (Himburg et al., 2014). Images were obtained using an Axiovert 200 microscope (Carl Zeiss, Thornwood, NY).
BrdU incorporation, cell cycle analysis and Annexin V analysis
For BrdU incorporation analysis, Grb10m/+ mice or Grb10+/+ mice were injected intraperitoneally with 2 mg BrdU suspended in PBS. At 24 hours, BM cells were collected and stained with anti-BrdU FITC (BD). For cell cycle analysis, BM cells were flushed into PBS plus 10% FBS and stained for surface c-kit, sca-1 and lineage markers for 30 minutes. Cells were then fixed with buffer III (BD) for 30 minutes, followed by incubation of permeabilization buffer (BD). Anti-Ki67 FITC (BD) was added to the cells and incubated for 30 minutes. 7AAD was added to stain for DNA content. For analysis of HSC apoptosis in Grb10m/+ mice or Grb10+/+ mice, BM cells were stained with anti-c-kit, anti-sca-1 and anti-lineage antibodies for 30 minutes, followed by application of the FITC Annexin V Apoptosis Detection Kit I (BD), as previously described (Quarmyne et al., 2015). For in vivo analyses, mice were irradiated with 700 cGy to allow sensitive detection of radiation effects on cell cycle or cell death in BM KSL cells. For in vitro cell cycle and apoptosis assays, BM c-kit+sca-1+lineage- (KSL) cells were sorted into 96-well plate using a BD FACS Aria sorter. KSL cells were irradiated at 300cGy in vitro. At certain time points after irradiation, cells were collected and cell cycle and apoptosis assays were performed.
Homing Assay
2 × 104 BM Sca-1+lin-cells (CD45.2+) were injected into lethally irradiated B6.SJL recipients (CD45.1+) and donor cell engraftment was measured by flow cytometry at 18 hours after transplant, as previously described (Quarmyne et al., 2015).
Senescence Assay
Grb10m/+ and Grb10+/+ mice were irradiated with 700 cGy TBI using a Cs 137 irradiator. At 24 hours following irradiation, BM cells were isolated from both groups in cold PBS containing 2% FBS. Senescent cells were stained for senescence activated β-galactosidase using the Quantitative Cellular Senescence Assay Kit (Cell Biolabs) and analyzed on a BD CANTO II (BD Biosciences).
mTOR and Akt inhibition studies
For in vivo inhibition of mTORC1, temsirolimus (CCI-779, Selleckchem) was reconstituted in ethanol at 10mg/ml and then diluted in 5% Tween-80 (Sigma-Aldrich) and 5% PEG (polyethylene glycol) 400 (Hampton Research). Grb10m/+ mice and Grb10+/+ mice were irradiated at 550cGy at day 0, then injected with CCI-779 (10mg/kg intraperitoneally) or vehicle every other day through day +10. At day +10, BM cells were isolated from irradiated Grb10m/+ mice and Grb10+/+ mice and stained for SLAM+KSL cells. For in vivo inhibition of Akt, MK2206 (Selleckchem) was reconstituted in 30% Captisol (Sigma-Aldrich). Grb10m/+ mice and Grb10+/+ mice were irradiated at 550 cGy at day 0, then orally administered with MK2206 (120 mg/kg) or vehicle every other day through day +10. At day +10, BM cells were isolated from irradiated Grb10m/+ mice and Grb10+/+ mice and stained for SLAM+KSL cells.
Quantitative real time PCR
RNA was isolated using the Qiagen RNeasy micro kit. RNA was reverse transcribed into cDNA using iScript cDNA synthesis kit. qRT-PCR analysis for STAT5b, LMX1a, Grb10, p16, p21, p18, p27, p57, CDK2, CDK4, Cyclin D and Cyclin E was performed using Taqman Gene Expression assays.
Proximity ligation assay
For detection of binding between Grb10 and c-kit, a proximity ligation assay (PLA) was performed using the Duolink In Situ Red Starter Kit (Sigma-Aldrich). BM KSL cells from Grb10 m/+ and Grb10 +/+ mice were sorted by FACS and treated with 100 ng/ml stem cell factor (SCF) or saline for 15 minutes at 37°C. Cel ls were then fixed in 4% paraformaldehyde (PFA) for 20 minutes on a slide and incubated with rabbit anti-Grb10 (Abcam) and goat anti-c-kit antibodies (R&D) overnight. PLA was performed by the addition of species-specific secondary antibodies (goat and rabbit) with PLUS and MINUS oligonucleotide probes from the Duolink kit. Binding of the PLUS and MINUS oliogonucleotide probes only occurs when the two proteins of interest are in close proximity and results in a high intensity dsRed fluorescent signal. Slides were then counterstained with DAPI to visualize cell nuclei and enable enumeration of total cells. Images were captured using an Axiovert 200 fluorescent microscope (Carl Zeiss, Thornwood, NY) and processed with ImageJ software. The DAPI channel was used to establish the boundaries of individual cells. The fluorescence intensity of the DuoLink signal was then quantified in the dsRed channel by integrating the fluorescence intensity within each cell boundary. Isolated single pixels on the image were removed to filter noise. Results were exported into Excel and analyzed by GraphPad Prism 6 (GraphPad Software).
Supplementary Material
Acknowledgments
This research was supported by NIAID grant AI067798 (JPC), NHLBI grant HL086998 (JPC) and the California Institute for Regenerative Medicine Leadership Award, LA1-08014 (JPC).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes 3 supplemental figures and 1 supplemental table.
AUTHOR CONTRIBUTIONS
J.P.C. conceived of the study; X.Y., H.A.H., N.J.C. and J.P.C. designed the experiments; X.Y., M.Q., K.P., E.T., Y.Z., T.F., J.K., L.Z. and P.L.D. performed the experiments and analyzed data; J.P.C. provided overall supervision; X.Y. and J.P.C. wrote the manuscript with input from all authors.
ACCESSION NUMBERS
All microarray data from this study have been uploaded into the GEO repository. These data can be accessed using the Accession Code GSE79744.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson D, Laborda J, Baladron V, Kassem M, Sheikh S, Jensen C. Dual role of delta-like 1 homolog (Dlk1) in skeletal muscle development and adult muscle regeneration. Development. 2013;140:3743–3753. doi: 10.1242/dev.095810. [DOI] [PubMed] [Google Scholar]
- Berg JS, Lin KK, Sonnet C, Boles NC, Weksberg DC, Nguyen H, Holt LJ, Rickwood D, Daly RJ, Goodell MA. Imprinted genes that regulate early mammalian growth are coexpressed in somatic stem cells. PLoS ONE. 2011;6:e26410. doi: 10.1371/journal.pone.0026410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, Shan H, Qu J, Sweezer EM, Place T, Kirby PA, et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal. 2008;1:ra5. doi: 10.1126/scisignal.1160940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalambous M, Smith FM, Bennett WR, Crew TE, Mackenzie F, Ward A. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc Natl Acad Sci, USA. 2003;100:8292–8297. doi: 10.1073/pnas.1532175100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley M, Garfield AS, Madon-Simon M, Charalambous M, Clarkson RW, Smalley MJ, Kendrick H, Isles AR, Parry AJ, Carney S, Oakey RJ, Heisler LK, Moorwood K, Wolf JB, Ward A. Developmental programming mediated by complementary roles of imprinted Grb10 in mother and pup. PLoS Biol. 2014;12:1–13. doi: 10.1371/journal.pbio.1001799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan PL, Himburg HA, Helms K, Russell JL, Fixsen E, Quarmyne M, Harris JR, Deoliviera D, Sullivan JM, Chao NJ, et al. Epidermal growth factor regulates hematopoietic regeneration after radiation injury. Nat Med. 2013;19:295–304. doi: 10.1038/nm.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron SR, Charalambous M, Radford E, McEwen K, Wildner H, Hind E, Morante-Redolat JM, Laborda J, Guillemot F, Bauer SR, et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature. 2011;475:381–385. doi: 10.1038/nature10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan G, DePinho RA. mTORC1 signaling governs hematopoietic stem cell quiescence. Cell Cycle. 2009;8:1003–1006. doi: 10.4161/cc.8.7.8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielchinsky Y, Laufer N, Weitman E, Abramovitch R, Garanot Z, Bergman Y, Pikarsky E. Pregnancy restores the regenerative capacity of the aged liver via activation of an mTORC1-controlled hyperplasia/hypertrophy switch. Genes Dev. 2010;24:543–548. doi: 10.1101/gad.563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J, Kobayashi M, Ramdas B, Chatterjee A, Ma P, et al. S6K1 regulates hematopoietic stem cell self-renewal and leukemia maintenance. J Clin Invest. 2016;126:2621–2625. doi: 10.1172/JCI84565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himburg HA, Muramoto G, Daher P, Meadows SK, Russell JL, Doan P, Chi JT, Salter AB, Lento WE, Reya T, et al. Pleiotrophin regulates the expansion and regeneration of hematopoietic stem cells. Nature Medicine. 2010;16:475–482. doi: 10.1038/nm.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himburg HA, Harris JA, Ito T, Daher P, Russell JL, Quarmyne M, Doan PL, Helms K, Nakamura M, Fixsen E, et al. Pleiotrophin regulates the retention and self-renewal of hematopoietic stem cells in the bone marrow vascular niche. Cell Rep. 2012;25:964–975. doi: 10.1016/j.celrep.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himburg H, et al. Pleiotrophin mediates hematopoietic regeneration via activation of Ras. J Clin Invest. 2014;124:4753–4758. doi: 10.1172/JCI76838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra EJ, von Oerthel L, van der Linden AJA, Schellevis RD, Scheppink G, Holstege FCP, Groot-Koerkamp MJ, van der Heide LP, Smidt MP. Lmx1a is an activator of Rgs4 and Grb10 and is responsible for the correct specification of rostral and medial mdDA neurons. Eur J Neuroscience. 2013;37:23–32. doi: 10.1111/ejn.12022. [DOI] [PubMed] [Google Scholar]
- Holt L, Siddle K. Grb10 and Grb14: enigmatic regulators of insulin action – and more. Biochem J. 2005;388:393–406. doi: 10.1042/BJ20050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper AT, Butler JM, Nolan DJ, Kranz A, Lida K, Kobayashi M, Kopp HG, Shido K, Petit I, Yanger K, et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell. 2009;4:263–274. doi: 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nakada D, Yilmaz O, Tothova Z, Joseph NM, Lim MS, Gilliland DG, Morrison SJ. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after PTEN deletion. Cell Stem Cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartsson J, Ronnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- Liu M, Bai J, He S, Villarreal R, Hu D, Zhang C, Yang X, Liang H, Slaga TJ, Yu Y, et al. Grb10 promotes lipolysis and thermogenesis by phosphorylation-dependent feedback inhibition of mTORC1. Cell Metabolism. 2014;19:1–14. doi: 10.1016/j.cmet.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Chang J, Parachoniak C, Pandika M, Aghi M, et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22:21–35. doi: 10.1016/j.ccr.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende N, Kuchen EE, Lesche M, Grinenko T, Kokkaliaris KD, et al. CCND1-CDK4-mediated cell cycle progression provides a competitive advantage for human hematopoietic stem cells in vivo. J Exp Med. 2014;212:1171–1183. doi: 10.1084/jem.20150308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014;20:833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Yang L, Huang H, Liang F, Ling C, Hu Y. mTORC1 is necessary but mTORC2 and GSK3β are inhibitory for AKT3-induced axon regeneration in the central nervous system. Elife. 2016;5:e14908. doi: 10.7554/eLife.14908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert R, Bartolomei M. Tissue specific regulation and function of Grb10 in growth and neuronal commitment. Proc Natl Acad Sci, USA. 2015;112:6841–6847. doi: 10.1073/pnas.1411254111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos MG, Guo P, Kofler NM, Pinho S, Gutkin MC, Tikhonova A, Aifantis I, Frenette PS, Kitajewski J, Rafii S, et al. Endothelial jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell Rep. 2013;4:1022–1034. doi: 10.1016/j.celrep.2013.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmyne M, et al. Protein tyrosine phosphatase-sigma regulates hematopoietic stem cell repopulating capacity. J Clin Invest. 2015;125:177–182. doi: 10.1172/JCI77866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian P, He XC, Paulson A, Li Z, Tao F, Perry JM, Guo F, Zhao M, Zhi L, Venkatraman A, et al. The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell. 2015;18:1–15. doi: 10.1016/j.stem.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai CR, et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to GAlert. Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FM, Holt LJ, Garfield AS, Charalambous M, Koumanov F, Perry M, Bazzani R, Sheardown SA, Hegarty BD, Lyons RJ, et al. Mice with disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol Cell Biol. 2007;27:5871–5886. doi: 10.1128/MCB.02087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol. 2002;192:245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- Vecchione A, Marchese A, Henry P, Rotin D, Morrione A. The Grb10/Nedd4 complex regulates ligand induced ubiquitination and stability of the insulin-like growth factor 1 receptor. Mol Cell Biol. 2003;23:3363–3372. doi: 10.1128/MCB.23.9.3363-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, Zhao M, Christenson MK, Sanchez R, Yu JY, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500:345–351. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Oakey RJ. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2006;2:e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. PTEN dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yu Y, Yoon S, Poulogiannis G, Yang Q, Ma X, Villen J, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharek SJ, Filmore CM, Lau AN, Gludish DW, Chou A, Ho JW, Zamponi R, Gazit R, Bock C, Jager N, et al. Lung stem cell self-renewal relies on BMI1-dependent control of expression at imprinted loci. Cell Stem Cell. 2011;9:272–281. doi: 10.1016/j.stem.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Perry JM, Marshall H, Venkatraman A, Qian P, He XC, Ahamde J, Li L. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat Med. 2014;20:1321–1326. doi: 10.1038/nm.3706. [DOI] [PubMed] [Google Scholar]
- Zheng L, et al. Biological pathway selection through Bayesian integrative modeling. Stat Appl Genet Mol Biol. 2014;13:435–457. doi: 10.1515/sagmb-2013-0043. [DOI] [PubMed] [Google Scholar]
- Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453:306–313. doi: 10.1038/nature07038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


