Abstract
Problem
Diagnostic neuropathology traditionally relies on subjective interpretation of visual data obtained from a brightfield microscope. This results in high variability, inability to multiplex, and unsatisfactory reproducibility even among experts. These diagnostic problems may affect patient outcomes and confound clinical decision-making. Furthermore, standard histological processing of pathological specimens results in auto-fluorescence and other artifacts, which have nearly blocked the implementation of fluorescent microscopy in diagnostic pathology. Thus, generation of objective and quantitative methodologies would augment the toolbox available to neuropathologists, which would ultimately enhance clinical decision making.
Objective
To develop image analysis methods to quantitatively validate anti-PTBP1 antibody for use in diagnostic neuropathology.
Method
We propose a computerized image analysis method to validate anti-PTBP1 antibody. Images were obtained from standard neuropathological specimens stained with anti-PTBP1 antibody. First, the noise characteristics of the images were modeled and images are de-noised according to the noise model. Next, images are filtered with sigma-adaptive Gaussian filtering for local normalization, and cell nuclei are detected and segmented with a k-means based deterministic approach.
Result
Experiments on 29 data sets from three cases of brain tumor (recurrent glioma, primary resections of glioblastoma harboring the EGFRVIII mutation, pilocytic astrocytoma) and reactive gliosis show statistically significant differences between the number of positively stained nuclei in images stained with and without anti-PTBP1 antibody (p values, t-test, are 40×10−4, 33×10−4, 6×10−4 and 46×10−3, respectively).
Conclusion
The experimental of analysis of specimens from three different brain tumor groups and one reactive gliosis group indicate the feasibility of using anti-PTBP1 antibody in diagnostic neuropathology and computerized image analysis provides a systematic and quantitative approach to explore feasibility.
Keywords: PTBP1, antibody validation, automated image analysis, neuropathology
1. INTRODUCTION
Diagnostic problems encountered by neuropathologists when evaluating the histopathology of a complex case could be solved by improving their diagnostic toolbox. Two branch points in neuropathological workflows require the generation of new image analysis tools and techniques. First, the ability to identify multiple biomarkers on the same cell would help pathologists identify distinct subtypes of cancers without exhausting tissue. A second problem exists in the objective interpretation of biomarker expression levels. Standard biomarker evaluation involves utilization of enzyme-based histochemical reactions, the most common of which is DAB precipitation by HRP-linked secondary antibody systems, which is optimally designed for binary designations of “positive” or “negative” or at best semi quantitative, subjective designations of “1+”, “2+”, “3+”. Both problems could be resolved by implementing modern fluorescence microscopy techniques, which permit multiplexing and are superior quantitative modalities relative to immunohistochemistry. Unfortunately, the implementation of fluorescence in routine diagnostic pathology is fraught with caveats. For example, erythrocytes fluoresce in the 500–600 nm spectrums, which are commonly used excitation spectra for epifluorescence. Furthermore, prolonged formalin fixation (a common practice in formalin fixed, paraffin embedded (FFPE) tissue sections) results in autofluorescence. Thus, fluorescent microscopy of routine neurosurgical biopsies would result in visually complex images. Our objective was to develop an automated image analysis technique that would overcome these challenges. We developed this technique by validating Poly-pyrimidine Tract-Binding Protein 1 (PTBP1) antibody in neuropathology specimens. We took advantage of the low autofluorescent background present in 300–500 nm spectrum of FFPE tissues, a naturally occurring idiosyncrasy of FFPE autofluorescence. Thus, staining with DAPI, which is characterized by a peak excitation/emission spectrum of 358 nm/461 nm, can be used as a tool to identify signals. While several histopathological image analysis methods exist in the literature [11–14], to the best of our knowledge, there is no work that uses multiplexed guided images with PTBP1 antibody with DAPI.
The main steps for the automated validation of anti-PTBP1 antibody are: 1) de-noising according to the type of noise in images, 2) local normalization with Sigma-Adaptive Gaussian (SAG) filtering, and 3) cell nuclei detection and segmentation with a k-means based deterministic approach. The contribution of this work is validating anti-PTBP1 antibody to use on routine neurosurgical biopsies by developing the computerized system that allows automated analysis of images, which removes fluorescent background from specimens stained with DAPI and anti-PTBP1 antibody. Quantitative evaluations are performed by using 29 data sets from reactive gliosis and three cases of brain tumors: 1) recurrent glioma, 2) primary resections of glioblastoma harboring the Epidermal Growth Factor Receptor (EGFRVIII) mutation, and 3) Pilocytic Astrocytoma (PA).
The remaining part of this paper is organized as follows: Description of data sets is given in Section II. Validation of anti-PTBP1 antibody is explained in Section III. Experimental results are shown in Section IV while conclusion and discussion is given in Section V.
2. DATA SETS
In this work, immunofluorescence images are used obtained from confocal photomicroscopy. The data sets are provided from 29 specimens (16 samples from patients with PA WHO Grade I, 4 samples of reactive gliosis from a patient with prior diagnosis of glioblastoma, 4 samples from recurrent high grade glioma in a patient with prior diagnosis of glioblastoma and 5 samples from a patient with EGFRVIII mutation positive glioblastoma) by using a laser scanning confocal fluorescence microscope (Zeiss LSM 700). All images were captured with a 20x objective since nuclei stained with/without anti-PTBP1 antibody or DAPI in these images can be seen clearly by pathologists. The pixel size is 0.312×0.312 micron2 (3.1991 pixels per micron) in the 8-bit uncompressed images of size of 2048×2048 pixels.
3. ANTIBODY VALIDATION BY IMAGE ANALYSIS
Due to the inherent variability amongst antibodies, quality control experiments are always required in diagnostic pathology [1–5]. Different studies suggest different proteins, which are likely suitable to use as glioma antibodies (such as ELTD1, F11R, LINGO1, SLIT3 etc.), with different validation approaches in the literature [6–11]. However, a method to quantitatively and objectively validate PTBP1 for gliomas (in human/animal) in formalin-fixed, paraffin embedded tissue evaluated by immunofluorescence confocal microscopy does not exist. Such automated testing is required to ultimately implement multiplexed image acquisition in solid-tissue biopsies. Therefore, we developed a methodology to validate PTBP1, a multifunctional RNA-binding protein, antibody using automated image analysis.
Automated validation of antibodies by image analysis is a very challenging task because of:
Tissue based problems: Cells may have irregular shapes/structures or may be clustered. Individual cells may not be distinguished easily.
Staining based problems: Cells may not stain well, which may cause incorrect number/shape/size of detected objects.
Antibody based problems: Quantity, quality and concentration of the antibody may be different in different data sets or even in different images of the same data set. These variations may cause unexpected/undesired results due to differences in images.
Microscope based problems: Quality of the camera and microscope being may be low. Therefore, images may be with low quality with unclear edges or noisy.
Target based problems: The variability of the target protein due to secondary modifications, localization, epitome availability etc.
Our approach is designed to overcome these problems by 1) noise modeling and reduction (Section 3.1), 2) intensity normalization (Section 3.2), 3) detection of nuclei using the images that are stained with DAPI (to get boundary and location information) and the images that are stained with anti-PTBP1 antibody (to get intensity values) (Section 3.3), and 4) segmentation of nuclei based on a deterministic approach (Section 3.4).
3.1. Noise Modeling and Reduction
There are different noise reduction methods, ranging from wavelet [12–15], iterative [16] and diffusion [17–21] methods to non-local mean [22–24] and graphical model frameworks [25–27] used for different types of noise e.g. uniform (Figure 1.a), Gaussian (Figure 1.b), impulsive (i.e., salt & pepper) (Figure 1.c) or speckle (Figure 1.d). To determine the most appropriate algorithm for noise reduction (i.e., to avoid poor de-noising), the type of noise in the image should be identified. Therefore, we chose three regions (at least 100×100 pixels), which have homogenous (or close to homogenous) intensity values, from images and analyzed their histograms to identify noise type.
Figure 1.
Noise types: Uniform noise (a); Gaussian noise (b); Impulsive noise (c); Speckle noise (d)
Figure 2 shows an example image stained with anti-PTBP1 antibody (Figure 2.a), three regions (with variance σ1 = 0.5, σ2 = 2.5 and σ3 = 1), which are surrounded with yellow, red and cyan rectangular, on the grayscale form of the image (Figure 2.b) and the histogram of the grayscale image (Figure 2.c). We have modeled the noise in the images as speckle noise [28] based on the shape of the histograms (Figure 2.d,e,f) of those three regions. Therefore, we implemented a speckle filtering algorithm to reduce the noise in our images.
Figure 2.
Image stained with anti-PTBP1 antibody (a); Three homogenous regions are shown with yellow, red and cyan rectangular on the grayscale form of the image (b); Histogram of the grayscale image (c); Histogram of the regions with yellow (d), red (e) and cyan (f) rectangular. (For better visualization the brightness and contrast has been increased by 40% in figures (a) and (b))
In our work, we reduce the noise by a Non-local Median based Speckle Filtering (NMSF), which is inspired by the Non-local Means Filtering (NMF) of Coupe et al. [29]. However, unlike the non-local means filtering, we used median function (non-linear filter) since the data in our images are non-linear and mean function may not always be the most efficient in this case.
The NMSF method (1) is applied when we consider a grayscale image u = (u(xi))xi∈Ω2, which is defined on a domain Ω2 ⊂ R2 and u(xi) ∈ R+ is the noisy at pixel xi ∈ Ω2 (please see [29] for details);
| (1) |
where, Bi refers to the square block that is centered at xi with size |Bi| = (2α + 1)2, α ∈ N; v (Bi) is the unobserved vector of true values of block Bi; u(Bi) is the vector gathering the gray level values of block Bi; NL(u)(Bi) is restored block of pixel xi; w(Bi, Bj) is the weight value used for restoring u(Bi) given u(Bj) and based on the similarity of blocks u(Bi) given u(Bj). The weight values are calculated by,
| (2) |
where u(Bi) = (u(1)(Bi),........u(p) (Bi))T is an image patch gathering the intensities of the block Bi; zik is a constant value for normalization by ensuring that ΣBj∈Δik w(Bik,Bj) = 1 and also
| (3) |
In the NMSF method, for each block Bik centered at pixel xik, a non-local means restoration is performed from blocks Bj. The restored value of the block Bik is the weighted average of all the blocks Bj in the search volume. For a pixel xi included in several blocks, several estimates are obtained and merged. The restored value of pixel xi is the mean of the different estimations stored in vector Ai. The algorithm can be summarized as:
Divide the image into blocks with overlapping supports (The block Bik contains P = 2α + 1 elements. Each block is centered on pixel xi, which constitutes a subset of Ω2 and pixels are equally distributed).
Calculate NL(u)(Bik) to perform a non-local means restoration of these blocks.
Restore the pixel intensities. For a pixel xi included in several blocks Bik, different estimates from several NL(u)(Bik) is computed and stored in a vector Ai. The restored intensity of pixel xi is the mean of the restored values NL(u)(Bik).
The noise reduction method is illustrated in Figure 3 with a Region of Interest (ROI) area, which is shown with the yellow rectangle in Figure 3.a. (The ROI part is magnified in Figure 3.b to increase visibility of individual nuclei). The ROI part after noise reduction (Figure 3.c), residual image (i.e. difference between the original and de-noised ROI area) (Figure 3.d) and the whole image after noise reduction (Figure 3.e) are shown as grayscale. To present better visualization, pseudo-color representation with the Hue-Saturation-Value (HSV) color model is given in Figure 3.f,g,h for the images shown in Figure 3.b,c,d respectively.
Figure 3.
Example grayscale image that shows ROI part as yellow rectangle on the image stained with DAPI but without anti-PTBP1 antibody from PA cases (a); ROI part magnified (b); ROI part after noise reduction (c); Residual image (d); The whole image after noise reduction (e); The ROI part, its de-noised form and the residual image are shown in HSV color space in (f),(g) and (h) respectively (To increase visualization in this figure (a–e), brightness and contrast has been increased (40%))
The effect of the noise reduction is demonstrated on a nucleus in Figure 4. The nucleus (Figure 4.a) in the original image has speckle and blurry edges, which are smoother (Figure 4.b) after de-noising. Difference between the original and de-noised image is shown in Figure 4.c.
Figure 4.
A nucleus in the original image (a); Shape and pattern of the nucleus after de-noising (b); Residual image data removed by the filtering (c)
The effect of noise reduction can be observed by comparing variance of 1) original image, 2) image de-noised with NMF, and 3) image de-noised with NMSF since there is no image obtained without noise in our work.
Figure 5 shows average variances, which are calculated from images in each data set, for three cases of brain tumor and reactive gliosis. It should be noted here that mean values of original images and de-noised images are the same although their variances change. Therefore, quantitative values in Figure 5 indicate that noise reduction with the NMSF method is more successful compared to the NMF method on our images.
Figure 5.
Effect of noise reduction methods is shown quantitatively for reactive gliosis (a), recurrent glioma (b), EGFRVIII (c), and PA (d)
3.2. Local Intensity Normalization with SAG Filtering
Another problem to tackle is the non-uniform staining of the images. Therefore, we applied a local intensity normalization method. The goal of the normalization process is to make the variance and mean of the grayscale images uniform. In our implementation, the spatial local normalization is performed by applying SAG filtering instead of a standard Gaussian filtering applied globally with a fixed sigma value.
We propose SAG filtering to solve two main problems of the traditional Gaussian smoothing: 1) It is not clear how to choose a fixed sigma that is fit to smooth each pixel with Gaussian filter. 2) Filtering with the fixed sigma (even if it is chosen carefully for an image) cannot smooth different images in different data sets (even different images in the same data set) well enough for the next step (Section 3.3).
Figure 6 illustrates a grayscale image (Figure 6.a) and normalized images obtained by standard Gaussian filtering applied globally with different sigma values (5 and 15) (Figure 6.b,c) to show variations.
Figure 6.
Grayscale image (a); Normalized image by Gaussian filtering with sigma 5 (b), and 15 (c) (to better visualize, the brightness has been increased by 40%)
The local SAG filtering based normalization is applied with the following equation:
| (4) |
where the term IN (x, y) and Ig (x, y) refers to the normalized and grayscale images, respectively. In (4), μ(x, y) and σ(x, y) correspond to the mean and variance that are estimated by filtering the image Ig (x, y) with a Gaussian filter whose sigma is selected adaptively for each image.
The adaptive sigma value of the Gaussian filter is calculated using the distance map obtained with Euclidean distance transform. To get the distance map, we use the binary image obtained from the grayscale image Ig (x, y) by applying the adaptive thresholding method described in [30].
The Euclidean distance transform is defined as a transformation, which assigns a number that is the distance between that pixel and the nearest zero pixel in the binary image (note that zero pixels correspond to background, non-zero pixels to nuclei in the binary image). Let maxD be the maximum value in the distance map, the adaptive sigma value of the Gaussian filter is obtained by:
| (5) |
where T is set to 100 experimentally. In (5), we used maxD since pixels with the maximum distance value should be smoothed as much as possible. However, inter-cell boundaries should not be blurred while simultaneously smoothing intra-cell pixels. Therefore, the threshold value T is used in (5) to handle over-smoothing problem.
Under-smoothing problem may occur when maxD is low; therefore, the sigma value is defined adaptively as follows:
| (6) |
where t is set to 50 experimentally. The pseudo-code of the proposed local SAG filtering based normalization method is given in Table 1.
Table 1.
Pseudo-code of the local SAG filtering based image normalization method
|
Figure 7 shows the image obtained with the proposed normalization approach from the image given in Figure 6.a.
Figure 7.

Normalized image
3.3. Nuclei Detection and Segmentation
In this stage, nuclei are detected and segmented on grayscale DAPI image by a k-means based deterministic method. The proposed method mimics the pathologist workflow. Positively stained nuclei in the images stained with anti-PTBP1 antibody (primary or secondary) appear brighter than other objects (Figure 8.a). Therefore, visual detection of these nuclei is performed according to their brightness. If a bright region in these images corresponds to a nucleus in the image stained with “DAPI and anti-PTBP1 antibody” or “DAPI but without anti-PTBP1 antibody” then the nucleus having the bright region is considered as a nucleus positively stained with anti-PTBP1 antibody by pathologists. Similarly, in our work, automated detection and segmentation of these nuclei stained positively is achieved by using i) intensity values from images stained with/without anti-PTBP1 antibody, and ii) location and shape of the nuclei from the images stained with “DAPI and anti-PTBP1 antibody” or “DAPI but without anti-PTBP1 antibody”.
Figure 8.
Example image stained with anti-PTBP1 antibody (primary) from PA (a); Image stained with DAPI from PA (b); Grayscale forms of the image stained with anti-PTBP1 antibody (c) and DAPI (d); Generated DAPI_PTBP1 image (e); Nuclei positively stained with anti-PTBP1 antibody (f) (To better visualize images in (a–c), brightness and contrast has been increased by 10%)
In the proposed method, the first step is to convert anti-PTBP1 (Figure 8.a) and DAPI stained tissue images (Figure 8.b) into grayscale (Figure 8.c, d) (these images are originally gray-scale but then artificially colored for better visualization). After noise reduction, a new image, called DAPI_PTBP1 (Figure 8.e), is generated by combining the shape information from the DAPI-stained image and texture information from the anti-PTBP1-stained image. In the final step, k-means based clustering is applied with k=6 [31–34]. The first cluster corresponds to background (black) and remaining five clusters categorize gray level values into five groups (dark gray, gray, bright, brighter and the brightest gray level value). The sixth cluster contains the nuclei that are positively stained with anti-PTBP1 antibody with the brightest gray level values (Figure 8.f).
3.4. Numerical Analyses of Segmented Nuclei
In order to measure the effect of the anti-PTBP1 antibody, the ratio between total numbers of nuclei segmented from the images stained with anti-PTBP1 antibody and images stained with “DAPI and anti-PTBP1 antibody” is compared to the ratio from the images stained without anti-PTBP1 antibody and images stained with “DAPI but without anti-PTBP1 antibody”. The following ratio is calculated:
| (7) |
where NwithPTBP1 refers to number of nuclei stained (positively) with anti-PTBP1 antibody, NwithDAPIandPTBP1 refers to number of nuclei stained (positively) with “DAPI and anti-PTBP1 antibody”. Similarly, the following ratio is calculated by using images stained with “DAPI but without anti-PTBP1 antibody” and images stained with anti-PTBP1 antibody:
| (8) |
where NnoPriPTBP1 refers to number of nuclei stained (positively) without anti-PTBP1 antibody, NwithDAPInoPriPTBP1 refers to number of nuclei stained (positively) with “DAPI but without anti-PTBP1 antibody”.
We can deduce that the antibody is effective, if the ratio calculated with (7) is greater than the ratio calculated with (8), which can be written as:
| (8) |
Figure 9 shows the flow chart of applied method for validation of anti-PTBP1 antibody.
Figure 9.
Flow chart of the applied method for validation of anti-PTBP1 antibody
4. EXPERIMENTAL RESULTS
Quantitative evaluations are based on the total number of nuclei that match bright regions in images with and without anti-PTBP1 antibody. Example numerical values are given in Table 2, which is obtained from the data set of 7 images from 1 patient diagnosed with reactive gliosis.
Table 2.
Total number of nuclei matching bright regions in images stained with and without anti-PTBP1 antibody on 7 images from 1 patient diagnosed with reactive gliosis.
| Image Number | Total Number of Nuclei That Match with Bright Regions in Images | |
|---|---|---|
| Image without anti-PTBP1 antibody | Image with anti-PTBP1 antibody | |
| 1 | 21 | 123 |
| 2 | 15 | 180 |
| 3 | 25 | 288 |
| 4 | 25 | 309 |
| 5 | 16 | 302 |
| 6 | 5 | 28 |
| 7 | 12 | 106 |
| Mean: | 17 | 190.9 |
| Standard deviation: | 7.2 | 111.2 |
Experimental results show that the total number of nuclei in images stained with anti-PTBP1 antibody is always more than those in the images stained without anti-PTBP1 antibody (p = 1×10−3 (t-test)).
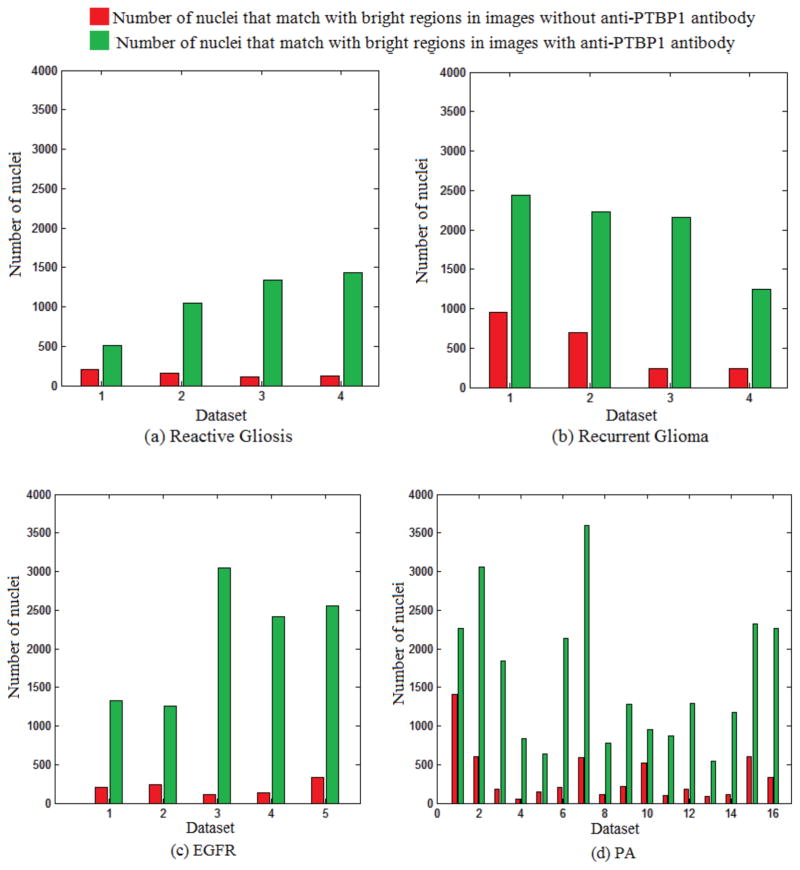
Figure 10 shows the total number of nuclei that match with bright regions in images stained with and without anti-PTBP1 antibody in data sets from recurrent glioma (Figure 10.a), reactive gliosis (Figure 10.b), EGFRVIII (Figure 10.c) and PA (Figure 10.d) data sets. The number of nuclei in images stained with anti-PTBP1 antibody is always higher than those in images stained without anti-PTBP1 antibody (Figure 10).
Figure 10.
Number of nuclei that match with bright regions in images with and without anti-PTBP1 antibody from reactive gliosis (a), recurrent glioma (b), EGFRVIII (c) and PA (d) data sets
The difference between numbers of nuclei is meaningful for those three cases of brain tumor and reactive gliosis. Table 3 shows p values (t-test), average (μ) and standard deviation (σ) of number of nuclei obtained from images stained with and without anti-PTBP1 antibody.
Table 3.
Result of t-test (p values), average (μ) and standard deviation (σ) of number of nuclei obtained from images stained with and without anti-PTBP1 antibody
| Case | p value | Images stained with anti-PTBP1 antibody | Images stained without anti-PTBP1 antibody | ||
|---|---|---|---|---|---|
| μ | σ | μ | σ | ||
| Reactive Gliosis | 40×10−4 | 1082.5 | 412.1 | 147.2 | 41.1 |
| Recurrent Glioma | 33×10−4 | 2019.2 | 527.4 | 531.2 | 351.8 |
| EGFRVIII | 6×10−4 | 2124 | 791.7 | 203.6 | 92.2 |
| PA | 46×10−3 | 1615.3 | 911.2 | 153.4 | 71.6 |
Quantitative values in Table 4 are obtained without noise reduction. Values of μ and σ given in Table 3 are lower than those μ and σ values in Table 4 due to the effect of the NMSF method.
Table 4.
P values (t-test), average (μ) and standard deviation (σ) of number of nuclei obtained from images without noise reduction
| Case | p value | Images stained with anti-PTBP1 antibody | Images stained without anti-PTBP1 antibody | ||
|---|---|---|---|---|---|
| μ | σ | μ | σ | ||
| Reactive Gliosis | 1×10−2 | 3321.5 | 1123.9 | 1188.7 | 476.1 |
| Recurrent Glioma | 1×10−3 | 3721.0 | 666.3 | 898.2 | 675.3 |
| EGFRVIII | 2×10−4 | 4113.2 | 1269.6 | 452.4 | 310.9 |
| PA | 3×10−4 | 2793.8 | 1191.7 | 981.7 | 552.9 |
Also, it is observed from numerical values of R WithPriAnti and R NoPriAnti that the anti-PTBP1 antibody works well (i.e., RWithPriAnti > RNoPriAnti, which can be seen in Figure 11).
Figure 11.
R WithPriAnti and R NoPriAnti values for each data set
5. CONCLUSION AND DISCUSSION
In this work, an automated image analysis method is proposed for validation of anti-PTBP1 antibody. The advantage of the proposed method is that it provides accurate, objective, repeatable and quantitative results. The method was tested on 29 data sets diagnosed with one of four cases: PA, WHO grade I, Second biopsy of a patient with a prior neurosurgical resection diagnosed as glioblastoma with the new resection showing reactive gliosis, Second biopsy of a patient with a prior neurosurgical resection diagnosed as glioblastoma with the new resection showing recurrent high grade glioma; and first surgery of a patient showing glioblastoma, WHO grade IV harboring EGFRVIII mutation.
The noise reduction method in this work is chosen according to the type of the noise in images. In our data sets, images show speckle type of noise (Figures 2.c, f). Therefore, the noise reduction approach is based on speckle filtering, which gives accurate results (Figures 3,4) because the numbers of nuclei detected with the proposed approach were confirmed by a neuropathologist who has more than 10 years of experience in traditional pathology. Figure 5 and Table 3 show the effect of noise reduction with the proposed NMSF technique.
Intensity normalization is provided by a local normalization approach. The proposed SAG filtering based normalization method assigns sigma value for each image automatically instead of using a constant sigma for the Gaussian filtering.
It is observed from numerical values in Table 2 and graphs in Figure 10 that images stained without anti-PTBP1 antibody always show less number of positively stained nuclei compared to images stained with anti-PTBP1 antibody. Comparisons of quantitative values show meaningful difference between those numbers of nuclei (Table 2). Ratios calculated using images stained with and without anti-PTBP1 and DAPI indicate the efficiency of the antibody (Figure 10). These data demonstrate and automated image analysis approach to antibody validation for PTBP1 that can easily be applied to other biomarkers showing nuclear localization.
The proposed approach will be tested on more data sets chosen from different cases in neuropathology as an extension of this work.
References
- 1.Puntmann V. How-to guide on biomarkers: biomarker definitions, validation and applications with examples from cardiovascular disease. Postgrad Med J. 2009;85(1008):538–545. doi: 10.1136/pgmj.2008.073759. [DOI] [PubMed] [Google Scholar]
- 2.Prensner JR, Chinnaiyan A, Srivastava S. Systematic, evidence-based discovery of biomarkers at the NCI. Clinic Exp Metastasis. 2012;29(7):645–652. doi: 10.1007/s10585-012-9507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pusztai L, Hatzis C, Andre F. Reproducibility of research and preclinical validation: problems and solutions. Nat Rev Clin Oncol. 2013;10(12):720–724. doi: 10.1038/nrclinonc.2013.171. [DOI] [PubMed] [Google Scholar]
- 4.Bucur O, Pennarun B, Stancu AL, Nadler M, Muraru MS, Bertomeu T, Khosravi FR. Poor antibody validation is a challenge in biomedical research: a case study for detection of c-FLIP. Apoptosis. 2013;18(10):1154–62. doi: 10.1007/s10495-013-0880-0. [DOI] [PubMed] [Google Scholar]
- 5.Begley C, Ellis L. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483(7391):531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 6.Towner RA, Jensen RL, Vaillant B, Colman H, Saunders D, Giles CB, Wren JD. Experimental validation of 5 in-silico predicted glioma biomarkers. Neuro-Oncology. 2013;15(12):1625–1634. doi: 10.1093/neuonc/not124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Towner RA, Jensen RL, Colman H, Vaillant B, Smith N, Casteel R, Saunders D, Gillespie DL, Mansat RS, Lupu F, Giles CB, Wren JD. ELTD1, a potential new biomarker for gliomas. Neurosurgery. 2013;72(1):77–91. doi: 10.1227/NEU.0b013e318276b29d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niclou SP, Fack F, Rajcevic U. Glioma proteomics: status and perspectives. J Proteomics. 2010;73(10):1823–1838. doi: 10.1016/j.jprot.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Deutsch EW, Ball CA, Berman JJ, Bova GS, Brazma A, Bumgarner RE, Campbell D, Causton HC, Christiansen JH, Daian F, Dauga D, Davidson DR, Gimenez G, Goo YA, Grimmond S, Henrich T, Herrmann BG, Johnson MH, Korb M, Mills JC, Oudes AJ, Parkinson HE, Pascal LE, Pollet N, Quackenbush J, Ramialison M, Ringwald M, Salgado D, Sansone SA, Sherlock G, Stoeckert CJ, Swedlow J, Taylor RC, Walashek L, Warford A, Wilkinson DG, Zhou Y, Zon YI, Liu AY, True YD. Minimum information specification for in situ hybridization and immunohistochemistry experiments (MISFISHIE) Nat Biotechnol. 2008;26(3):305–312. doi: 10.1038/nbt1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pradidarcheep W, Labruyere WT, Dabhoiwala NF, Lamers WH. Lack of specificity of commercially available antisera: better specifications needed. J Histochem Cytochem. 2008;56(12):1099–1111. doi: 10.1369/jhc.2008.952101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pong WW, Walker J, Wylie T, Magrini V, Luo J, Ryan J, et al. F11R is a novel monocyte prognostic biomarker for malignant glioma. PLoS ONE. 2013;8(10):e77571. doi: 10.1371/journal.pone.0077571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pizurica A, Philips W, Lemahieu I, Acheroy M. A versatile wavelet domain noise filtration technique for medical imaging. IEEE Trans Med Imag. 2003;22(3):323–331. doi: 10.1109/TMI.2003.809588. [DOI] [PubMed] [Google Scholar]
- 13.Luisier F, Vonesch C, Blu T, Unser M. Fast interscale wavelet denoising of poisson-corrupted images. Signal Processing. 2010;90(2):415–427. [Google Scholar]
- 14.Bal U. Dual tree complex wavelet transform based denoising of optical microscopy images. Biomed Opt Express. 2012;3(12):3231–3239. doi: 10.1364/BOE.3.003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B, Fadili J, Starck JL, Olivo-Marin JC. Multiscale variance-stabilizing transform for mixed-poisson-gaussian processes and its applications in bioimaging. IEEE International Conference on Image Processing (ICIP’07); San Antonio, TX. Sept 16-Oct 19 2007; 2007. pp. 233–236. [DOI] [Google Scholar]
- 16.Lanza A, Morigi S, Sgallari F, Wen YW. Image restoration with poisson-gaussian mixed noise. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization. 2014;2(1):12–24. [Google Scholar]
- 17.Perona P, Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell. 1990;12(7):629–639. [Google Scholar]
- 18.Ling J, Bovik AC. Smoothing low-snr molecular images via anisotropic median-diffusion. IEEE Trans Med Imag. 2002;21(4):377–384. doi: 10.1109/TMI.2002.1000261. [DOI] [PubMed] [Google Scholar]
- 19.Kroon DJ, Slump CH, Maal TJ. Optimized anisotropic rotational invariant diffusion scheme on cone-beam ct. Medical Image Computing and Computer-Assisted Intervention (MICCAI’10) Lecture Notes in Computer Science. 2010;6363(1):221–228. doi: 10.1007/978-3-642-15711-0_28. [DOI] [PubMed] [Google Scholar]
- 20.You YL, Kaveh M. Fourth-order partial differential equations for noise removal. IEEE Trans Image Process. 2000;9(10):1723–1730. doi: 10.1109/83.869184. [DOI] [PubMed] [Google Scholar]
- 21.Rajan J, Kannan K, Kaimal M. An improved hybrid model for molecular image denoising. J Math Imaging Vision. 2008;31(1):73–79. [Google Scholar]
- 22.Danielyan A, Wu YW, Shih PY, Dembitskaya Y, Semyanov A. Denoising of two-photon fluorescence images with block-matching 3d filtering. Methods. 2014;68(2):308–316. doi: 10.1016/j.ymeth.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Bindilatti AA, Mascarenhas ND. A nonlocal poisson denoising algorithm based on stochastic distances. IEEE Signal Process Lett. 2013;20(11):1010–1013. [Google Scholar]
- 24.Yang J, Fan J, Ai D, Wang X, Zheng Y, Tang S, Wang Y. Local statistics and non-local mean filter for speckle noise reduction in medical ultrasound image. Neurocomputing. 2016;195(1):88–95. doi: 10.1016/j.neucom.2015.05.140. [DOI] [Google Scholar]
- 25.Shafiee MJ, Haider SA, Wong A, Lui D, Cameron A, Modhafar A, Fieguth P, Haider MA. Apparent ultra-high b-value diffusion-weighted image reconstruction via hidden conditional random fields. IEEE Trans Med Imag. 2015;34(5):1111–1124. doi: 10.1109/TMI.2014.2376781. [DOI] [PubMed] [Google Scholar]
- 26.Sanches JM, Nascimento JC, Marques JS. Medical image noise reduction using the sylvester-lyapunov equation. IEEE Trans Image Process. 2008;17(9):1522–1539. doi: 10.1109/TIP.2008.2001398. [DOI] [PubMed] [Google Scholar]
- 27.Yoon JW. Statistical denoising scheme for single molecule fluorescence microscopic images. Biomed Signal Proces. 2014;10(1):11–20. [Google Scholar]
- 28.Martin P, Rothberg S. Introducing speckle noise maps for laser vibrometry. Optics and Lasers in Engineering. 2009;47(3):431–442. http://dx.doi.org/10.1016/j.optlaseng.2008.06.010. [Google Scholar]
- 29.Coupe P, Hellier P, Kervrann C, Barillot C. Nonlocal means-based speckle filtering for ultrasound images. IEEE Transactions on Image Processing. 2009;18(10):2221–2229. doi: 10.1109/TIP.2009.2024064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng ML, Tan YP. Contrast adaptive binarization of low quality document images. IEICE Electron Express. 2004;1(16):501–506. [Google Scholar]
- 31.Sonka M, Hlavac V, Boyle R. Image Processing, Analysis, and Machine Vision. 4. Cengage Learning, Publisher, Global Engineering; Stamfort, USA: 2000. Chapter 2.4-Color Images. [Google Scholar]
- 32.Ayech MW, Ziou D. Terahertz image segmentation using k-means clustering based on weighted feature learning and random pixel sampling. Neurocomputing. 2016;175(1):243–264. http://dx.doi.org/10.1016/j.neucom.2015.10.056. [Google Scholar]
- 33.Sarrafzadeh O, Dehnavi AM. Nucleus and cytoplasm segmentation in microspopic images using k-means clustering and region growing. Journal of Advanced Biomedical Research. 2015;4(1):174–184. doi: 10.4103/2277-9175.163998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frandsen PB, Calcott B, Mayer C, Lanfear R. Automatic selection of partitioning schemes for phylogenetic analyses using iterative k-means clustering of site rates. BMC Evolutionary Biology. 2015;15(1):13–39. doi: 10.1186/s12862-015-0283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]