Abstract
We aim to establish the effect of environmental diversity in evaluating nanotoxicity to bacteria. We assessed the toxicity of 4 nm polyallylamine hydrochloride-wrapped gold nanoparticles to a panel of bacteria from diverse environmental niches. The bacteria experienced a range of toxicities as evidenced by the different minimum bactericidal concentrations determined; the sensitivities of the bacteria was A. vinelandii = P. aeruginosa > S. oneidensis MR-4 > A. baylyi > S. oneidensis MR-1. Interactions between gold nanoparticles and molecular components of the cell wall were investigated by TEM, flow cytometry, and computational modeling. Binding results showed a general trend that bacteria with smooth LPS bind more PAH AuNPs than bacteria with rough LPS. Computational models reveal that PAH migrates to phosphate groups in the core of the LPS structure. Overall, our results demonstrate that simple interactions between nanoparticles and the bacterial cell wall cannot fully account for observed trends in toxicity, which points to the importance of establishing more comprehensive approaches for modeling environmental nanotoxicity.
Graphical abstract
Introduction
Due to the unique physicochemical properties that arise from their high surface-area-to-volume ratio, nanomaterials are increasingly used in consumer products. It is inevitable that, at some stage of the manufacturing, use, and disposal of such products, some of the nanomaterials they contain may be released into the environment. Therefore, there is a growing focus to understand the behavior of engineered nanomaterials in the environment and determine their potential environmental impacts.1,2 One can probe the potential environmental impact through the use of bacterial models, which, as decomposers, occupy an important trophic level; decomposers recycle nutrients that can be used by primary producers.3 Therefore, any effects on bacteria may impact organisms in other trophic levels, making bacteria a good diagnostic for overall environmental health. Often, only one bacterial model is used in nanoparticle toxicity studies,4–6 but this can lead to results that may not be generalizable across bacteria from different environments. Therefore, we have assembled a panel of Gram-negative bacteria with sequenced genomes that occupy different environmental niches for use in nanotoxicity studies.
When evaluating nanoparticle toxicity to bacteria, direct interactions of nanoparticles to the bacterial surface play a role in the toxicity, with several studies demonstrating a correlation between amount of NPs bound to bacteria and observed toxicities.7–10 It has been shown that bound NPs can rupture the bacterial cell membrane,11,12 lead to alterations in the membrane potential,13 release ions that are localized right at the bacterial surface,14 and generate reactive oxygen species at the cell membrane.15 In previous work, we have shown that a main component of the Gram-negative bacterial surface, lipopolysaccharides (LPS), are important in facilitating the binding of nanoparticles with the surface of the model bacterium, S. oneidensis MR-1.16 There are two broad classes of LPS, designated as either rough or smooth. Rough LPS have a lipid A region that anchors the LPS into the membrane and an oligosaccharide portion that is bound to the lipid A. By contrast, smooth LPS have both lipid A and core oligosaccharide regions, with the addition of an O-antigen, a polysaccharide domain bound to the core oligosaccharide, elongating the overall LPS structure. Based on the clear role of LPS in binding nanoparticles, it makes sense to generate a panel that focuses on variation in LPS structure. Such a panel would exclude Gram-positive bacteria, which are also important environmental organisms, but allows us to focus on specific surface chemistry differences between the bacteria used.
The five bacteria that make up the bacterial panel introduced in this manuscript include Azotobacter vinelandii UW, Acinetobacter baylyi ADP1, Shewanella oneidensis MR-1, Shewanella oneidensis MR-4, and Pseudomonas aeruginosa PAO1. They are a mix of Gramnegative bacteria that have smooth or rough LPS on their surface. Differences between the bacteria in the panel are highlighted in Table 1. In the environment, A. vinelandii has an important role in the nitrogen cycle since it is capable of fixing nitrogen even in the presence of atmospheric oxygen.17 The smooth LPS of A. baylyi likely has branched O-antigens since that is characteristic of the genus Acinetobacter;18 this bacterium is capable of great metabolic diversity, notably in its ability to metabolize aromatic compounds that are often products of plant degradation.19 S. oneidensis MR-1 has an important environmental role in geochemical nutrient cycling since it is capable of reducing a wide variety of metals.20 Similarly, S. oneidensis MR-4 is also capable of dissimilatory reduction of many different metals.21 Finally, P. aeruginosa PAO1 is an obligate aerobe that can adapt to live in many different environments due to its metabolic diversity. P. aeruginosa is often used as a biofilm formation model; biofilms may serve as a sink for NPs entering the environment, making it likely that P. aeruginosa would encounter nanomaterials that are released into the environment.22
Table 1.
The bacteria in the panel come from a range of habitats and have different respiration abilities. The panel includes bacteria with either rough or smooth LPS presented on their surface. Each bacterium in the panel has an important role in the environment.
| Bacterial Strain | Habitat | Respiration | LPS type | Environmental Role |
|---|---|---|---|---|
| Azotobacter vinelandii UW | Soil48 | Obligate aerobe48 | Smooth49 | Nitrogen cycle |
| Acinetobacter baylyi ADP1 | Soil, sediment, aquatic18 | Obligate aerobe18 | Smooth18 | Metabolize aromatic compounds |
| Shewanella oneidensis MR-1 | Soil, marine20 | Facultative anaerobe20 | Rough25 | Geochemical nutrient cycle |
| Shewanella oneidensis MR-4 | Soil, marine20 | Facultative anaerobe20 | Rough21 | Geochemical nutrient cycle |
| Pseudomonas aeruginosa PAO1 | Ubiquitous50 | Obligate aerobe50 | Smooth51 | Metabolic diversity |
Since these bacteria occupy diverse environmental niches and have different surface compositions, we expect that they will be representative of bacteria in the environments that nanoparticles may end up in. These differences should also make them suitable for showing a range of responses to nanoparticle exposure so that a particular nanoparticle is not deemed non-toxic because a single bacterial species happens to be tolerant to it. The motivation for noting the different LPS structures on each bacterial species’ cell surface is that the differences in LPS length and composition may impact their interaction with the NPs used in this study. The saccharide portions of the LPS structures of P. aeruginosa,23,24 S. oneidensis MR-1,25 and S. oneidensis MR-421 have been elucidated (Fig. S1), but the LPS structures of the other bacteria in the panel are not as well characterized. The charges of these LPS structures as well as other LPS structures used in simulations in this manuscript are given (Table 2).
Table 2.
The number of phosphates and charge of the LPS core polysaccharide structure are provided for several bacterial strain variants (first three rows) and model structures (remaining four rows). O-antigens were entirely absent in all rough model structures and only two O-antigens are included in the “smoother” structure. The PAH used in the simulations has a charge of +10 leading to the total charges listed in the final column. Negative and positive charges were neutralized in the simulations using the corresponding number of sodium cations or chloride anions, respectively.
| Bacterial Strain | Number of Phosphate Units | Charge of LPS | Charge of LPS + PAH |
|---|---|---|---|
| S. oneidensis MR-1 | 5 | −7 | +3 |
| S. oneidensis MR-4 variant 1 | 4 | −6 | +4 |
| S. oneidensis MR-4 variant 2 | 6 | −5 | +5 |
| Rough P. aeruginosa PAO1 variant | 6 | −12 | −2 |
| “Smoother” P. aeruginosa PAO1 variant | 6 | −14 | −4 |
| Rough S. typhimurium | 4 | −10 | 0 |
| Rough E. coli | 5 | −9 | +1 |
To demonstrate the use of the bacterial panel in this manuscript, each species is exposed to 4-nm-diameter polyallylamine hydrochloride (PAH)-coated gold nanoparticles (AuNPs). AuNPs were used in this study due to their chemical inertness, size/shape tunability, and ease of characterization.26,27 Polyelectrolyte coating is an industrially-relevant modification of materials as these functionalized materials have many applications in diverse fields,28–30 and this particular coating is known to interact with bacterial surfaces and cause membrane disruption as its toxicity mechanism.7,31 While there is limited work studying PAH AuNPs with the bacteria in this panel,32 in previous work, the toxicity of PAH AuNPs was investigated for S. oneidensis MR-1 using a colony counting method.7 In this manuscript, the effects of the PAH AuNPs to each bacterium are noted by determining the minimum bactericidal concentration (MBC). The MBC is defined as the lowest nanoparticle concentration that kills at least 99% of the bacteria. This is a facile method to demonstrate the different responses that each bacterium has to NP exposure. To test the hypothesis that extent of NP binding to the bacterial cell surface correlates with toxicity, transmission electron microscopy (TEM) is used to visualize and flow cytometry is used to quantify PAH AuNP binding to each species. In parallel, a molecular dynamics simulation was used to calculate relative association energies of the PAH that caps the nanoparticles for different LPS structures to determine if the presentation of negatively charged phosphate groups facilitates the interaction of LPS with the amine groups of PAH. Taken together, the results demonstrate that a simple hypothesis related to the molecular character of the LPS is not sufficient to explain the nanoparticle association and toxicity results. This is a benefit to using such a panel, as it identifies instances where the biological complexity can mask simple, expected trends. Indeed, this panel does reveal which bacterial strains are most critical for follow-on work and facilitates the formulation of further hypotheses. While AuNPs are the focus of this work, this panel can be adapted for use with a range of nanomaterials.
Materials and Methods
Materials
Magnesium sulfate (MgSO4), sucrose, sodium molybdate dihydrate (Na2MoO4·2H2O), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), gold (III) chloride trihydrate (HAuCl4·3H2O), sodium citrate tribasic dihydrate (C6H5Na3O7·2H2O), polyallylamine hydrochloride (PAH, MW 17.5 kDa), sodium borohydride (NaBH4), and sodium chloride (NaCl) were purchased from Sigma-Aldrich (Milwaukee, WI). Potassium phosphate dibasic trihydrate (K2HPO4·3H2O) was purchased from Mallinckrodt (Phillipsburg, NJ). Potassium phosphate monobasic (KH2PO4) was obtained from J.T. Baker (Center Valley, PA). Dulbecco’s phosphate-buffered saline was purchased from Corning (Aurora, CO), LB broth and agar were obtained from BD Difco (Franklin Lakes, NJ). SYTO9 nucleic acid stain was obtained from Molecular Probes (Waltham, MA). Calcium chloride (CaCl2) and ferrous sulfate (FeSO4) were purchased from Fisher Scientific (Rockford, IL). Absolute anhydrous 99.5% ethanol was obtained from Pharmco-Aaper (Brookfield, CT). All chemicals were used as received. Deionized water (18.2 MΩ) was purified using a Milli-Q Millipore water purification system (Billerica, MA).
Shewanella oneidensis
MR-1 was obtained from Jeffrey Gralnick (Dept of Microbiology, University of Minnesota). Shewanella oneidensis MR-4 was obtained from Daad Saffarini (Dept of Biological Sciences, University of Wisconsin - Milwaukee). Acinetobacter baylyi (ATCC® 33305™), Azotobacter vinelandii (ATCC® 13705™), and Pseudomonas aeruginosa (ATCC® 47085™) were purchased from the American Type Culture Collection (Manassas, VA).
PAH-coated AuNPs (PAH AuNPs)
PAH AuNPs were synthesized by polyelectrolyte wrapping of as-synthesized citrate-capped AuNPs (see ESI for synthesis of citrate-capped AuNPs).33 To the approximately 3.2 L of as-synthesized citrate-capped AuNPs, 32.0 mL of 100 mM NaCl and 100.0 mL of a PAH solution (MW 17.5 kDa) (10 mg/mL in 1 mM NaCl) was added with vigorous stirring. The solution was stirred overnight and concentrated to around 30 mL by diafiltration cassettes (Tangential Flow Filtration Capsules, 50K MWCO, VWR). The concentrated PAH AuNPs were purified by centrifugation at 13,000×g for 55 min.
AuNP characterization
PAH AuNPs were characterized post-synthesis by UV-vis extinction spectroscopy, TEM, and ζ-potential measurement. The size and ζ-potential for the PAH AuNPs were also characterized in the exposure medium using UV-vis extinction spectroscopy and ζ-potential measurement. UV-vis extinction spectra post-synthesis were obtained on a Cary 500 UV-vis-NIR spectrophotometer and the measurements taken in exposure medium were obtained on an Ocean Optics USB2000 spectrophotometer. For TEM studies, 5 μL of a dilute solution of AuNPs was drop-cast onto a TEM grid (Ted Pella, Redding, CA), and the AuNP sample images were taken with a JEOL 2100 TEM. ζ-potential measurements were obtained using a Brookhaven ZetaPALS instrument.
Bacterial Culture Conditions
Bacteria were stored at −80 °C until ready for use. For S. oneidensis MR-1, S. oneidensis MR-4, A. baylyi, and P. aeruginosa, the appropriate bacterial stock was plated on a sterilized Luria-Bertani (LB) agar plate and incubated at 30 °C. Two colonies were inoculated in 10 mL of LB broth and incubated overnight. For A. vinelandii, plates with Burk’s medium adapted from Newton, et al34 were used and two colonies were inoculated in 10 mL of Burk’s medium. The bacteria at late log phase were centrifuged at 750×g for 10 min and washed with Dulbecco’s phosphate-buffered saline (DPBS) before resuspension in HEPES buffer (2 mM HEPES, 25 mM NaCl, pH=7.4) to the appropriate cell density.
Minimum Bactericidal Concentration Determination
The cells were diluted in HEPES buffer to a cell density of 2×106 cells/mL. Cells were either exposed to PAH-AuNPs (2.81, 0.28, or 0.028 ppm) or to free PAH (21.16, 2.116, or 0.2116 ppm) for 10 minutes by mixing 180 μL of bacterial suspension with 20 μL of PAH AuNP or free PAH suspension. Free PAH controls were performed since the PAH AuNP suspension contained free PAH that was left over from the wrapping process. The concentrations of free PAH present in the AuNP suspensions were determined using a fluorescamine assay as previously described (see ESI for fluorescamine assay experimental details)35 and were used to distinguish the impact of free polyelectrolyte from polyelectrolyte presented on the AuNP surface. Following the 10 minute exposure, six 10 μL drops of each treatment were dropped onto a dried, UV-sterilized LB agar plate (Burk’s medium plates were used for A. vinelandii). Once the drops absorbed into the agar, the plates were incubated upside-down in a 30 °C incubator overnight. The PAH AuNP and free PAH concentration that killed at least 99% of the bacteria were recorded.
Transmission Electron Microscopy Analysis
Before taking images of nanoparticle-exposed bacteria with the TEM, the samples had to be embedded in epoxy resin.7,36 At an optical density of 0.8 in HEPES, the bacteria were exposed to 0.281 ppm PAH AuNPs for 10 min and then washed three times in 0.1 M cacodylate buffer. The cells were fixed using 2.5% glutaraldehyde in 0.1 M cacodylate buffer. This step proceeds for 50 min, flipping the pellet after 25 min to ensure fixation. The pellet is then washed (without resuspension) three times in 0.1 M cacodylate buffer.
To dehydrate the cells, ethanol was used at increasing concentrations in water (30%, 50%, 70%, 80%, 95%, and 100% ethanol). They were rinsed three times with propylene oxide before being incubated with a 2:1 propylene oxide:resin mix for 2 hours uncovered. Then they were incubated with 1:1 propylene oxide:resin overnight followed by 1:1 propylene oxide:resin for 4 hours and pure resin overnight. After replacement with fresh resin, the samples were incubated at 40 °C for 24 hours and then 60 °C for 48 hours. The samples were cut into ~70-nm-thick sections using a LEICA EM UC6 ultramicrotome and stained with uranyl acetate and lead citrate to improve image contrast. The sections were placed on 200 mesh copper grids that have Formvar and carbon supports, and images were taken using a Tecnai T12 transmission electron microscope using an operating voltage of 120 kV.
Flow Cytometry Analysis
After the bacteria reached the late log phase, they were washed in DPBS and resuspended in HEPES buffer. Bacterial suspensions at 4 ×108 cells/mL were exposed to 2.81 ppm PAH AuNPs for 10 min and then incubated with 3.34 mM SYTO9 dye at room temperature for 15 min. The samples were analysed with a Becton Dickenson LSRII SORP flow cytometer with a 20 mW, 488 nm laser, using a control set of bacteria not exposed to NPs to draw the gates. SYTO9 fluorescence was used to distinguish bacteria from other debris in the sample, and light scattering was used to determine which bacteria had associated AuNPs. In total, each sample was done in triplicate, collecting 20,000 events in each run.
Computational Models and Simulation
The use of molecular dynamics simulations to characterize the large-scale association and relaxation of LPS with the PAH presented on the AuNP surface can yield insight into the underlying chemistry by resolving which sites, or sets of sites, induce interactions.37,38 Representative model structures of the LPS molecules found on the surface of P. aeruginosa, Salmonella typhimurium, and Escherichia coli have been prepared. While these three structures are not perfect matches for the bacteria in this panel (largely because not all the LPS structures are well-known), they represent a range of LPS structures and should still yield insight about critical interaction features, since they exhibit different overall charges and have differing numbers of phosphate units in their structure. Several force fields have been developed for prediction of interfacial properties of biological materials and their interactions with inorganic and organic nanostructures.39–42 We employed the CHARMM36 force field because it is a transferable potential that has been widely used and benchmarked, and it presented no challenges to the numerical convergence in the current studies. These structures have been energy minimized and equilibrated in the presence of 13,000 – 25,000 explicit (TIP3P) water molecules (depending on the size of the system) through equations of motion driven by the CHARMM36 force field. The mixture of PAH, LPS molecules, and water is neutralized through the addition of counter-ions that corresponds to the number of sodium cations or chloride anions needed to neutralize the “Charge of LPS + PAH” column of Table 2. For simplicity, we use a 10-mer PAH construct as it provides a balance between the non-chain like monomer and computationally expensive long-chain polymers with hundreds or more monomeric units. The chemical and molecular structure of the selected LPS have been obtained from known properties of the LPS from the chosen bacteria listed above. Each trajectory was then propagated for 19–20 nanoseconds at a cost of 65–75 hours of computer time on the XSEDE Bridges regular memory nodes with 2.3 GHz Intel Xeon EP-Series CPUs and 128 GB memory per CPU. In all cases, the PAH approaches the LPS molecule, allowing us to consider the time to approach and the location of the approach as figures of merit (or observables).
A computational study was performed to investigate PAH binding using a rough LPS model and a LPS construct with “smoother” character. The LPS models used here vary according to the corresponding incorporation of O-antigens. The addition of two O-antigen units to the rough P. aeruginosa LPS was constructed using the CHARM-GUI43 and models a smoother construct (Fig. 1) useful for our systematic study. The interaction between PAH and the LPS of P. aeruginosa, E. coli, and S. typhimurium (structures shown in Fig. S5), was observed through similar molecular dynamics trajectories.
Figure 1.
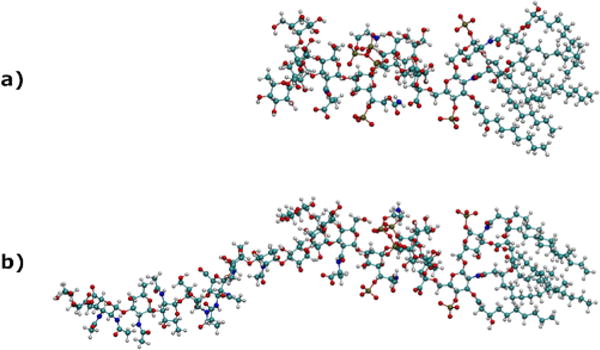
Ball-and-stick renderings of representative structures of a) rough LPS and b) rough LPS with two added O-antigen units onto the P. aeruginosa LPS structure.
The strength of the association of PAH to each LPS was evaluated from trajectory simulations to better understand specific interactions between the polyelectrolytes and LPS with different structures. Specifically, the changes in the interaction energies between the LPS and PAH are calculated after the complete simulation trajectories are obtained. The effects of the water molecules and ions on these energies are subtracted. Consequently, the reported energies include only the contributions from the interaction between the polyelectrolytes and LPS molecules.
Results and Discussion
Synthesis and Characterization of AuNPs
We first verified the size of the citrate-capped AuNPs before and after functionalization with PAH using several methods. Citrate-capped AuNPs in solution were validated after synthesis by UV-Vis spectroscopy44 (Fig. S2), which demonstrated that nanoparticle diameter was approx. 4 nm. After functionalization with PAH, TEM analysis indicated that the PAH AuNPs possessed a core diameter of 4.2 ± 1.2 nm (n>200), and a representative TEM image is shown (Fig. 2). The ζ-potential of the PAH AuNPs was 46.59 ± 2.63 mV. Taken together, these results demonstrate that positively charged PAH AuNPs were synthesized with uniform size distribution. In the exposure medium, the size of the PAH AuNPs were determined to be 12 ± 2 nm by UV-vis extinction spectroscopy, indicating there was some affiliation of the nanoparticles to each other during the exposure. The ζ-potential of the PAH AuNPs was 34 ± 2 mV, which shows a slight reduction in ζ-potential in HEPES buffer, but the magnitude is large enough for the particles to remain stable in suspension.
Figure 2.
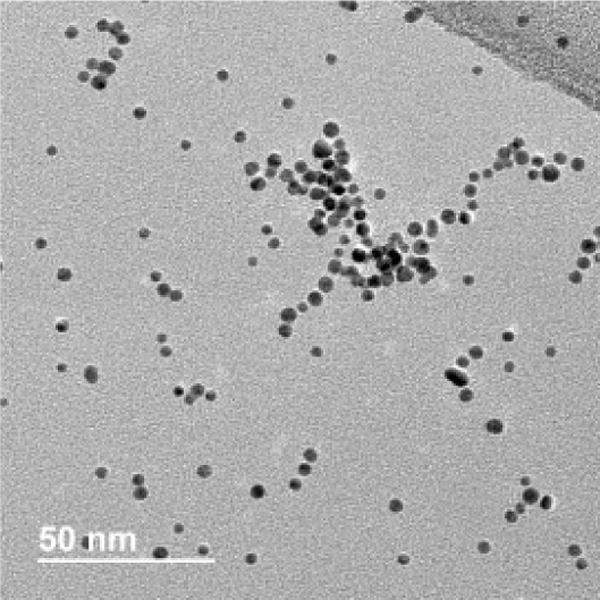
A representative TEM image of PAH AuNPs. Core diameter was determined to be 4.2 ± 1.2 nm (n>200).
Minimum Bactericidal Concentration (MBC) Determination
To test their toxicity, the MBCs were determined for PAH AuNPs and free PAH (Table 3). Each of the bacteria had different sensitivities to the toxicants used in this study. The sensitivities of each bacterium to PAH AuNPs are A. vinelandii = P. aeruginosa > S. oneidensis MR-4 > A. baylyi > S. oneidensis MR-1, which was tolerant to all concentrations of PAH AuNPs used in this study. The trend identified by MBC shows that the toxicity cannot be as easily predicted simply based on the smooth or rough character of bacterial LPS as originally hypothesized, since the observed sensitivities do not follow a pattern correlated with LPS type. For three of the bacteria (A. baylyi, S. oneidensis MR-1, S. oneidensis MR-4), the toxicity of the PAH AuNPs is explained by the free PAH that is present in those suspensions, which is consistent with previous findings.35 For A. vinelandii and P. aeruginosa, a nanospecific effect is seen for the PAH AuNPs since the concentration of free PAH required to kill 99% of these bacteria was higher than the concentration present in the toxic PAH AuNP suspensions. The wide range of sensitivities to both PAH AuNPs and PAH demonstrates the importance of using a bacterial panel when assessing nanoparticle properties that impact toxicity. From this experiment, the two bacteria exhibiting a nanoparticle effect have been identified to be used for follow-on work to study the mechanism of toxicity for this nanoparticle type, whereas the other three bacteria were merely experiencing the toxicity of free PAH.
Table 3.
Minimum bactericidal concentration values observed for each bacterium after exposure to PAH AuNPs and to free PAH. The value in parentheses after the MBC for PAH AuNPs indicates the amount of free PAH present in that concentration of PAH AuNPs as determined by the fluorescamine assay.35 (Ex: 0.281 ppm of PAH AuNPs contains a free PAH concentration of 2.12 ppm). The MBCs were determined in at least triplicate measurements.
| Bacteria | MBCPAH AuNPs (ppm) | Free PAH present at PAH AuNP MBC concentration (ppm) | MBCFree PAH (ppm) |
|---|---|---|---|
| A. vinelandii UW | ≤0.0281 | ≤0.212 | 2.12 |
| P. aeruginosa PAO1 | ≤0.0281 | ≤0.212 | >21.2 |
| S. oneidensis MR-4 | 0.281 | 2.12 | 2.12 |
| A. baylyi ADP1 | 2.09 | 15.8 | 14.8 |
| S. oneidensis MR-1 | ≥2.81 | ≥21.2 | ≥21.2 |
Nanoparticle Association with Bacteria
Post-exposure to PAH AuNPs, the binding was visualized using TEM, with examples where binding is visible shown in Fig. 3a–e. Dark field TEM was utilized to confirm the presence of the diffracting AuNPs as described previously (see ESI for dark field TEM images).45 The images show that bound PAH AuNPs cover just a small area of the bacterial cell envelope, with upwards of tens of nanoparticles bound to any particular bacterium. The binding was quantified using flow cytometry (Fig. 3f–g). Based on flow cytometry data, at 2.81 ppm PAH AuNP, binding was seen for all bacteria except S. oneidensis MR-4, which showed minimal binding. The order of binding from greatest to least is A. vinelandii (12 ± 1%), A. baylyi (6.2 ± 0.8%), P. aeruginosa (5.3 ± 0.7%), S. oneidensis MR-1 (4.2 ± 0.5%), and S. oneidensis MR-4 (0.3 ± 0.1%). These data show that, in general, the bacteria with smooth LPS exhibit higher AuNP binding than those with rough LPS, although for P. aeruginosa and S. oneidensis MR-1, these binding amounts are very similar. We speculate that this is because the O-antigen of smooth LPS generally has a larger number of negatively-charged sites for cationic nanoparticles to interact with than rough LPS.46
Figure 3.
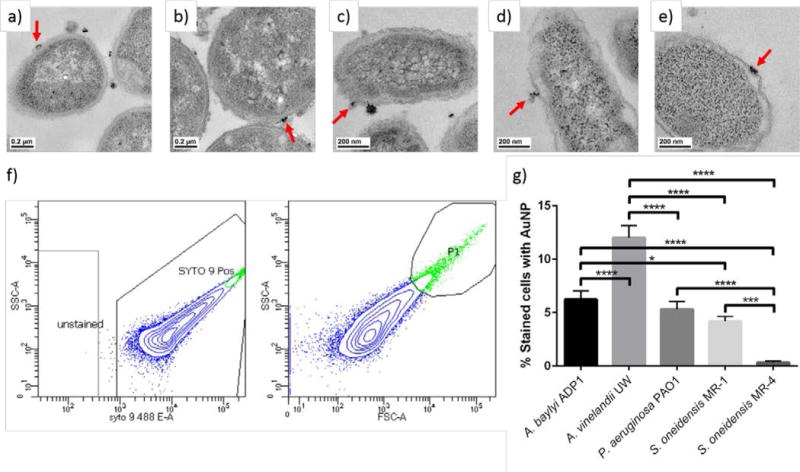
Transmission electron micrographs showing association of PAH AuNPs with a) A. baylyi ADP1, b) A. vinelandii UW, c) P. aeruginosa PAO1, d) S. oneidensis MR-1, and e) S. oneidensis MR-4. The red arrows show an example of PAH AuNP attachment to the bacterial cell wall. Representative flow cytometry data for f) A. baylyi exposed to 2.81 ppm PAH AuNPs. The left plot was used to identify cells based on the presence of SYTO9 stain, which is the boxed region of events labeled “SYTO 9 Pos”. The right plot contains only the cells present in the boxed region of the left plot, and the events with both high side scattering and forward scattering were the population of cells with bound AuNPs. This gate was drawn using the maximum scattering seen in cells that were not exposed to PAH AuNPs. The blue dots correspond to cells stained with SYTO9 and the green events are stained bacterial cells that are bound to AuNPs. From flow cytometry, the percentage of cells that were bound to AuNPs are shown for the bacterial species after exposure to g) 2.81 ppm PAH AuNPs. *p<0.05, ***p<0.001, and ****p<0.0001.
Comparing the binding data with toxicity data reveals some interesting observations. Namely, there are instances where there are similar binding amounts but different observed toxicities (i.e. P. aeruginosa and S. oneidensis MR-1) and where there is similar toxicity but a different amount of binding (A. vinelandii and P. aeruginosa). It is often assumed that direct nanoparticle interactions with the cell envelope drive toxicity, and these simple relationships between molecular components of the cell wall and nanoparticle properties can and have been identified.7 Once you start incorporating a wider range of organisms, some of these simple relationships start to be masked by an increasing biological complexity. This indicates that there are more complex mechanisms involved in this interaction, and identifying these other mechanisms will be important for each nanoparticle/bacterial interaction and can lead to insight into that biological complexity.
Computational model results
While the experiments performed cannot be done at the same time-scale as the computational models, simulations are used here to derive some molecular-level insight about the interaction between the PAH on the nanoparticle surface (modeled as a 10-mer) and LPS. Representative snapshots of the motion of PAH toward LPS (from P. aeruginosa) are shown in Fig. 4. In the first simulation, the electrostatic association energies of PAH with rough LPS extracted from P. aeruginosa, S. typhimurium, and E. coli are monitored. The configuration of these rough LPS are shown in Fig. S5. The number of phosphates in the core region of the LPS from S. typhimurium is one unit less than that of E. coli and two units less than the P. aeruginosa LPS structure, allowing consideration of the impact of the core phosphate on association with PAH. Overall charge differences between the structures also allows for consideration of the impact of charge.
Figure 4.
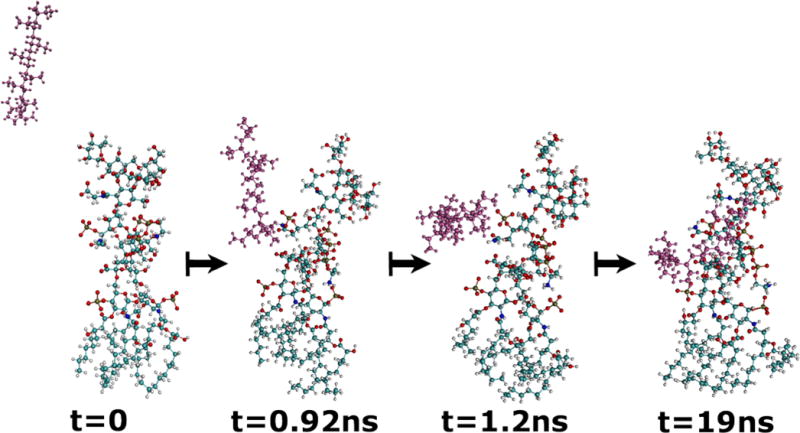
Snapshots of LPS from P. aeruginosa (in color-coded atoms) and PAH (with atoms in magenta) during a 19 ns simulation of the two molecules in explicit water (not shown). The PAH can be seen migrating toward the phosphate groups of the LPS.
The electrostatic association energies are shown in Fig. 5. The PAH molecule was seen to move towards phosphate units of the core region in the trajectories; the distances between the phosphorus atoms and PAH through the 19 ns simulation are shown in the ESI (Figs. S6–S8). These graphs suggest that the total charge of the LPS and the number of the phosphate units are important parameters in determining PAH association. The E. coli LPS has one unit less total negative charge compared to that of S. typhimurium (Table 2). Consequently, we observe a slower association of PAH to E. coli LPS. However, the E. coli LPS has one phosphate unit more than the LPS of S. typhimurium. The combination of these two competing factors is one possible reason for the observation of nearly equal association energy values at the end of the simulations for E. coli and S. typhimurium LPS. On the other hand, the LPS of P. aeruginosa has a higher negative charge and more phosphate units, leading to a quicker association of PAH to LPS that is also stronger at the end of the simulation than the other two bacteria.
Figure 5.
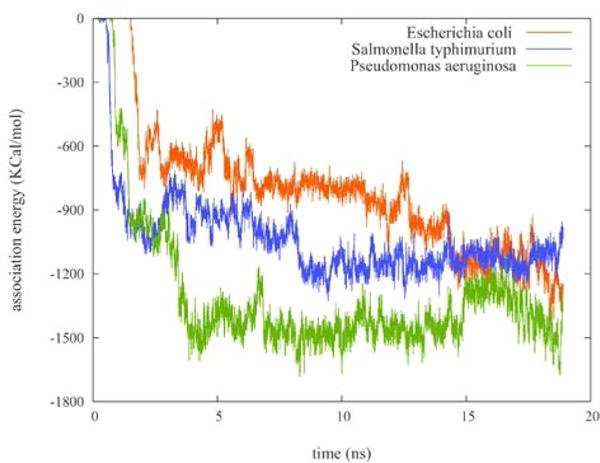
Electrostatic energies of the LPS/PAH association during a 19 ns simulation of 10-mer PAH interacting with the rough LPS of P. aeruginosa (green), E. coli (red), and S. typhimurium (blue) shown in Fig. S5.
We also investigated the different electrostatic association energies of PAH with increasingly smooth LPS character. The degree of PAH association to the rough LPS-exhibiting P. aeruginosa was compared to that of a smoother construct of P. aeruginosa with two added O-antigen units and is shown in Fig. 6. The LPS structure of P. aeruginosa with two added O-antigen units has a total charge of -14. The changes in the LPS/PAH electrostatic association energies and the distance between a selected phosphorus atom and the PAH center of mass are shown in Fig. 6. The traces in Fig. 6 suggest that PAH associates to the O-antigen sections of the smoother LPS molecule at early stages of the simulation and finally moves toward the phosphate units in the core region. Moreover, the addition of two O-antigen units does not significantly slow down the overall movement of PAH towards the core region of LPS. Typically, smooth LPS has many more than the two repeats of O-antigen monomer used here. In the case of the B-band in P. aeruginosa, for example, there can be greater than 50 repeats. Future computational work will build toward this more complex LPS structure. Since the B-band LPS structure of P. aeruginosa PAO1 has many more negatively charged sites—because many of its sugars are amino-derivatized uronic acid or fucose moieties,46—this band is expected to offer more binding sites for cationic particles than the more hydrophobic A-band. Therefore, smooth LPS should have many more binding sites that extend further into solution than those of rough LPS. This is likely why we see more binding to the bacteria with smooth LPS in our experimental work.
Figure 6.
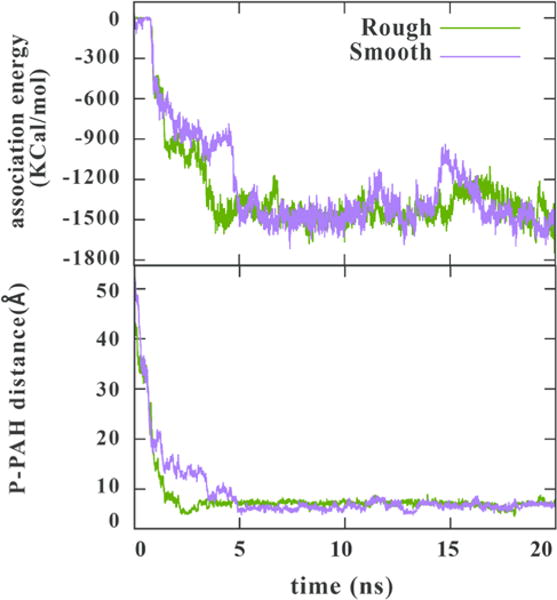
Electrostatic energies of the LPS/PAH association (top) and the distance between a selected phosphorus on LPS to the PAH center of mass (bottom) during a 20 ns simulation of 10-mer PAH interacting with the rough (green) and smoother (yellow) LPS constructs from P. aeruginosa shown in Fig. 2. (Note that the electrostatic energy trace for rough LPS is recapitulated from Fig. 5 to facilitate interpretation.)
Conclusions
This manuscript exploits a set of bacteria which represent a diverse array of environments that nanoparticles may be released into. These bacteria also have important ecological roles, making any effects felt by them impactful on overall environmental health. We demonstrated the use of this bacterial panel in monitoring the toxicity of a model nanoparticle, PAH AuNPs. While we observed increased PAH AuNP binding for bacteria with smooth LPS compared to those with rough LPS, the resulting toxicity did not follow this same trend. We expected that the toxicity observed would correlate with the binding of these NPs to the bacteria, a process mediated by the bacterium’s LPS, which is the major surface structure, making up 75% of the Gram-negative bacterial surface for some bacteria.47 In reality, the situation is more complex, which demonstrates the importance of using a bacterial panel for nanotoxicity studies. Regardless of the care in controlling for many variables, biology can introduce complexity to otherwise simple relationships.
The increased complexity of the biological panel presented here can be used for several applications. Due to the different LPS present on the surface of these bacteria, this panel is a good candidate for investigating bacterial surfaces. Indeed, the results obtained from the molecular dynamics simulation yield early insight into the interactions of polyelectrolyte-wrapped NPs with bacteria by taking the sugar sequences of the LPS into account. This panel is also good for an initial screen of nanoparticle toxicity; in using this bacterial panel, we can identify which bacteria are experiencing an effect specific to nanoparticles, which merit further investigation. While we would expect different results than those presented here if different NPs were used, this Gram-negative bacterial panel can be adapted for use with a range of nanomaterials.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation under the Center for Sustainable Nanotechnology, CHE-1503408. J.T.B. was supported by the University of Minnesota Biotechnology Training Grant Program through the National Institutes of Health. K.M.L. was supported by a University of Minnesota UROP award. We are grateful to Tian (Autumn) Qiu for her work with the fluorescamine assay. The authors gratefully acknowledge Dr. Michael P. Schwartz, Dr. Joel A. Pedersen, and Dr. Franz M. Geiger for helpful discussion. The TEM work in this study was carried out in the Characterization Facility, University of Minnesota, which receives partial support from NSF through the MRSEC program. We thank Fang Zhou for microtome sectioning of the resin-embedded bacteria samples for TEM analysis. The authors are grateful for the University of Minnesota’s University Flow Cytometry Resource for flow cytometric analysis.
The computing resources necessary for this research were provided in part by the National Science Foundation through XSEDE resources on Bridges under grant number TG-CTS090079. Additional computing resources were provided by the Maryland Advanced Research Computing Center (MARCC).
Footnotes
Electronic Supplementary Information (ESI) available: Known LPS structures from the bacterial panel, citrate-capped AuNP synthesis, quantification of free PAH, TEM of PAH AuNP binding to bacteria, flow cytometry gating method and supplemental data, and simulation of distance between LPS phosphorus and PAH center of mass. See DOI: 10.1039/x0xx00000x
References
- 1.Murphy CJ, Vartanian AM, Geiger FM, Hamers RJ, Pedersen J, Cui Q, Haynes CL, Carlson EE, Hernandez R, Klaper RD, Orr G, Rosenzweig Z. ACS Cent Sci. 2015;1:117–123. doi: 10.1021/acscentsci.5b00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eigenheer R, Castellanos ER, Nakamoto MY, Gerner KT, Lampe AM, Wheeler KE. Environ Sci Nano. 2014;1:238–247. [Google Scholar]
- 3.Naeem S, Hahn DR, Schuurman G. Nature. 2000;403:762–764. doi: 10.1038/35001568. [DOI] [PubMed] [Google Scholar]
- 4.Borcherding J, Baltrusaitis J, Chen H, Stebounova L, Wu CM, Rubasinghege G, Mudunkotuwa IA, Caraballo JC, Zabner J, Grassian VH, Comellas AP. Environ Sci Nano. 2014;1:123–132. doi: 10.1039/C3EN00029J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostermeyer AK, Mumuper CK, Semprini L, Radniecki T. Environ Sci Technol. 2013;47:14403–14410. doi: 10.1021/es4033106. [DOI] [PubMed] [Google Scholar]
- 6.Gunsolus IL, Hang MN, Hudson-Smith NV, Buchman JT, Bennett JW, Conroy D, Mason SE, Hamers RJ, Haynes CL. Environ Sci Nano. 2017;4:636–646. [Google Scholar]
- 7.Feng ZV, Gunsolus IL, Qiu TA, Hurley KR, Nyberg LH, Frew H, Johnson KP, Vartanian AM, Jacob LM, Lohse SE, Torelli MD, Hamers RJ, Murphy CJ, Haynes CL. Chem Sci. 2015;6:5186–5196. doi: 10.1039/c5sc00792e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L, Yang J, Xie J, Luo Z, Jiang J, Yang YY, Liu S. Nanoscale. 2013;5:3834–3840. doi: 10.1039/c3nr34254a. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Greden K, Alvarez PJJ, Gregory KB, Lowry GV. Environ Sci Technol. 2010;44:3462–3467. doi: 10.1021/es9031198. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Chen Z, Chen Y, Xu J, Li J, Jiang X. J Am Chem Soc. 2013;135:12940–12943. doi: 10.1021/ja4058635. [DOI] [PubMed] [Google Scholar]
- 11.Goodman CM, McCusker CD, Yilmaz T, Rotello VM. Bioconjug Chem. 2004;15:897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]
- 12.Tu Y, Lv M, Xiu P, Huynh T, Zhang M, Castelli M, Zengrong L, Huang Q, Fan C, Fang H, Zhou R. Nat Nanotechnol. 2013;8:594–601. doi: 10.1038/nnano.2013.125. [DOI] [PubMed] [Google Scholar]
- 13.Pelgrift RY, Friedman AJ. Adv Drug Deliv Rev. 2013;65:1803–1815. doi: 10.1016/j.addr.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Bondarenko O, Ivask A, Käkinen A, Kurvet I, Kahru A. PLoS One. 2013;8:e64060. doi: 10.1371/journal.pone.0064060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster HA, Ditta IB, Varghese S. Appl Microbiol Biotechnol. 2011;90:1847–1868. doi: 10.1007/s00253-011-3213-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson KH, Gunsolus IL, Kuech TR, Troiano JM, Melby ES, Lohse SE, Hu D, Chrisler WB, Murphy CJ, Orr G, Geiger FM, Haynes CL, Pedersen JA. Environ Sci Technol. 2015;49:10642–10650. doi: 10.1021/acs.est.5b01841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Setubal JC, dos Santos P, Goldman BS, Ertesvåg H, Espin G, Rubio LM, Valla S, Almeida NF, Balasubramanian D, Cromes L, Curatti L, Du Z, Godsy E, Goodner B, Hellner-Burris K, Hernandez JA, Houmiel K, Imperial J, Kennedy C, Larson TJ, Latreille P, Ligon LS, Lu J, Maerk M, Miller NM, Norton S, O’Carroll IP, Paulsen I, Raulfs EC, Roemer R, Rosser J, Segura D, Slater S, Stricklin SL, Studholme DJ, Sun J, Viana CJ, Wallin E, Wang B, Wheeler C, Zhu H, Dean DR, Dixon R, Wood D. J Bacteriol. 2009;191:4534–4545. doi: 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dijkshoorn L, Nemec A, Vaneechoutte M, De Baere T, Pantophlet RA, Williams PA, Kay CM, Averhoff B, Graf I, Ornston LN, Schlimpert S, Buchan A, Parke D, Craven SH, Ezezika OC, Momany C, Neidle EL, Gerischer U, Jerg B, Fischer R, Gutnick DL, Bach H, Tomaras AP, Dorsey CW, McQueary C, Actis LA, Seifert H, Wisplinghoff H, Towner KJ, editors. U Gerischer. Caister Academic Press; Norfolk, UK: 2008. [Google Scholar]
- 19.de Berardinis V, Durot M, Weissenbach J, Salanoubat M. Curr Opin Microbiol. 2009;12:568–576. doi: 10.1016/j.mib.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Hau HH, Gralnick JA. Annu Rev Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 21.Vinogradov E, Kubler-Kielb J, Korenevsky A. Carbohydr Res. 2008;343:2701–2705. doi: 10.1016/j.carres.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrow JB, Arango Pinedo C, Holbrook RD. J Environ Qual. 2010;39:1934–1941. doi: 10.2134/jeq2009.0455. [DOI] [PubMed] [Google Scholar]
- 23.Pier GB. Int J Med Microbiol. 2007;297:277–295. doi: 10.1016/j.ijmm.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rocchetta HL, Burrows LL, Lam JS. Microbiol Mol Biol Rev. 1999;63:523–553. doi: 10.1128/mmbr.63.3.523-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinogradov E, Korenevsky A, Beveridge TJ. Carbohydr Res. 2003;338:1991–1997. doi: 10.1016/s0008-6215(03)00325-2. [DOI] [PubMed] [Google Scholar]
- 26.Zhao P, Li N, Astruc D. Coord Chem Rev. 2013;257:638–665. [Google Scholar]
- 27.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Angew Chemie Int Ed Engl. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gole A, Murphy CJ. Chem Mater. 2005;17:1325–1330. [Google Scholar]
- 29.Richardson JJ, Cui J, Björnmalm M, Braunger JA, Ejima H, Caruso F. Chem Rev. 2016;116:14828–14867. doi: 10.1021/acs.chemrev.6b00627. [DOI] [PubMed] [Google Scholar]
- 30.Gentile P, Carmagnola I, Nardo T, Chiono V. Nanotechnology. 2015;26:422001. doi: 10.1088/0957-4484/26/42/422001. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Kong Y, Kundu S, Cirillo JD, Liang H. J Nanobiotechnology. 2012;10:19. doi: 10.1186/1477-3155-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu TA, Bozich JS, Lohse SE, Vartanian AM, Jacob LM, Meyer BM, Gunsolus IL, Niemuth NJ, Murphy CJ, Haynes CL, Klaper RD. Environ Sci Nano. 2015;2:615–629. [Google Scholar]
- 33.Gole A, Murphy CJ. Chem Mater. 2004;16:3633–3640. [Google Scholar]
- 34.Newton JW, Wilson PW, Burris RH. J Biol Chem. 1953;204:445–451. [PubMed] [Google Scholar]
- 35.Qiu TA, Torelli MD, Vartanian AM, Rackstraw NB, Buchman JT, Jacob LM, Murphy CJ, Hamers RJ, Haynes CL. Anal Chem. 2017;89:1823–1830. doi: 10.1021/acs.analchem.6b04161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrand AM, Schlager JJ, Dai L, Hussain SM. Nat Protoc. 2010;5:744–757. doi: 10.1038/nprot.2010.2. [DOI] [PubMed] [Google Scholar]
- 37.Cui Q, Hernandez R, Mason SE, Frauenheim T, Pedersen JA, Geiger F. J Phys Chem B. 2016;120:7297–7306. doi: 10.1021/acs.jpcb.6b03976. [DOI] [PubMed] [Google Scholar]
- 38.Wu EL, Engström O, Jo S, Stuhlsatz D, Yeom MS, Klauda JB, Widmalm G, Im W. Biophys J. 2013;105:1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cournia Z, Smith JC, Ullmann GM. J Comput Chem. 2005;26:1383–1399. doi: 10.1002/jcc.20277. [DOI] [PubMed] [Google Scholar]
- 40.Emami FS, Puddu V, Berry RJ, Varshney V, Patwardhan SV, Perry CC, Heinz H. Chem Mater. 2014;26:2647–2658. [Google Scholar]
- 41.Heinz H, Lin TJ, Mishra RK, Emami FS. Langmuir. 2013;29:1754–1765. doi: 10.1021/la3038846. [DOI] [PubMed] [Google Scholar]
- 42.Lin TJ, Heinz H. J Phys Chem C. 2016;120:4975–4992. [Google Scholar]
- 43.Jo S, Cheng X, Lee J, Kim S, Park S-J, Patel DS, Beaven AH, Lee K Il, Rui H, Park S, Lee HS, Roux B, Mackerell AD, Jr, Klauda JB, Qi Y, Im W. J Comput Chem. 2017;38:1114–1124. doi: 10.1002/jcc.24660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haiss W, Thanh NTK, Aveyard J, Fernig DG. Anal Chem. 2007;79:4215–4221. doi: 10.1021/ac0702084. [DOI] [PubMed] [Google Scholar]
- 45.Klein ND, Hurley KR, Feng ZV, Haynes CL. Anal Chem. 2015;87:4356–4362. doi: 10.1021/acs.analchem.5b00124. [DOI] [PubMed] [Google Scholar]
- 46.Lam JS, Graham LL, Lightfoot J, Dasgupta T, Beveridge TJ. J Bacteriol. 1992;174:7159–7167. doi: 10.1128/jb.174.22.7159-7167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Brun AP, Clifton LA, Halbert CE, Lin B, Meron M, Holden PJ, Lakey JH, Holt SA. Biomacromolecules. 2013;14:2014–2022. doi: 10.1021/bm400356m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rediers H, Vanderleyden J, De Mot R. Microbiology. 2004;150:1117–1119. doi: 10.1099/mic.0.27096-0. [DOI] [PubMed] [Google Scholar]
- 49.Gaona G, Nuñez C, Goldberg JB, Linford AS, Nájera R, Castañeda M, Guzmán J, Espín G, Soberón-Chávez G. FEMS Microbiol Lett. 2004;238:199–206. doi: 10.1016/j.femsle.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 50.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GKS, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock REW, Lory S, Olson MV. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 51.Hong CS, Shitashiro M, Kuroda A, Ikeda T, Takiguchi N, Ohtake H, Kato J. FEMS Microbiol Lett. 2004;231:247–252. doi: 10.1016/S0378-1097(04)00009-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


