SUMMARY
Alternative gene splicing gives rise to N-methyl-D-aspartate (NMDA) receptor ion channels with defined functional properties and unique contributions to calcium signaling in a given chemical environment in the mammalian brain. Splice variants possessing the Exon 5-encoded motif at the amino terminal domain (ATD) of the GluN1 subunit are known to display robustly altered deactivation rates and pH sensitivity, however the underlying mechanism for this functional modification is largely unknown. Here, we show through electron cryo-microscopy (cryo-EM) that the presence of the Exon 5 motif in GluN1 alters the local architecture of heterotetrameric GluN1-GluN2 NMDA receptors and creates contacts with the ligand-binding domains (LBDs) of the GluN1 and GluN2 subunits, which are absent in NMDA receptors lacking the Exon 5 motif. The unique interactions established by the Exon 5 motif are essential to the stability of the ATD/LBD and LBD/LBD interfaces that are critically involved in controlling proton sensitivity and deactivation.
Keywords: NMDA receptors, cryo-EM, alternative splicing, electrophysiology
INTRODUCTION
Excitatory neurotransmission is fundamental to the development and function of the mammalian brain, and is mediated mainly by the activity-dependent release of glutamate followed by activation of postsynaptic ionotropic glutamate receptors (iGluRs) including N-methyl-D-aspartate (NMDA) receptors (Traynelis et al., 2010). Calcium influx mediated by NMDA receptors is pivotal to cellular signaling and plasticity and is controlled by diverse subtypes of NMDA receptors defined by multiple patterns of heterotetramerization of GluN1, GluN2(A–D), and GluN3(A–B) subunits. Alternative splicing of the GluN1 subunit occurs both within the carboxy-terminal end (Exons 21 and 22) and the amino terminal domain (ATD) at the extracellular region (Exon 5) (Paoletti et al., 2013). A common route to modulation of protein function is alternative splicing of the mRNA transcript, and numerous examples of altered function through alternative splicing exist among a wide variety of ion channel families including Shaker (Hoshi et al., 1991) and BK (Chen et al., 2005) potassium channels, voltage-gated sodium (Farmer et al., 2012) and calcium (Castiglioni et al., 2006) channels, acid-sensing sodium channels (Bassler et al., 2001), and transient receptor potential channels (Gracheva et al., 2011; Zhou et al., 2013), as well as iGluRs (Lerma et al., 2001; Schiffer et al., 1997) (Palmer et al., 2005; Penn et al., 2012; Sommer et al., 1990). One of the hallmark functions in NMDA receptors is that the splice variants of the GluN1 subunit containing the twenty-one amino acid residues encoded by Exon 5 in the ATD have significantly reduced sensitivity to proton and faster deactivation speed (Figure 1) (Adams et al., 2014; Rumbaugh et al., 2000; Traynelis et al., 1995; Vance et al., 2012), indicating that those splice variants contribute to excitatory postsynaptic current and elicit calcium signaling differently both under normal conditions and in the acidified environment created locally by high frequency neuronal firing, stroke, and seizure (Kaku et al., 1993). The Exon 5 sequence is conserved throughout vertebrate GluN1 subunits as far back as cartilaginous fishes and hemichordates (consurf.tau.ac.il), which suggests that the location and composition of Exon 5 is evolutionarily advantageous for the maintenance of proper neurological functions. Despite the importance of this classical paradigm in basic neuroscience and pathology, the molecular mechanism underlying functional alteration by Exon 5 has been enigmatic due to the lack of a structure of the intact NMDA receptor with Exon 5. Here, we show through cryo-electron microscopy (cryo-EM) that the presence of the Exon 5 motif in GluN1 alters the local architecture of heterotetrameric GluN1-GluN2 NMDA receptors and creates interdomain contacts with the ligand-binding domains (LBDs) of the GluN1 and GluN2 subunits. These unique interactions likely influence the stability of the ATD/LBD and LBD/LBD interfaces, which are critically involved in controlling proton sensitivity and deactivation. Our results also suggest that a fundamental mechanism of alternative splicing in ion channels could serve a broad function indeed, by introducing or removing interaction networks at precise locations in the protein structure as a means to modulate function.
Figure 1. Inclusion of Exon 5 in GluN1 alters ion channel properties in NMDA receptors.
Two-electrode voltage clamp recording from cRNA injected Xenopus oocytes (A) and fast-perfusion whole cell patch clamp recording from DNA transfected HEK293 cells (B) of the GluN1-4a-GluN2B (−Exon 5) and GluN1-4b-GluN2B (+Exon 5) NMDA receptors illustrating reduced proton sensitivity and faster deactivation speed elicited by Exon 5, respectively. Error bars in (A) represent ±SD for data obtained from 15 and 17 different oocytes for the −Exon 5 and +Exon 5, respectively. The recording traces in (B) are fit (magenta and red) by a double exponential equation.
RESULTS
Cryo-EM structure identifies the Exon 5-encoded motif at domain and subunit interfaces
To define the structure and location of the Exon 5-encoded motif, we performed single particle cryo-EM analysis on the intact rat GluN1-GluN2B NMDA receptor containing the Exon 5 sequence (hereafter referred to as GluN1b-GluN2B; Figure 2). Our initial study on the GluN1b-GluN2B NMDA receptor bound to glycine and glutamate showed conformational variability in the ATD, which resulted in limited resolution (~6.5 Å) and only weak density for the Exon 5-encoded region. To improve the resolution, we minimized conformational flexibility by incorporating an inter-GluN2B ATD disulfide bond with the GluN2B S214C mutation (GluN1b-GluN2BEMX) and prepared the samples with glycine and glutamate bound in the LBDs, Ifenprodil in the ATD, and MK-801 in the TMD ion channel. This modified construct formed the GluN1-GluN2B NMDA receptors that retained functional properties including activation by glycine and glutamate, and inhibition by an allosteric modulator (ifenprodil or zinc), antagonists (5,7 dichlorokynurenic acid or D-AP5) or channel blockers (magnesium and MK-801) (Figures 2 and S1). 3D classification resulted in one reconstruction that resolves to ~4.6 Å resolution as estimated by Fourier shell correlation (FSC) using the 0.143 cut-off (Figures 3 and Figure S2), enabling us to visualize density for bulky aromatic residues and Asn-linked glycosylation and permitting building of molecular models with confidence (Figure S3). The cryo-EM structure unambiguously shows the heterotetrameric assembly of the GluN1b-GluN2B NMDA receptors. The bi-lobed structures of the GluN2B ATD, the GluN1 LBD, and the GluN2B LBD in the current cryo-EM structure are in the closed conformation, occupied by Ifenprodil, glycine, and glutamate, consistent with crystal structures of the isolated domains (Jespersen et al., 2014; Karakas et al., 2011). At the TMD, the ion channel pore is blocked by MK-801 as illustrated by the cryo-EM density (Figure S3C). The cryo-EM structure shows ordered density for eight residues (GluN1b Tyr204-Lys211) encoded by Exon 5 at the ATD/LBD inter-domain interface, which ‘caps’ the GluN1b-GluN2B subunit interface of the LBD through interactions between GluN1b Lys190 and GluN2B Tyr507, and between GluN1b Lys211 and GluN1b Asp786 (Figure 3B). The regions surrounding GluN1b Lys190, GluN1b Lys211, and GlwuN2B Tyr507 are devoid of missense variants in humans, suggesting they play important roles in protein function and may be under strong selection (Lek et al., 2016; Ogden et al., 2017). Remarkably, the region between GluN1b Glu186 and Lys190, which is not encoded by Exon 5, has an altered architecture compared to that in the GluN1a-GluN2B NMDA receptor lacking Exon 5 (Figure 3C) (Karakas and Furukawa, 2014). Specifically, those five residues are extended toward the ATD-LBD linker of the GluN2B subunit rather than turning back to GluN1 β7 as in GluN1a, thereby creating the GluN1b-specific contacts with the GluN2B subunit, including those between GluN1b Lys190 and GluN2B Tyr507, and between GluN1b Lys211 and GluN2B Tyr507 (Figure 3B). The structure of the region immediately surrounding the Exon 5 motif is similar to that of the allosterically inhibited receptor (PDB code: 4PE5), the non-active receptor (PDB code: 5FXH and 5FXI), and the active receptor (PDB code: 5FXG); therefore, the pattern of interactions between the Exon 5 motif and other domains is likely independent of the functional states of the receptors (Karakas and Furukawa, 2014; Tajima et al., 2016).
Figure 2. Construct design and protein purification.
(A) GluN1b and GluN2B constructs used in the current structural study (GluN1b-GluN2BEMX). The glycosylation knock-out mutations are to improve the expression level (green ovals). Free cysteines in GluN2B were mutated to serines (orange ovals). Cross-link mutations were incorporated to reduce conformational heterogeneity and to improve the quality of 3D reconstruction (gray ovals). (B) SDS-PAGE (8%) gel and the chromatograph from size-exclusion chromatography (Superose-6) showing purity and size-homogeneity of the GluN1b-GluN2BEMX sample.
Figure 3. Cryo-electron microscopy structure identifies the Exon 5 encoded motif at domain and subunit interfaces.
(A) Cryo-EM structure of the intact GluN1b-GluN2B NMDA receptor at ~4.6 Å. The GluN1b subunits are colored magenta and gold while the GluN2B subunits are colored cyan and blue. (B) Highlighted region from (A), illustrating cryo-EM density for residues of the Exon 5 motif at the ATD/LBD interface. (C) Overlay of models from the GluN1a-GluN2B and GluN1b-GluN2B intact NMDA receptors. Note the large shift in position of GluN1 Lys190 between the GluN1a and GluN1b isoforms.
The interactions between the Exon 5 motif and LBDs observed in the cryo-EM structure are physiological
To assess the physiological relevance of the observed inter-subunit arrangement around the Exon 5 motif, we engineered cysteine mutants into otherwise wild type GluN1-4b and GluN2B NMDA receptors with no C-terminal domain (deltaCTD; see Methods) and tested for spontaneous disulfide bond formation at the following subunit interfaces: I) the beginning region of the Exon 5 motif and the GluN2B ATD/LBD linker (GluN1b Ser191 or Lys192 and GluN2B Gln401); II) the beginning region of the Exon 5 motif and the GluN2B LBD (GluN1b Ser191 or Lys192 and GluN2B Gly771); and III) the end region of the Exon 5 motif and the GluN2B LBD (Figure 4A). We expressed and purified these mutant GluN1-4b-GluN2B deltaCTD NMDA receptors and conducted Western blot analysis under non-reducing conditions to detect band shifts indicative of disulfide bond formation. This showed disulfide crosslinks between single cysteine mutants of the Exon 5 motif and GluN2B that lead to formation of GluN1b-GluN2B deltaCTD heterodimers that can be recognized by antibodies against both GluN1 and GluN2B (Figure 4B). When only one of the subunits contains the cysteine modification or the experiment is performed in the presence of reducing agent, no bands representing heterodimers were observed, indicating that dimer formation occurs through specific disulfide formation between the engineered pairs (Figure 4B). Thus, this series of disulfide bond experiments under physiological conditions are consistent with the subunit interactions observed in the cryo-EM structure. The disulfide bond formation above had minimal effect on pH sensitivity and deactivation speed.
Figure 4. Assessment of interdomain and intersubunit interactions mediated by the Exon 5 motif.
(A) Model of the GluN1b (magenta) and GluN2B (cyan) subunits with key residues labeled. We identified three groups of interactions at this site (I–III) through crosslinking and Western blot analysis. (B) Results of Western blots after crosslinking experiments for our series of mutants. Single cysteine mutants were unable to form dimers, while pairs of mutants were detected at masses indicating disulfide bond formation, which could be eliminated by application of a reducing agent.
Interactions between the Exon 5 motif and the LBDs control proton sensitivity
Based on the current cryo-EM structure, we explored the underlying molecular mechanism for reduced proton sensitivity by Exon 5. We conducted mutagenesis in the context of the full length GluN1-4b-GluN2B NMDA receptor, including disrupting interactions between the Exon 5 motif and the GluN1 LBD and/or between the Exon 5 motif and the GluN2B LBD, charge reversal of the three negatively charged residues encoded by Exon 5, and truncations of the disordered region (Figure 5A–C). Potency of proton inhibition was measured by detecting whole-cell macroscopic currents at various pH points using two-electrode voltage clamp (TEVC) in Xenopus laevis oocytes (Figure S4). Disrupting the interaction between the Exon 5 motif and the GluN1 LBD (by mutating GluN1-4b Lys190Ala or GluN1-4b Asp786Ala) or between the Exon 5 motif and the GluN2B LBD (by mutating GluN1-4b Lys211Ala or GluN2B Tyr507Ala) increased the IC50 values for proton inhibition from pH 7.04 to pH ~7.3 and ~7.4, respectively, but did not restore proton sensitivity to the level observed for the Exon 5-lacking GluN1-4a-GluN2B NMDA receptor (IC50 = pH 7.72) (Figure 5A and Table 1). When we disrupted both of the two inter-domain interactions above with the GluN1-4b-Lys190Ala/Lys211Ala mutations, and similarly with GluN1-4b-Asp786Ala/GluN2B-Tyr507Ala mutations, the IC50 values for these respective mutant combinations shifted to pH 7.65 and 7.66, which is statistically indistinguishable from the wild type GluN1-4a-GluN2B NMDA receptor (Figure 5A). Contrary to the large effect observed by disrupting the Exon 5 – LBD interactions, charge reversal of negatively charged residues (Figure 5B) and truncations of the disordered region (Figure 5B and Table S1) had little or no effect on receptor proton sensitivity. The effect of truncation is observed only in the most extreme construct (ΔExon5-C) together with GluN1-4b-Lys211Ala. These results imply that the Exon 5 motif reduces proton sensitivity of the ion channel by affecting the subunit interface between the GluN1b LBD and the GluN2B LBD. To further test this hypothesis, we disrupted the GluN1b-GluN2B LBD heterodimer interface with the GluN2B Leu781Ala mutation and observed a significant increase in proton sensitivity (Figure 5D). Overall, these results agree with the concept that the GluN1-GluN2B LBD dimer interface is a critical region for controlling proton sensitivity and that the Exon 5 motif reduces proton sensitivity by stabilizing the GluN1-GluN2B LBD dimer interface by interacting with both the GluN1 LBD and the GluN2B LBD. Disrupting the similar LBD subunit interface in the GluN1-GluN2A NMDA receptors was also shown to result in increased proton sensitivity (Gielen et al., 2008), indicating that the GluN1-GluN2 LBD interface is the common locus for controlling the proton sensitivity in both GluN2A- and GluN2B-containing NMDA receptors.
Figure 5. Interactions between the Exon 5 motif and the LBDs control proton sensitivity.
Here, we employed four types of mutations in the context of TEVC experiments to explore several potential effects of the Exon 5 motif: (A) disruption the direct Exon 5 – LBD interactions, (B) Reversal of negative charges to positives, (C) alter local structure of the Exon 5 motif by truncations (detailed in Table S1), and (D) disruption of the heterodimeric interface between the GluN1 and GluN2B LBDs. Shown in gray lines and two-tone spheres are the Exon 5 motif and the GluN1b-GluN2B LBD heterodimers, respectively. The lightning bolt and scissors indicate disruption of interactions and truncations, respectively. The areas of engineered mutations are highlighted by transparent yellow ovals. IC50 values of proton inhibition for the wild type GluN1-4a-GluN2B and GluN1-4b-GluN2B NMDA receptors and the mutant GluN1-4b-GluN2B NMDA receptors were estimated by recording macroscopic current at various pHs by TEVC from at least five oocytes per construct. (*: p < 0.05, **: p < 0.01; ***: p < 0.001; ns: not significant, as determined by one-way ANOVA with Tukey’s test. Bar graphs represent mean ± SEM). All of the TEVC recordings look similar to the ones in Figure 1A.
Table 1. Electrophysiological data for all tested mutants.
pH inhibition data were collected from two-electrode voltage clamp (TEVC) experiments, while τ values are from whole-cell patch clamp (WCPC) experiments recorded with 5 ms application of agonist. The number of replicates for WCPC or TEVC experiments are listed as nwcpc and ntevc, respectively. Sequential numbering refers to the specific data point labeled in Figure S6.
| Construct Number |
Construct | τfast | τslow | τfast (%) | τweighted | nwcpc | pH IC50 | ntevc |
|---|---|---|---|---|---|---|---|---|
| 1 | GluN1a WT, GluN2B WT | 113.1 ± 12.1 | 741.5 ± 52.7 | 70 ± 2 | 289.2 ± 21.3 | 12 | 7.72 ± 0.02 | 15 |
| 2 | GluN1b WT, GluN2B WT | 71.31 ± 3.83 | 436.5 ± 36.7 | 88 ± 2 | 106.2 ± 5.53 | 44 | 7.04 ± 0.01 | 17 |
| 3 | GluN1b(K190A), GluN2B WT | 73.21 ± 7.48 | 467.4 ± 21.6 | 87 ± 1 | 124.0 ± 7.90 | 10 | 7.33 ± 0.01 | 5 |
| 4 | GluN1b WT, GluN2B(Y507A) | 66.59 ± 4.53 | 427.8 ± 80.9 | 86 ± 8 | 102.9 ± 8.50 | 9 | 7.28 ± 0.04 | 6 |
| 5 | GluN1b(K211A), GluN2B WT | 94.85 ± 19.5 | 615.6 ± 89.4 | 81 ± 6 | 147.8 ± 12.3 | 8 | 7.40 ± 0.04 | 7 |
| 6 | GluN1b(D786A), GluN2B WT | 73.22 ± 9.00 | 733.1 ± 121.8 | 86 ± 2 | 162.5 ± 33.0 | 8 | 7.35 ± 0.03 | 10 |
| 7 | GluN1b(K190A, K211A), GluN2B WT | 128.9 ± 47.7 | 391.0 ± 87.2 | 82 ± 11 | 108.8 ± 13.6 | 8 | 7.65 ± 0.02 | 5 |
| 8 | GluN1b(D786A), GluN2B(Y507A) | 103.3 ± 12.1 | 581.1± 60.3 | 84 ± 1 | 175.1 ± 11.2 | 7 | 7.66 ± 0.04 | 5 |
| 9 | GluN1b(D197, D200, D205K), GluN2B WT | 70.32 ± 8.04 | 249.1 ± 85.2 | 95 ± 2 | 85.6 ± 9.69 | 10 | 7.21 ± 0.09 | 5 |
| 10 | GluN1b(ΔEx5-A), GluN2B WT | 42.70 ± 4.15 | 222.7 ± 20.1 | 92 ± 1 | 57.07 ± 4.93 | 11 | 6.89 ± 0.03 | 12 |
| 11 | GluN1b(ΔEx5-B), GluN2B WT | 46.56 ± 4.35 | 502.3 ± 150.4 | 95 ± 1 | 64.19 ± 5.17 | 11 | 6.84 ± 0.06 | 7 |
| 12 | GluN1b(ΔEx5-C), GluN2B WT | 51.13 ± 18.0 | 336.2 ± 43.8 | 88 ± 1 | 87.19 ± 25.2 | 6 | 7.21 ± 0.02 | 8 |
| 13 | GluN1b(ΔEx5-C, K211A), GluN2B WT | 147.9 ± 46.7 | 921.2 ± 162.4 | 79 ± 5 | 303.4 ± 38.3 | 9 | 7.57 ± 0.02 | 6 |
| 14 | GluN1b WT, GluN2B(L781A) | 221.6 ± 22.2 | 526.3 ± 91.2 | 51 ±12 | 340.0 ± 27.1 | 7 | 8.76 ± 0.18 | 7 |
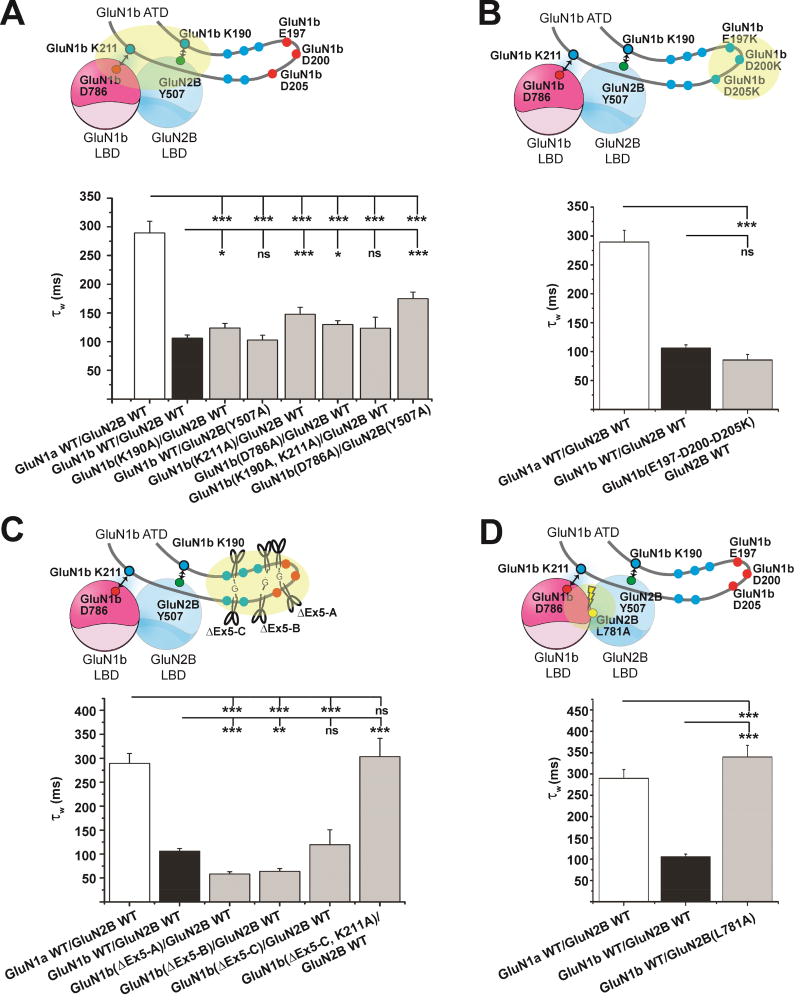
Structure and proper placement of the Exon 5 motif at the ATD-LBD interface is critical for controlling deactivation rates
Another hallmark of Exon 5-containing NMDA receptors is their faster deactivation rate compared to NMDA receptor isoforms without Exon 5 (Rumbaugh et al., 2000; Vance et al., 2012). We first assessed if the Exon 5-LBD interactions important for reducing proton-sensitivity are also involved in facilitating faster deactivation by monitoring the effects of our Exon 5 mutations on deactivation rate. This was achieved by performing whole-cell patch clamp (WCPC) experiments coupled with fast perfusion (5 ms glutamate application; Figure S5 and Table 1), which matches the time scale of synaptic transmission, followed by measuring current decay as described by two exponential components (τfast and τslow). Disruption of the Exon 5 – LBD interactions by the point mutations at GluN1-4b Lys190, GluN1-4b Lys 211, GluN1-4b Asp786, and GluN2B Tyr507 had little or no effect in slowing down the NMDA receptor deactivation time course compared to the wild type GluN1-4b-GluN2B NMDA receptor as monitored by a weighted average of the two time constants (τweighted) (Figure 6A). Furthermore, conversion of negatively charged residues (Figure 6B) or truncation of the flexible region of the Exon 5 motif (Figure 6C and Table S1) resulted in only minimal effects. However, these minor, but statistically significant effects correlate with the proton sensitivity as assessed by a linear regression analysis of time constants (τweighted, τfast, τslow) and proportion of τfast against pH IC50 (Figure S6). Notably, as a function of pH IC50, not only do τweighted and τfast increase (Figure S6A and S6B), but the relative contribution to deactivation by τfast decreases (Figure S6C). To completely reverse the effect of Exon 5, its structure and positioning needs to be sufficiently disrupted by a combination of the truncation ΔExon5-C, and perturbation of the Exon 5 – LBD interaction by the GluN1b Lys211Ala mutation (Figure 6C). Thus, the major factor controlling the deactivation rate is the proper structural arrangement and placement of the Exon 5 motif within the ATD-LBD interface, which becomes difficult to achieve if the GluN1-GluN2B LBD interface is perturbed by the GluN2B Leu781Ala mutation (Figure 6D). Consequently, the effect of the Exon 5 motif is nearly eliminated and the deactivation rate becomes equivalent to the Exon 5-lacking GluN1-4a-GluN2B NMDA receptor (Figure 6D).
Figure 6. Structure and proper placement of the Exon 5 motif at the ATD-LBD interface is critical for controlling deactivation rates.
The same set of the mutant GluN1-4b-GluN2B NMDA receptors in Figure 4 were tested for deactivation rates in HEK 293T cells by WCPC. No pronounced effects were observed by disrupting the Exon 5 – LBD interactions (A) or reversing residue charges (B). (C) To reverse the effect of Exon 5, a combination of truncation and disruption of the Exon 5 – LBD interaction (GluN1b ΔEx5-C/K211A) is necessary. (D) Disruption of the GluN1b-GluN2B LBD heterodimer interface by GluN2B Leu781Ala overrode the effect of Exon 5. In all cases, WCPC recordings on transfected HEK293 cells were done on at least six cells per construct. (*: p < 0.05, **: p < 0.01; ***: p < 0.001; ns: not significant, as determined by one-way ANOVA with Tukey’s test. Bar graphs represent mean ± SEM). Representative WCPC recordings are shown in Figure S5.
DISCUSSION
Taken together, our structural and functional data show that the ATD-LBD interface is an important locus for regulating ion channel function including proton sensitivity and deactivation rate in NMDA receptors. The Exon 5 motif acts as a ligand naturally incorporated into the primary sequence in order to control the GluN1-GluN2B LBD inter-subunit interface, which in turn regulates ion channel activity at the TMD. Our current and previous structures demonstrate that the interaction between the ATD and the LBD is more extensive in NMDA receptors compared to that of the non-NMDA iGluRs (Karakas and Furukawa, 2014; Lee et al., 2014; Tajima et al., 2016). Therefore, a small structural element such as the Exon 5 motif can modulate the ATD-LBD interface and alter functions of NMDA receptors while the equivalent region in non-NMDA iGluRs has too few potential interactions for the ATD to be able to modulate the LBD and vice versa. Consistently, there is little or no ATD-mediated modulation of functions such as proton sensitivity and deactivation rates in non-NMDA iGluRs. Extending the same analogy of the Exon 5 motif as a tethered ligand, it stands to reason that polyamines such as spermine and spermidine bind and modulate a similar if not the identical domain interface and elicit Exon 5-like effects, including reduced proton sensitivity (Mony et al., 2011) and faster deactivation rates (Rumbaugh et al., 2000). A similar mechanism may also hold true for a set of subtype-specific compounds such as PYD-106, which are predicted to bind to the ATD-LBD interface (Khatri et al., 2014). This fine-tuning capability of the ATD-LBD interface in NMDA receptors may yet be employed to develop context-dependent therapeutic compounds that will only exhibit activity in the acidified environment created by seizure and stroke.
STAR METHODS
Requests for further information or additional resources should be addressed to the corresponding author, Hiro Furukawa, via email at furukawa@cshl.edu.
CULTURED CELLS
The proton concentration-response relationship was measured using two-electrode voltage clamp (TEVC) recordings on defolliculated Xenopus laevis oocytes injected with 0.5 ng cRNAs encoding GluN1 and GluN2B at a 1:2 weight ratio. The oocytes were incubated in 96 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM HEPES, at a final pH of 7.5, supplemented with 50 µM D,L-2-amino-5-phosphonovaleric acid (D,L-AP-5) for 18–24 hrs at 18 °C, with the exception of GluN2B-Leu781Ala mutants, which required an injection of 5 ng cRNA and 48–60 hrs incubation time in the presence of 50 µM D,L-AP-5 and 50 µM 5,7-dichlorokynurenic acid (DCKA).
Whole-cell patch-clamp recordings were conducted in HEK (293T) cells co-transfected with individual pCI-neo vectors containing GluN1, GluN2, and enhanced GFP using the calcium phosphate precipitation method (Chen and Okayama, 1987) at a 1:1:1 or a 1:1:0.5 ratio. Cells were grown on glass coverslips coated with poly-D-lysine (100 µg/mL) in DMEM + 1X GlutaMax™ (Gibco®) with 10% FBS and 1% penicillin/streptomycin. After 4–5 hrs, the transfection media was replaced with fresh media supplemented with 200 µM D,L-AP-5 and 200 µM 7-chlorokynueric acid (7CKA) to inhibit receptor activation and reduce cell death. After 18–36 hrs, single cells were selected for recordings based on eGFP fluorescence.
EXPERIMENTAL DETAILS
Cryo-EM/Single particle analysis
The GluN1b-GluN2B NMDA receptor proteins were purified as previously (Tajima et al., 2016). The purified GluN1b-GluN2B NMDA receptor proteins at 3 mg/ml in 0.002% Lauryl Maltose Neopentyl Glycol (LMNG) were mixed with 0.1% digitonin and placed on an EM grid (C-flat Holey Carbon copper grid glow discharged for 45 s at 15 mA). The sample was plunge-frozen in liquid ethane using a Vitrobot (FEI) at a relative humidity of 85% at 15 °C for 3 s blot time. Movies were collected on an FEI Titan-KRIOS microscope operating at 300 kV coupled with a Gatan K2 Summit direct electron detector at 22.5K magnification (0.655 Å per super resolution pixel) with 96 total electrons in 70 frames over a 21 second exposure and with a defocus range of ~−1.5 µm and ~−3.0 µm. Super-resolution movie frames were corrected for magnification distortion, binned two-fold, and aligned with exposure-weighting by Unblur (Grant and Grigorieff, 2015). Contrast transfer function (CTF) parameters were estimated using CTFFIND4 (Rohou and Grigorieff, 2015). Particles were picked semi-automatically and 2D classified by IMAGIC. Particles representing ‘clean’ 2D classes were selected and subjected to refinement using FrealignX (Grigorieff, 2016) via cisTEM (cistem.org) starting from the manually built initial model from the 2D classes. To avoid over-refinement, the resolution limit for particle refinement was always set at least 2 Å lower than the resolution of the reconstructed structure as defined by the 0.143 cut-off of the Fourier shell correlation (FSC). The highest resolution used for refinement was 7 Å. 3D classification into four classes resulted in one major class. The refinement was done with and without imposition of C2 symmetry. The quality of the cryo-EM density of the Exon 5–motif did not change between the two refinements. The GluN1a-GluN2B crystal structure (PDB ID: 4PE5 (Karakas and Furukawa, 2014)) was docked into the cryo-EM density followed by rigid-body fitting of the individual ATD and LBDs of both GluN1 and GluN2B into the cryo-EM map using COOT (Emsley and Cowtan, 2004) The resulting model was manually modified to fit into the density using COOT and refined against the cryo-EM map using Phenix real space refinement (Afonine et al., 2013). Summary of data collection and refinement statistics are shown in Figure S2. In order to calculate the FSC, maps calculated from two halfs of the data were masked with a circular mask of 100 Å radius. The FSC between these was calculated and adjusted to take into account the volume in the mask which exceeds the expected volume of 380 kDa (Sindelar and Grigorieff, 2012). This scales the FSC to approximate the FSC that would be obtained with a tight mask, but eliminates artifacts that a tight mask can create. A similar correction was performed for the FSC between the final map and the model, using an adjustment that was scaled to take into account the fact that the model contains no noise.
Inter-subunit crosslink and western blot
The full-length wild type GluN1-4b and GluN2B without C-terminal domain (ends at Gly852; deltaCTD) N-terminally tagged with Twin-strep tag were subjected to crosslink mutagenesis in Figure 4. Recombinant baculoviruses harboring every combination of GluN1-4b and GluN2B deltaCTD were made as described previously (Tajima et al., 2016) and were used to infect Sf9 cells for protein expression. The mutant GluN1-4b-GluN2B deltaCTD NMDA receptors were purified as in Tajima et al, without the final step involving Superose-6 size exclusion chromatography. The purified sample was mixed with SDS-PAGE loading buffer with and without β-mercaptoethanol and subjected to 7% SDS-PAGE followed by transfer to a nitrocellulose membrane at 100 V for 1 hour. After the transfer, the blots were blocked by 5% skim milk (10% for GluN2B antibody) in a buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.1% Tween 20 (TBST) for 1 h. After washing the membrane with TBST, the membranes were incubated in the primary antibody solutions prepared by 2,000-fold dilution of the anti-GluN1 antibody in TBST+0.5% milk or anti-GluN2B antibody in TBST+1% skim milk at 4 °C overnight. The membranes were washed with TBST and incubated in the secondary antibody solution (anti-mouse-HRP conjugate at 1/10,000 dilution in TBST+0.5%). After a 1 h incubation, the membranes were washed with TBST and developed with enhanced chemiluminescence (GE Healthcare).
Electrophysiology
TEVC recordings were performed using agarose-tipped microelectrodes (0.4–1.0 MΩ) filled with 3 M KCl at a holding potential of −60 mV at room temperature (23 °C). The recording solutions contained 100 mM NaCl, 2.5 mM KCl, 10 mM HEPES, 10 mM Tricine, and 0.3 mM BaCl2 at various pHs adjusted with HCl. 100 µM glycine and L-glutamate were applied to induce a current response. The data were acquired and analyzed by the program Pulse (HEKA).
Whole-cell patch clamp recording microelectrodes were pulled from thin-walled filamented borosilicate glass (World Precision Instruments) using a Flaming/Brown horizontal puller (Sutter Instruments). Electrodes were filled with an internal pipette solution containing 110 mM D-gluconate, 110 mM CsOH, 30 mM CsCl, 5 mM HEPES, 5 mM BAPTA, 4 mM NaCl, 2 mM MgCl2, 2 mM NaATP, 0.3 mM NaGTP, and 0.5 mM CaCl2, with the final pH adjusted to 7.35 using CsOH. When filled with internal solution, the micropipette tips had resistances of 3–4 MΩ when placed in the extracellular recording solution containing 150 mM NaCl, 3 mM KCl, 10 mM HEPES, 30 mM D-mannitol, 1 mM CaCl2, and 10 µM EDTA, with the final pH adjusted to 7.4 with NaOH. For recordings, this bath solution was supplemented with either 30 µM glycine or 30 µM glycine plus 1 mM L-glutamate. Recordings were performed at −40 mV at room temperature using an AxoPatch 200B amplifier, with currents digitized at 40 kHz using a DigiData 1322A digitizer (Molecular Devices) controlled by Clampex 10.3 in the pClamp software package (Molecular Devices). Anti-alias filtering was performed with an 8-pole Bessel filter at 8 kHz (−3 dB, Frequency Devices). A two-barrel theta tube was used for rapid solution exchange using a piezoelectric translator. Data was collected and averaged using the pClamp software package, and curves were fit with the program ChanneLab. The plots and statistics presented above were generated in Origin 7.5.
Supplementary Material
KEY RESOURCES TABLE
| Resource | Source | Identifier |
|---|---|---|
| Lauryl Maltose Neopentyl Glycol (LMNG) | Anatrace | NG310 |
| Digitonin | Sigma Aldrich | D5628 |
| Threo Ifenprodil hemitartrate | Tocris | 2892 |
| (+)-MK-801 maleate | Tocris | 0924 |
| D,L-2-amino-5-phosphonovaleric acid (DL-AP-5) | Tocris | 0105 |
| 5,7-dichlorokynurenic acid (DCKA) | Tocris | 0286 |
| 7-chlorokynurenic acid (7CKA) | Tocris | 0237 |
| Sf-900 III Serum free medium | Gibco | 12658027 |
| DMEM media | Gibco | 11995040 |
| Gibco Glutamax | ThermoFisher | 35050061 |
| Xenopus laevis oocytes | Harvested in-house | N/A |
| HEK(293T) cells | ATCC | CRL-1573 |
| Sf9 cells | Gibco | 12659017 |
| Unblur | Grant and Grigorieff, 2015 | grigoriefflab.janelia.org/unblur |
| CTFFind4 | Rohou and Grigorieff, 2015 | grigoriefflab.janelia.org/ctffind4 |
| FrealignX | Grigorieff, 2016 | grigoriefflab.janelia.org/frealign |
| cisTEM | cisTEM | cistem.org |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix | Adams et al., 2010 | www.phenix-online.org |
| Pulse | HEKA | www.heka.com |
| Clampex 10.3/pClamp suite | Molecular Devices | www.moleculardevices.com/systems/axon-conventional-patch-clamp/ |
| ChanneLab | Synaptosoft | www.synaptosoft.com/Channelab/ |
| OriginLab 7.5 | Origin Lab | www.originlab.com |
HIGHLIGHTS.
-
-
A cryo-EM structure at 4.6 Å identified the alternatively-spliced Exon 5 motif.
-
-
This motif lies at the interface of the GluN1 ATD, GluN1 LBD, and GluN2B LBD.
-
-
Interactions at this site alter receptor proton sensitivity and deactivation.
-
-
The Exon 5 motif regulates ion channel activity via the GluN1-GluN2 LBD interface.
Acknowledgments
We thank Zhiheng Yu, Chuan Hong, Rick Huang, and M. Jason de la Cruz at the Janelia EM facility and Noriko Simorowski for excellent technical support. We also thank Pierre Paoletti for critical reading of this manuscript. This work was supported by NIH grants NS036654 (SFT), NS093753 (MCR), MH085926 and GM105730 (HF), Stanley Institute of Cognitive Genomics (HF), and Burroughs Wellcome Fund (HF). The structural coordinates related to this work have been deposited in the Protein Data Bank with the entry code 6CNA. The cryo-EM map can be found in the EMDB under accession code EMD-7529.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
M.C.R., M.J.M., E.K., J.Z., H.F. and S.T. conceived and performed experiments involving electrophysiology. T.G., E.K., H.F., and N.G. solved the cryo-EM structure. M.C.R. and H.F. wrote the manuscript.
Declaration of Interests
Authors declare no conflict of interests.
References
- Adams DR, Yuan H, Holyoak T, Arajs KH, Hakimi P, Markello TC, Wolfe LA, Vilboux T, Burton BK, Fajardo KF, et al. Three rare diseases in one Sib pair: RAI1, PCK1, GRIN2B mutations associated with Smith-Magenis Syndrome, cytosolic PEPCK deficiency and NMDA receptor glutamate insensitivity. Molecular genetics and metabolism. 2014;113:161–170. doi: 10.1016/j.ymgme.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Headd JJ, Terwilliger TC, Adams PD. Computational Crystallography Newsletter. 2013;4:43–44. [Google Scholar]
- Bassler EL, Ngo-Anh TJ, Geisler HS, Ruppersberg JP, Grunder S. Molecular and functional characterization of acid-sensing ion channel (ASIC) 1b. J Biol Chem. 2001;276:33782–33787. doi: 10.1074/jbc.M104030200. [DOI] [PubMed] [Google Scholar]
- Castiglioni AJ, Raingo J, Lipscombe D. Alternative splicing in the C-terminus of CaV2.2 controls expression and gating of N-type calcium channels. J Physiol. 2006;576:119–134. doi: 10.1113/jphysiol.2006.115030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Molecular and cellular biology. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tian L, MacDonald SH, McClafferty H, Hammond MS, Huibant JM, Ruth P, Knaus HG, Shipston MJ. Functionally diverse complement of large conductance calcium- and voltage-activated potassium channel (BK) alpha-subunits generated from a single site of splicing. J Biol Chem. 2005;280:33599–33609. doi: 10.1074/jbc.M505383200. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Farmer C, Cox JJ, Fletcher EV, Woods CG, Wood JN, Schorge S. Splice variants of Na(V)1.7 sodium channels have distinct beta subunit-dependent biophysical properties. PLoS One. 2012;7:e41750. doi: 10.1371/journal.pone.0041750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. Structural Rearrangements of NR1/NR2A NMDA Receptors during Allosteric Inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva EO, Cordero-Morales JF, Gonzalez-Carcacia JA, Ingolia NT, Manno C, Aranguren CI, Weissman JS, Julius D. Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats. Nature. 2011;476:88–91. doi: 10.1038/nature10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. Journal of structural biology. 2015;192:204–208. doi: 10.1016/j.jsb.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N. Frealign: An Exploratory Tool for Single-Particle Cryo-EM. Methods Enzymol. 2016;579:191–226. doi: 10.1016/bs.mie.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi T, Zagotta WN, Aldrich RW. Two types of inactivation in Shaker K+ channels: effects of alterations in the carboxy-terminal region. Neuron. 1991;7:547–556. doi: 10.1016/0896-6273(91)90367-9. [DOI] [PubMed] [Google Scholar]
- Jespersen A, Tajima N, Fernandez-Cuervo G, Garnier-Amblard EC, Furukawa H. Structural insights into competitive antagonism in NMDA receptors. Neuron. 2014;81:366–378. doi: 10.1016/j.neuron.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku DA, Giffard RG, Choi DW. Neuroprotective effects of glutamate antagonists and extracellular acidity. Science. 1993;260:1516–1518. doi: 10.1126/science.8389056. [DOI] [PubMed] [Google Scholar]
- Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011;475:249–253. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri A, Burger PB, Swanger SA, Hansen KB, Zimmerman S, Karakas E, Liotta DC, Furukawa H, Snyder JP, Traynelis SF. Structural determinants and mechanism of action of a GluN2C-selective NMDA receptor positive allosteric modulator. Molecular pharmacology. 2014;86:548–560. doi: 10.1124/mol.114.094516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Lu W, Michel JC, Goehring A, Du J, Song X, Gouaux E. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma J, Paternain AV, Rodriguez-Moreno A, Lopez-Garcia JC. Molecular physiology of kainate receptors. Physiol Rev. 2001;81:971–998. doi: 10.1152/physrev.2001.81.3.971. [DOI] [PubMed] [Google Scholar]
- Mony L, Zhu S, Carvalho S, Paoletti P. Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. The EMBO journal. 2011;30:3134–3146. doi: 10.1038/emboj.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden KK, Chen W, Swanger SA, McDaniel MJ, Fan LZ, Hu C, Tankovic A, Kusumoto H, Kosobucki GJ, Schulien AJ, et al. Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology. PLoS Genet. 2017;13:e1006536. doi: 10.1371/journal.pgen.1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CL, Cotton L, Henley JM. The molecular pharmacology and cell biology of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors. Pharmacol Rev. 2005;57:253–277. doi: 10.1124/pr.57.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nature reviews Neuroscience. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- Penn AC, Balik A, Wozny C, Cais O, Greger IH. Activity-mediated AMPA receptor remodeling, driven by alternative splicing in the ligand-binding domain. Neuron. 2012;76:503–510. doi: 10.1016/j.neuron.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Prybylowski K, Wang JF, Vicini S. Exon 5 and spermine regulate deactivation of NMDA receptor subtypes. J Neurophysiol. 2000;83:1300–1306. doi: 10.1152/jn.2000.83.3.1300. [DOI] [PubMed] [Google Scholar]
- Schiffer HH, Swanson GT, Heinemann SF. Rat GluR7 and a carboxy-terminal splice variant, GluR7b, are functional kainate receptor subunits with a low sensitivity to glutamate. Neuron. 1997;19:1141–1146. doi: 10.1016/s0896-6273(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Sindelar CV, Grigorieff N. Optimal noise reduction in 3D reconstructions of single particles using a volume-normalized filter. J Struct Biol. 2012;180:26–38. doi: 10.1016/j.jsb.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Keinanen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakmann B, Seeburg PH. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, Furukawa H. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature. 2016 doi: 10.1038/nature17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Hartley M, Heinemann SF. Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science. 1995;268:873–876. doi: 10.1126/science.7754371. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacological reviews. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KM, Hansen KB, Traynelis SF. GluN1 splice variant control of GluN1/GluN2D NMDA receptors. J Physiol. 2012;590:3857–3875. doi: 10.1113/jphysiol.2012.234062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Suzuki Y, Uchida K, Tominaga M. Identification of a splice variant of mouse TRPA1 that regulates TRPA1 activity. Nat Commun. 2013;4:2399. doi: 10.1038/ncomms3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.