Abstract
Summary: Protael is a JavaScript library for creating interactive visualizations of biological sequences and various associated data. It allows users to generate high-quality vector graphics (SVG) and integrate it into web pages.
Availability and implementation: Protael distribution, documentation and examples are freely available at http://protael.org; source code is hosted at https://github.com/sanshu/protaeljs.
Contact: adam@godziklab.org
1 Introduction
Predictive methods of bioinformatics address the gap between the explosive growth of sequence data and experimental characterization of biomolecules. The number and accuracy of prediction methods are increasing but the individual algorithms are often implemented as separate programs and webservers. From a user point of view this creates a need for ‘shopping around’ and then often manual integration of the results into a comprehensive bioinformatics characterization of a protein. At the same time, servers, which automatically combine results of different prediction algorithms, require graphical tools to integrate different types of protein-related data and predictions into a single visualization. While there are many excellent biological sequence visualization tools, most of them are tightly coupled with the specific data sources and websites and cannot be easily reused by other resource. At the time of writing we are aware of several reusable visualization libraries for the web, including pViz (Mukhyala et al., 2014), RSCB Protein Feature Viewer (http://andreasprlic.github.io/proteinfeatureview/), BioJS/FeatureViewer (Garcia et al., 2014) and Mason (Jaschob et al., 2015) for feature annotations and SnipViz (Jaschob et al., 2014) and JSAV (Martin, 2014) for multiple sequence alignment.
To enable fully customizable visualization of protein sequence features, predictions, annotations, various posttranslational modifications and alignments with known structures (including their local structural features) we developed Protael—a reusable and extendable library to display heterogeneous linear protein-related data on the web. The library is written in JavaScript and uses Cascading Style Sheets (CSS) to define styles of the graphics elements. The visualization content is fully controlled by a single JSON object. Protael is compatible with all modern browsers on desktop and mobile systems, and does not require any additional plugin installation. Protael visualization is currently focused on proteins but it can also be used and customized for nucleic acids. Generated SVG visualization allows unrestricted scaling and provides vector images. Protael is used to visualize protein data in Cancer3D (Porta-Pardo et al., 2015), XtalPred (Jahandideh et al., 2014), PDBFlex (http://pdbflex.org/) webservers and to display alignments of nucleotide sequences in bNAber (Eroshkin et al., 2014).
2 Implementation
Protael is a reusable library, which produces interactive and customizable SVG graphics completely on the client machine. Basic usage requires little knowledge of HTML and JavaScript, and is as simple as including links to Protael script and dependencies, and writing couple of lines of code. For more complex applications Protael provides numerous ways of customization and adding new features.
Protael uses open source libraries for generating visualization and controls, including SnapSVG (http://snapsvg.io/), JQuery (http://jquery.com) and JQueryUI (http://jqueryui.com). The Protael starter project is available on the Protael website and is set up in a way that provides auto-loading of the required versions of libraries from the content delivery networks.
All input for Protael is included in a JSON object, which contains information about the sequence and annotations to be displayed and can be provided via AJAX request to a server or loaded from a file. Documentation about JSON configuration is available at http://protael.org/docs/.
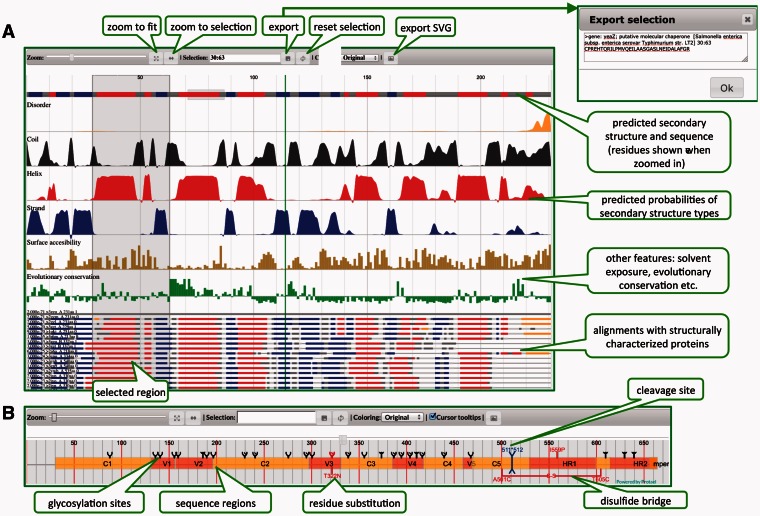
Protael interface contains a control bar that provides functions such as zoom, export of selected sequence regions, exporting publication quality vector images, applying different residue-based coloring schemes, etc. (see Fig. 1) Flexible data format allows users to provide arbitrary number of additional object properties, which are converted to HTML5 data-* attributes and could be later used for custom styling and actions.
Fig. 1.
Protael interface and examples of visualizations: (A) menu elements, visualization of predictions of local structural features, sequence features and alignments with known structures, (B) visualization of post-translational modifications, sequence regions, substitutions and cleavage sites (engineered HIV immunogen is used as an example)
2.1 Protael visualization
Protael is capable of displaying multiple data types, such as sequence annotations, substitutions, posttranslational modifications, cleavage sites, disulfide bridges, various types of quantitative data and alignments with known structures. Sequence features, annotations, quantitative data and aligned sequences can occupy multiple data tracks. Users can specify scale for quantitative data and assign tooltips and external URLs (links) to displayed elements.
2.2 Main sequence
Main sequence area shows the sequence itself and some of its features such as predicted transmembrane helices, signal peptides, low complexity regions, posttranslational modifications and disulfide bridges. This area has fixed topmost position and cannot be moved (other visualization tracks could be dragged vertically to form a more compact chart).
2.3 Sequence features and annotations
The area below the main sequence contains feature and annotation tracks (ftracks), if the appropriate data is provided in the JSON object. Each track can be styled separately. It is possible to override styling for individual feature by providing data in JSON object or via JavaScript call. Depending on the ‘allowOverlap’ parameter value in JSON object, overlapped features will be drawn either on top of the each other or on different sub-levels to avoid visual overlapping. Examples of feature tracks include annotated domains (represented as rectangles). Protael also allows distinct coloring and labeling of sequence regions, showing posttranslational modifications, cleavage sites and residue substitutions (Fig 1B).
2.4 Quantitative data
The features tracks area is followed by the quantitative data tracks (qtracks) area. Almost any kind of quantitative data, both discrete and continuous, could be displayed here. Examples of such data include probability graphs from predictions of secondary structure types, predicted structural disorder probability, evolutionary conservation indices, predicted surface accessibility, etc. Quantitative data can currently be presented as five different chart types: line, area, spline, spline-area and column chart. Several coloring options are available including solid color and gradient fills.
2.5 Multiple sequence alignment
The last (bottom) visualization level is used to display sequences aligned with the main sequence. Developer can choose between two display options for alignments—either as lines (useful for creating more compact view of up to 100 sequences) or as strings representing residue type in one-letter code (practical for up to 30 sequences). The coloring of aligned sequences, as well as the main sequence, is also controlled via JSON object. For example one can color aligned sequences according to the predicted or observed secondary structure type, or highlight only specific residues (for instance mutation or binding sites).
Funding
The development of Protael is funded by funded in part by the NIAID-NIH contract HHSN272201200026C (CSGID).
Conflict of Interest: none declared.
References
- Eroshkin A.M., et al. (2014) bNAber: database of broadly neutralizing HIV antibodies. Nucleic Acids Res., 42, D1133–D1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia L., et al. (2014) FeatureViewer, a BioJS component for visualization of position-based annotations in protein sequences. F1000Research. 3, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahandideh S., et al. (2014) Improving the chances of successful protein structure determination with a random forest classifier. Acta Crystallogr. D Biol Crystallogr. 70, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaschob D., et al. (2014) SnipViz: a compact and lightweight web site widget for display and dissemination of multiple versions of gene and protein sequences. BMC Res. Notes, 7, 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaschob D., et al. (2015) Mason: a JavaScript web site widget for visualizing and comparing annotated features in nucleotide or protein sequences. BMC Res. Notes. 8, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C. (2014) Viewing multiple sequence alignments with the JavaScript Sequence Alignment Viewer (JSAV). F1000Research, 3, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhyala K., Masselot A. (2014) Visualization of protein sequence features using JavaScript and SVG with pViz.js. Bioinformatics, 30, 3408–3409. [DOI] [PubMed] [Google Scholar]
- Porta-Pardo E., et al. (2015) Cancer3D: understanding cancer mutations through protein structures. Nucleic Acids Res., 43, D968–D973. [DOI] [PMC free article] [PubMed] [Google Scholar]