Abstract
Summary: PRIMUS is a pedigree reconstruction algorithm that uses estimates of genome-wide identity by descent to reconstruct pedigrees consistent with observed genetic data. However, when genetic data for individuals within a pedigree are missing, often multiple pedigrees can be reconstructed that fit the data. We report a major expansion of PRIMUS that uses mitochondrial (mtDNA) and non-recombining Y chromosome (NRY) haplotypes to eliminate many pedigree structures that are inconsistent with the genetic data. We demonstrate that discordances in mtDNA and NRY haplotypes substantially reduce the number of potential pedigrees, and often lead to the identification of the correct pedigree.
Availability and Implementation: We have implemented PRIMUS updates in PERL and it is available at primus.gs.washington.edu.
Contact: debnick@uw.edu or jennifer.e.below@uth.tmc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Correctly determining pedigree structures is key to identifying the causes of genetic disorders (Riordan et al., 1989). In some cases, reported pedigree structures are inconsistent with observed genetic sharing (Kerr et al., 2013), which may result in a loss of power and failure to find the disease causing variant(s) (Boehnke and Cox, 1997). Cryptic relatedness within datasets and sample swaps are frequently observed. Pedigree reconstruction can find cryptic relationships and correctly fit them into a pedigree structure (Staples et al., 2014). Early methods for checking pedigrees applied pairwise relationship prediction approaches that use co-dominant genetic markers (Epstein et al., 2000; Sun et al., 2002); however, pedigree reconstruction can more accurately predict relationships, find the correct pedigree, and identify cryptic pedigrees (Staples et al., 2014). Pedigree Reconstruction and Identification of a Maximum Unrelated Set (PRIMUS; Staples et al., 2014) uses estimates of genome-wide identity by descent (IBD) to reconstruct pedigree structures that fit the IBD estimates.
Missing genetic data for a set of people in a family often leads to the identification of multiple pedigrees that fit the data. We show that by using inconsistencies in the inheritance patterns of the human mitochondria (mtDNA) and non-recombining Y chromosome (NRY) haplotypes captured by genotyping arrays or by high-throughput sequencing (Lippold et al., 2014), improves the accuracy of pedigree reconstruction. We describe the utilization of mtDNA and NRY data for pedigree reconstruction in PRIMUS (v1.8.0) to reduce the number of generated pedigrees and improve the chance of identifying the correct pedigree structure.
2 Methods
2.1 Identifying discordant mtDNA and NRY haplotypes
PRIMUS supports standard PLINK format. Human NRY and mtDNA can be encoded as chromosome 24 and 26, respectively. For a pair of individuals, PRIMUS calculates the concordance of mtDNA and NRY haplotypes. The percent concordance of a haplotype is calculated as the percentage of matching mtDNA and NRY nucleotide positions across the total number of variable mtDNA and NRY positions and excludes positions with missing calls. A ‘discordant’ prediction between the NRY or mtDNA haplotype of two individuals occurs when concordance is below a user definable cutoff (a 99% default works well for many datasets), otherwise the NRY or mtDNA haplotype is predicted to be ‘concordant.’ Discordant status indicates that a pair of individuals has not co-inherited the mtDNA or NRY haplotypes from a recent common ancestor. Therefore, PRIMUS eliminates any pedigree structure in which two individuals with discordant mtDNA or NRY haplotypes shares a recent common maternal or paternal lineage, respectively.
By default, PRIMUS only eliminates pedigree structures that are inconsistent with the discordant mtDNA and NRY predictions, which we demonstrate are very informative and reliable. For example, in both sequencing and genotyping datasets, we observed a nearly 100% haplotype concordance between individuals who have inherited mtDNA or NRY from a recent common ancestor (i.e. fewer than four generations of separation; Supplemental Table S1); therefore, individuals with discordant predictions are very unlikely to be related through a recent common ancestor of the sex that corresponds to the discordant mtDNA or NRY prediction.
Although discordant mtDNA and NRY haplotypes are very useful in rejecting genetically inconsistent pedigrees, concordant haplotypes are less so. Because recombination does not influence mtDNA and NRY haplotypes, a single haplotype can be passed unchanged through a family for generations. Therefore, distant relatives can have concordant mtDNA and NRY haplotypes while they share little or no autosomal DNA with detectable IBD (Supplemental Fig. S1). If PRIMUS requires all concordant mtDNA and NRY predictions to be represented by a recent common ancestor, then distant, concordant ancestral mtDNA and NRY haplotypes can cause PRIMUS to reject the correct pedigree structure. Therefore, by default concordant mtDNA and NRY haplotypes are not used to rule out pedigree structures.
2.2 mtDNA and NRY checking
PRIMUS uses mtDNA and NRY discordance to improve pedigree reconstructions by checking whether the discordance is consistent with the expected mtDNA and NRY inheritance patterns within the pedigree. For example, if half-siblings are genotyped and have discordant mtDNA prediction, then their parent in common must be the father. To check whether this discordant prediction is consistent with a pedigree structure, PRIMUS finds the shortest first-degree-relative inheritance path connecting two individuals, A and B. Discordant predictions require an interruption in the transmission of the haplotype in the pedigree. For example, if A and B are males and have a discordant NRY, then there must be a female in the inheritance path connecting A and B. The logic that applies to NRY inheritance paths through males applies to mtDNA inheritance paths through females, except that the sex of A and B does not matter unless one is the direct ancestor of the other; in which case, the direct ancestor must be female. We illustrate valid and invalid inheritance paths in a pedigree in Supplemental Figure S2.
3 Results
PRIMUS is the current state-of-the-art program for reconstructing pedigrees and is the only program that lists all non-inbred pedigree structures that fit genetic data using up to 3rd degree relationships. To illustrate the use of mtDNA and NRY haplotypes during pedigree reconstruction, we compare reconstructions by PRIMUS with and without mtDNA and NRY haplotypes with simulated and real data. We modified the pedigree simulations described by Staples et al. (2014), to explore the effects of using mtDNA and NRY on the reconstruction of pedigrees with different structures, genotypes and combinations of missing samples. We selected pedigrees of size 20 and masked the genotypes for 0–50% of individuals in the pedigree. We modified the simulations by permuting the sex of the individuals, while maintaining the biologic integrity of the pedigree. For each simulation, we assigned a unique NRY haplotype to each male founder and a unique mtDNA haplotype to all founders and propagated these genotypes through the pedigree. We obtained mtDNA and NRY haplotypes from the phase 1 release of unrelated CEU and TSI haplotypes (Staples et al., 2013) with individual-level call rates >90% from the 1000 Genomes Project (Genomes Project, et al., 2012). These haplotypes consisted of 2832 mtDNA and 8665 NRY loci with quality score ≥30, call rates >95%, and with a minor allele frequency >1% for inclusion. We randomly assigned a unique haplotype to each of the founders in each pedigree and propagated the haplotypes through the pedigrees.
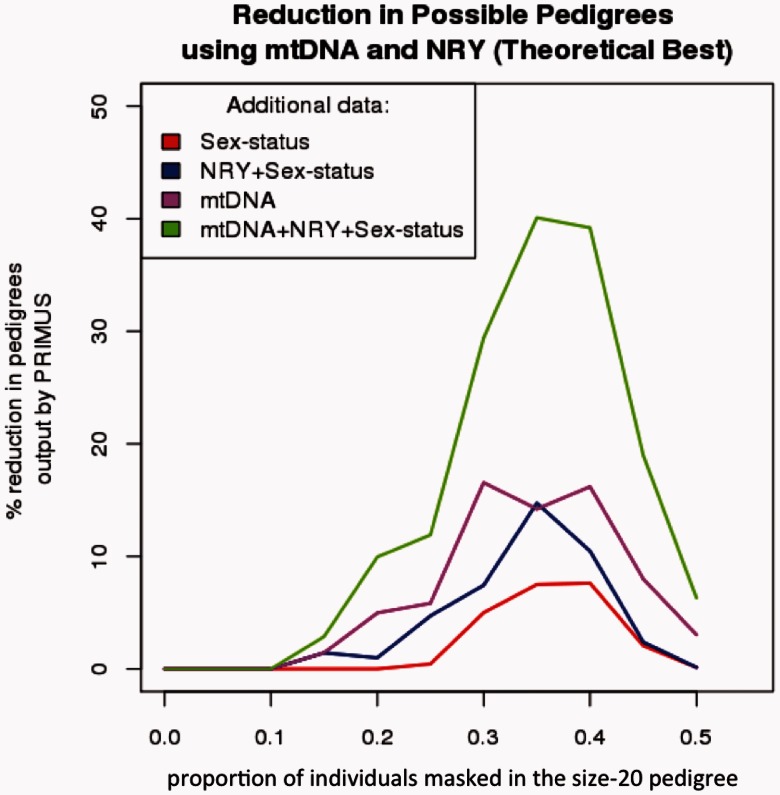
We considered pedigree reconstruction performance on the autosomal data alone as the baseline and compared this to the performance when we added additional information such as sex status, mtDNA, and NRY. We see a moderate reduction in the number of possible pedigrees with mtDNA and NRY individually, but the synergistic effects of mtDNA and NRY with sex status exceeded the combined individual improvements (Fig. 1). Our results show the largest improvement in pedigrees with more missing samples (35–40%), where a 37% reduction in the mean number of genetically consistent pedigrees is seen (Fig. 1). The improvement declines beyond 40% missing samples because the pedigrees become too sparse for the discordant mtDNA and NRY haplotypes to rule out possible pedigrees. We also see a substantial improvement in ranking the correct pedigree structure (Supplemental Fig. S3). In fact, when mtDNA and NRY are combined with individual sex status, we see a 4.5-fold increase over sex status alone in the number of simulations that reconstructed to the only true pedigree.
Fig. 1.
A summary of the percent reduction in the average number of possible pedigrees when data from mtDNA, NRY, sex or all of these are applied. The addition of either mtDNA or NRY data outperforms the addition of only sex status. The greatest reduction in the number of possible pedigrees is obtained when mtDNA, NRY and sex status are combined, eliminating nearly 40% of the incorrect pedigrees
The improvements in pedigree specificity using mtDNA and NRY genotypes are remarkable. As shown in Supplemental Figure S4, a pedigree that reconstructed to 58 possible pedigrees using autosomal DNA and sex status, resolved to the single correct pedigree with the addition of mtDNA data. The new implementation of PRIMUS identified this pedigree by eliminating the 57 pedigrees that were inconsistent with the mtDNA data.
We validated our results on real data by reconstructing the pedigrees within the Jackson Heart Study (JHS) cohort. We received Illumina Infinium HumanExome BeadChip genotype data for 2790 individuals. We estimated IBD proportions for these individuals using the method described in Staples et al. (2014). After cleaning (Supplemental Methods), there were 23 500 autosomal SNPs used for the IBD calculation, and 126 NRY SNPs and 172 mtDNA SNPs used for haplotype comparison. Using PRIMUS, we reconstructed the JHS pedigrees with and without mtDNA/NRY haplotype data. Inclusion of the mtDNA/NRY haplotypes reduced the number of pedigrees generated for the JHS family networks. We saw up to a 67% reduction in the number of generated pedigrees (Supplemental Fig. S5), with an average reduction of 18%. This improvement of specificity did not negatively impact sensitivity; PRIMUS with mtDNA and NRY identified the expected pedigrees at the same rate as PRIMUS without mtDNA and NRY. These results fall well within the range of improvement we saw in simulations with 1000 Genomes data. Therefore, with both simulated and real data, NRY and mtDNA provide a substantial improvement in automated pedigree reconstruction.
Supplementary Material
Acknowledgements
We thank Adam Gordon, Colleen Davis, and Phil Green for their input.
Funding
This work was supported by the National Science Foundation Graduate Research Fellowship [DGE-0718124 to J.S.]; and the National Human Genome Research Institute and National Institute of Health Heart, Lung, and Blood Institute for the UW Center for Mendelian Genomics [HG006493 to D.A.N]. The Jackson Heart Study is supported by contracts HHSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, HHSN268201300050C from the National Heart, Lung, and Blood Institute and the National Institute on Minority Health and Health Disparities.
Conflicts of Interest: none declared.
References
- Boehnke M., Cox N.J. (1997) Accurate inference of relationships in sib-pair linkage studies. Am. J. Hum. Genet., 61, 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein M.P., et al. (2000) Improved inference of relationships for pairs of individuals. Am. J. Hum. Genet, 67, 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C., et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr S.M., et al. (2013) Pedigree and genotyping quality analyses of over 10 000 DNA samples from the Generation Scotland: Scottish Family Health Study. BMC Med. Genet., 14, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold S., et al. (2014) Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences. Invest. Genet., 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan J.R., et al. (1989) Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science, 245, 1066–1073. [DOI] [PubMed] [Google Scholar]
- Staples J., et al. (2013) Utilizing graph theory to select the largest set of unrelated individuals for genetic analysis. Genet. Epidemiol., 37, 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples J., et al. (2014) PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet., 95, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., et al. (2002) Enhanced pedigree error detection. Hum. Hered., 54, 99–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.