Abstract
Syndecan-1 is a transmembrane proteoglycan expressed prominently by lung epithelium and has pleiotropic functions such as regulating cell migration, proliferation, and survival. Loss of syndecan-1 expression by lung cancer cells is associated with higher-grade cancers and worse clinical prognosis. We evaluated the effects of syndecan-1 in various cell-based and animal models of lung cancer and found that lung tumorigenesis was moderated by syndecan-1. We also demonstrate that syndecan-1 (or lack thereof) alters the miRNA cargo carried within exosomes exported from lung cancer cells. Analysis of the changes in miRNA expression identified a distinct shift toward augmented procancer signaling consistent with the changes found in lung adenocarcinoma. Collectively, our work identifies syndecan-1 as an important factor in lung cancer cells that shapes the tumor microenvironment through alterations in miRNA packaging within exosomes.
Lung cancer is the second most common cancer type and the leading cause of cancer-related death worldwide, accounting for nearly 30% of all cancer-related deaths.1 More than 200,000 cases of lung cancer are diagnosed in the United States each year, and an estimated 150,000 deaths in the United States each year are due to lung cancer. Several host and environmental factors that predispose to the development of lung cancer have been identified.2 However, cancer cells can also evolve to acquire a more aggressive phenotype through intrinsic changes in the cells themselves and through the microenvironment in which they reside.3
Syndecan-1 is a member of a family of four cell-surface heparan sulfate proteoglycans (syndecans 1 through 4). Syndecan-1 is primarily expressed on epithelial cells and can regulate cell proliferation, migration, adhesion, and survival.4, 5, 6, 7, 8, 9 Previous studies have demonstrated that a loss of syndecan-1 from epithelial cells induces a malignant phenotype in solid-organ cancers of the skin and gut.10, 11, 12 Similarly, syndecan-1 is a positive prognostic marker in lung cancer. Indeed, higher syndecan-1 levels in lung cancer cells are associated with a better survival rate, and the expression of syndecan-1 decreases with higher histologic grade of lung cancer.13, 14, 15
Syndecan-1 was recently found to regulate exosome biogenesis.16 Exosomes are a relatively newly appreciated method of intercellular communication that carry bioactive proteins, lipids, and RNAs as cargo to target neighboring and distal cells to exert control over their cellular phenotype.17, 18 Moreover, exosomes play important roles in shaping the tumor microenvironment and in controlling tumorigenesis.19 miRNAs, in particular, are enriched in exosomes and can regulate the malignant transformation of cells.20 Exosome delivery of miRNAs controls the tumor immunologic response,21 angiogenesis,22 and metastasis.23, 24, 25, 26, 20
Tumor growth and progression involve an intricate interplay between malignant cells and the microenvironment in which the cancer resides.2, 3 Because syndecan-1 regulates exosome production, we postulated that the loss of syndecan-1, as occurs in high-grade lung adenocarcinomas, reshapes the tumor microenvironment by changing the exosome cargo to promote tumorigenesis. Indeed, our results demonstrate that syndecan-1 regulates the miRNA profile within exosomes secreted from lung adenocarcinoma cells. Moreover, it was shown that syndecan-1 deficiency changes the exosome cargo to deliver more protumorigenic signals, which also coincides with increased tumor burden in several murine models of lung cancer.
Materials and Methods
Analysis of TCGA Data Set
Clinical data sets and SDC1 mRNA expression (z-scores; RNASeq version 2 RNAseq by expectation maximization) from lung adenocarcinoma patients (TCGA, provisional) were downloaded with cBioportal.27, 28 Data from the lower quartile of SDC1 mRNA expression were separated from those from the remaining samples, and an overall survival Kaplan-Meier curve was generated.
Cells
A549 cells were transduced with γ-retroviral vectors to stably express human syndecan-1 shRNA (A549shRNA.Sdc1) or scrambled control shRNA (A549shRNA.scr) as we previously described.6 Lewis lung carcinoma (LLC) cells are a murine cell line that lacks endogenous syndecan-1 expression. Therefore, cells that overexpressed murine syndecan-1 or control cells were also generated by γ-retroviral transduction. The transfer plasmid used to generate the γ-retrovirus also expressed Green fluorescent protein via an internal ribosomal entry site sequence, and Green fluorescent protein–positive cells were selected by fluorescence-activated cell sorting. Cell surface expression levels of human (A549) or murine (LLC) syndecan-1 were initially characterized and then periodically monitored by staining with B-A38 (Bio-Rad, Hercules, CA) or 281-2 (BD Pharmingen, San Jose, CA), respectively, and analyzed with a Guava benchtop flow cytometer (EMD Millipore, Billerica, MA).
Anchorage-Independent Assay
A549 cells were cultured in 0.4% agarose with growth medium using previously published protocols.11 Cell–agarose mixtures were incubated for up to 28 days at 37°C and 5% CO2, and fed two times a week by adding growth medium on top of the agarose layer.
In Vivo Models of Lung Cancer
LLC cells (106 cells/mouse) were injected s.c. into pouches on the right flank of C57BL/6J mice. After 3 weeks, the mice were euthanized, and tumors were removed, measured, and either fixed with 4% formalin or snap frozen in liquid nitrogen for further RNA or protein analysis.
A549 cells (106 cells/mouse) were injected retro-orbitally into severe combined immunodeficiency mice (The Jackson Laboratory, Sacramento, CA). Lungs were collected 4 weeks after injection, and the right lungs were fixed and prepared for histologic examination. The left lungs were immediately dissociated into a single-cell suspension using the gentleMACS Dissociator (Miltenyi Biotec, San Diego, CA). Green fluorescent protein–positive cells were counted with the Guava benchtop flow cytometer (EMD Millipore).
LSL-Kras mice (The Jackson Laboratory) were crossed with syndecan-1–deficient (Sdc1−/−) mice to generate LSL-Kras/wild type and LSL-Kras/Sdc1−/− mice. The LSL-Kras mouse has a lox-stop-lox sequence before an activated form of Kras (G12D). Mice were given Cre recombinase via intranasal instillation of adenovirus (5 × 106 particles) to activate the Kras transgene and induce tumorigenesis, followed by sacrifice at 14 weeks.
All animal procedures were approved by the Institutional Animal Care and Use Committees at the University of Washington (Seattle, WA) and Cedars-Sinai Medical Center (Los Angeles, CA).
Exosome and RNA Isolation
A549 cells were plated in a T-25 flask at 2 × 106 density and cultivated in Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin at 37°C in 5% CO2 overnight. Then, cells were washed three times with phosphate-buffered saline and cultured in Dulbecco's minimal essential medium supplemented with 10% exosome-depleted fetal bovine serum (System Biosciences, Inc., Palo Alto, CA) for 48 hours. Exosomes were isolated from cell culture media using ExoQuick-TC (System Biosciences, Inc.), and RNA was purified using the miRNeasy micro kit (Qiagen, Valencia, CA) per the manufacturer's protocol. RNA was sent to the Clinical Microarray Core at the University of California–Los Angeles (Los Angeles, CA), and RNA sequencing was performed using the HiSeq 3000 system (Illumina, San Diego, CA).
miRNA Sequencing Data Analysis
miRNA sequencing short reads were investigated using FastQC software version 0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) for quality control and aligned to the human reference genome (GRCh38) using Bowtie2 software version 2.2.3.29 Aligned sequences were mapped to mature miRNAs with annotation from miRBase software version 21.30 The relative expression abundances of miRNAs were calculated (in cpm), and the miRNAs with mean abundance values of <5 cpm in both control and knockdown conditions were filtered out. The read counts of remaining miRNAs were used to perform the differential expression analysis using edgeR software version 3.10.5.31 The P values of multiple tests were adjusted using the Benjamini-Hochberg method, and the significance level was designated as a false discovery rate of <0.01. The miRNA sequencing data from A549 and EKVX exosomes were uploaded onto the NIH–Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo; accession numbers GSE106277 and GSE106304).
miRPath software version 332 was used to determine the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways regulated by the miRNAs differentially expressed in exosomes. Down- and up-regulated miRNAs that had an over-fourfold difference in expression between conditions were separately entered into miRPath software version 3, and verified target genes were identified with DIANA-TarBase version 7.0 (TarBase), a database of manually curated miRNA–gene interactions that provides a list of genes that have been experimentally validated as targets for miRNA.33 miRNAs without any experimentally identified targets in TarBase were eliminated from further analysis.
Proliferation Assay
A549 cells were plated in a 12-well plate at 100,000 cells/well density and cultured overnight in Dulbecco's minimal essential medium supplemented with penicillin/streptomycin and exosome-depleted fetal bovine serum. The next day, exosomes from A549shRNA.scr and A549shRNA.Sdc1 cells were added (50 μg/mL), and A549 cells were cultured for 48 hours. Subsequently, cells were trypsinized and replated in a 96-well plate at a density of 5000 cells/well. Cell proliferation was measured using the CyQUANT Cell Proliferation Assay Kit (Invitrogen, Waltham, MA) at 0 and 24 hours after replating.
Analysis of Genes Differentially Expressed in Adenocarcinoma Tumors
The microarray data from Feng et al34 (https://www.ncbi.nlm.nih.gov/geo; accession number GSE43767) comparing the transcriptome of normal lungs (n = 15) to that of lung adenocarcinoma specimens (n = 69) was analyzed. Genes differentially expressed between the two conditions were identified with Gene Expression Omnibus 2R, and all genes with a false discovery rate of <0.01 were sorted into groups that were up- and down-regulated in adenocarcinoma conditions. These groups of genes were inputted into the WebGestalt online analytical toolkit (http://www.webgestalt.org, last accessed April 15, 2016), and KEGG pathway analysis was performed.
Additionally, miRvestigator was used to evaluate the putative miRNAs that target the differentially regulated genes in adenocarcinoma cells.35 A list of miRNAs that were significantly (P < 0.05) predicted to target genes that were either up- or down-regulated in adenocarcinoma tumors compared with normal controls was generated. This list was compared with the miRNAs that were differentially expressed in exosomes released from A549shRNA.Sdc1 cells versus controls.
Results
Syndecan-1 Expression Correlates with Survival in Lung Cancer
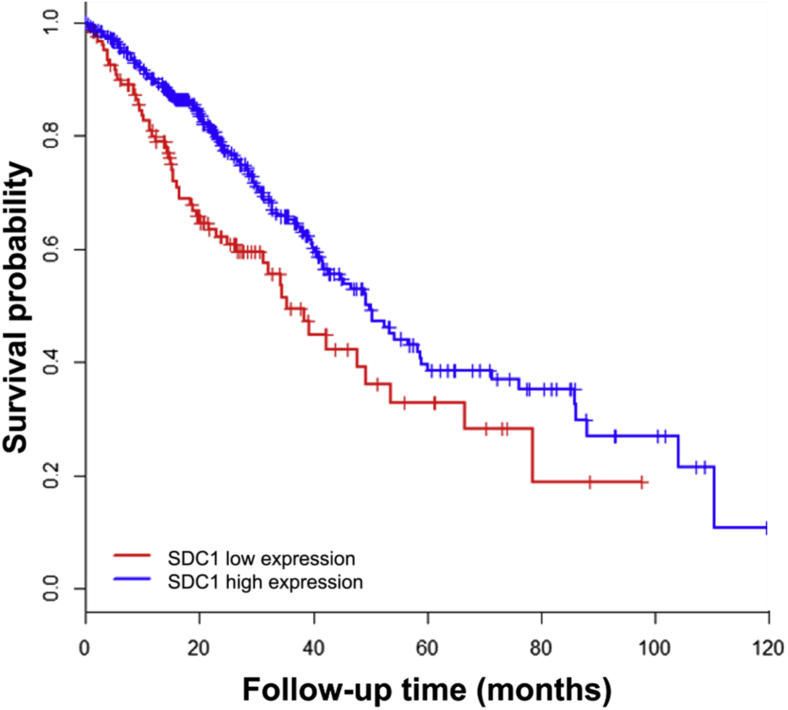
The data from the TCGA repository were compared with determine whether syndecan-1 expression is correlated with survival in patients with adenocarcinomas. Using a database of information on 496 patients with lung adenocarcinomas, the data set was rank-ordered by syndecan-1 expression levels, and the lower quartile of patients was defined as low expression (124 patients) and the remaining patients as high expression (372 patients). The median survival in the group with low syndecan-1 expression was found to be 35.2 versus 49.8 months in the group with high syndecan-1 expression (Figure 1). These results replicate those from previous publications demonstrating that syndecan-1 expression by lung cancer cells is a positive prognostic indicator.13, 14
Figure 1.
Kaplan-Meier overall survival curve based on low and high syndecan-1 (SDC1) expression in lung adenocarcinoma tumors. Clinical outcomes data and syndecan-1 expression levels in patients with lung adenocarcinomas were downloaded from the TCGA data repository. The data were segregated into a low-expression group (lower quartile based on syndecan-1 expression; 124 patients) versus the remaining 372 patients grouped as high expression. Overall survival was determined between the low and high syndecan-1 expression groups. P = 0.0029.
Lung Cancer Is More Tumorigenic in the Absence of Syndecan-1
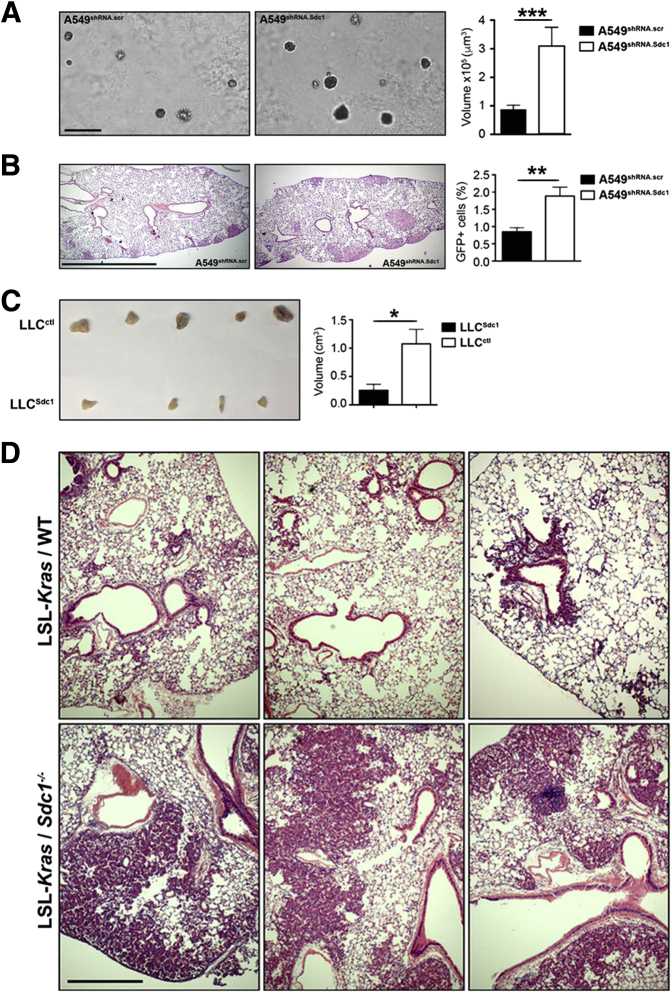
Our data (Figure 1) and those from previous studies demonstrate associations between the loss of syndecan-1 expression and lung cancer and a worse clinical phenotype.13, 14 Therefore, it was tested whether changes in syndecan-1 expression function in cancer progression. A549 cells, which are a NCI60 cell line validated to represent in vivo tumors,36 were cultured in an anchorage-independent assay, and it was found that A549shRNA.Sdc1 cells grew larger colonies than did A549shRNA.scr cells (Figure 2A). To test the growth in vivo, A549 cells were injected retro-orbitally into severe combined immunodeficiency mice, and lungs were harvested 4 weeks after injection. Larger tumors were found in the lungs of mice injected with A549shRNA.Sdc1 compared with mice injected with A549shRNA.scr (Figure 2B).
Figure 2.
Lung adenocarcinomas proliferate faster when lacking syndecan-1. A: A549shRNA.scr and A549shRNA.Sdc1 cells were cultured in 0.4% agarose for 4 weeks. Colony volume was calculated (4/3πr3). B: A549shRNA.scr and A549shRNA.Sdc1 cells stably expressing Green fluorescent protein (GFP) were injected i.v. into severe combined immunodeficiency mice. After 4 weeks, lungs were processed for histologic examination and flow cytometry of GFP-positive cells. C: Lewis lung carcinoma cells expressing murine syndecan-1 (LLCSdc1) and control cells (LLCctl) were injected into a pouch s.c. into C57BL/6J mice. Tumor growth was allowed for 3 weeks, tumors were removed, and their volume was quantified. D: LSL-Kras mice crossed with either wild-type (WT) or Sdc1−/− mice were intranasally instilled with 5 × 106 adenoviral particles that express Cre recombinase. Lungs were harvested 14 weeks after infection and analyzed for tumor growth. Each panel represents an independent mouse. n = 3 experiments (50 colonies counted per experiment; A); n = 3(B); n = 4 LLCSdc1 (C); n = 5 LLCctl (C). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bars: 200 μm (A); 2 mm (B); 500 μm (D).
Next, LLC cells, a murine lung cancer cell line, were used in an s.c. tumor growth model. LLCs have low expression of syndecan-1 (S.Y.S. and P.C., unpublished data), so LLC cell lines that overexpressed syndecan-1 and a vector control line that has low syndecan-1 expression were generated. Consistent with the A549 data, elevated syndecan-1 expression markedly reduced tumor growth in LLC cell lines that overexpressed syndecan-1, producing significantly smaller tumors than did control cells in vivo (Figure 2C).
In addition, the LSL-Kras mouse was crossed with Sdc1−/− mice to generate LSL-Kras/wild type and LSL-Kras/Sdc1−/− mice. Intranasal instillation of an adenovirus to transduce Cre recombinase in the lung epithelium activates Kirsten ras and induces lung tumor formation.37 Once again, reduced syndecan-1 expression conferred a more tumorigenic phenotype, with significantly larger tumors in the lungs of LSL-Kras/Sdc1−/− compared with LSL-Kras/wild-type mice (Figure 2D). Together, these data demonstrate that the loss of syndecan-1 expression in lung cancer cells confers a more protumor phenotype both in vitro and in vivo.
Syndecan-1 Regulates the miRNA Profile within Exosomes Released by Lung Cancer Cells
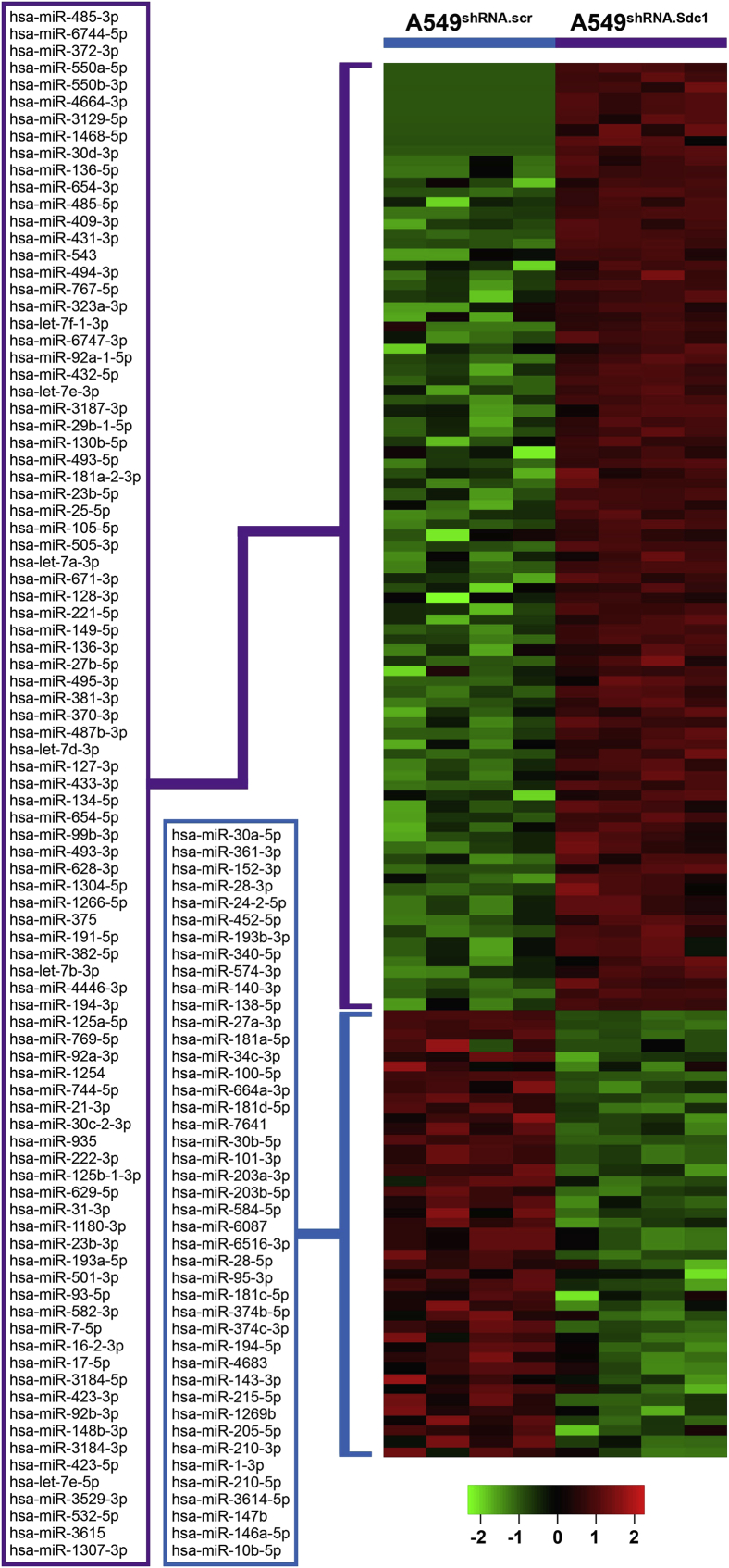
We postulated that syndecan-1 regulates the lung tumor microenvironment by altering the expression pattern of miRNAs contained within exosomes released by cancer cells. The miRNAs contained within exosomes collected from A549shRNA.scr and A549shRNA.Sdc1 cells were profiled, and 91 miRNAs that were less abundant and 43 miRNAs that were more abundant in cells that express syndecan-1 were identified (Figure 3 and Supplemental Table S1). These data demonstrate that syndecan-1 alters the exosome miRNA expression profile.
Figure 3.
Syndecan-1 expression by lung cancer cells alters miRNA content in exosomes. Exosomes were isolated from conditioned medium and processed for RNA sequencing. The heat map depicts the differentially expressed miRNAs in exosomes from A549shRNA.scr and A549shRNA.Sdc1 cells (false discovery rate < 0.01). Supplemental Table S2 contains the list of miRNAs.
The KRAS oncogene has been shown to regulate miRNA packaging into exosomes.38, 39 Therefore, whether the effect of syndecan-1 on miRNA packaging is dependent on the oncogenotype was evaluated. A549 cells are a lung adenocarcinoma cell line that contains a mutant KRAS gene, whereas EKVX cells are adenocarcinoma cell lines that have wild-type KRAS but a mutant p53 tumor-suppressor gene.40 miRNAs were profiled in exosomes isolated from EKVX cells that had suppressed syndecan-1 expression and were compared with those in appropriate control lines (Supplemental Table S2). Comparing the results from the A549 and EKVX cell lines, a number of miRNAs were found in common to be either up-regulated or down-regulated in exosomes depending on syndecan-1 expression level (Supplemental Table S3). The amounts of identical miRNAs found in both cell lines suggest that syndecan-1 controls exosome loading of a subset of miRNAs, independent of the oncogenotype of the tumor cell.
Loss of Syndecan-1 Expression Skews the miRNA Cargo within Exosomes toward Increased Signaling through Protumorigenic Pathways
To understand the impact of these changes in miRNA expression, miRNA targets were identified by TarBase, the manually curated database of experimentally verified miRNA–mRNA interactions, and KEGG pathway analysis of the miRNA targets was performed using miRPath software version 3.32, 33 Down- and up-regulated miRNAs respectively augment or suppress signaling through the functional pathways being targeted. Therefore, the pathways targeted by miRNAs that were less abundant in A549 cells lacking syndecan-1 expression were evaluated (Table 1 and Supplemental Table S4). The most significant pathways regulated by these miRNAs, which are all expected to have increased signaling, were distinctly procarcinogenic. For example, this group of lower-expressed miRNAs will augment signaling in pathways such as cell cycle, non–small cell lung cancer, and epidermal growth factor receptor, which are all protumorigenic. Conversely, the pathways regulated by miRNAs more abundant in the exosomes from cells deficient in syndecan-1 were also evaluated (Table 2 and Supplemental Table S5). The top pathway was extracellular matrix–receptor interactions, which is interesting considering that syndecan-1 regulates cell–matrix binding.41 Moreover, a loss of cell–matrix binding is necessary for anchorage-independent growth and cancer metastasis.2
Table 1.
Significantly Enriched Functional Pathways Altered by Down-Regulated miRNAs
| KEGG pathway | FDR |
|---|---|
| Fatty acid biosynthesis | 5.63 × 10−12 |
| Cell cycle | 6.08 × 10−10 |
| Viral carcinogenesis | 1.04 × 10−9 |
| Bladder cancer | 1.97 × 10−7 |
| Proteoglycans in cancer | 6.24 × 10−6 |
| Hepatitis B | 6.24 × 10−6 |
| Central carbon metabolism in cancer | 1.46 × 10−5 |
| Neurotrophin-signaling pathway | 1.46 × 10−5 |
| Chronic myeloid leukemia | 1.46 × 10−5 |
| Non-small cell lung cancer | 1.90 × 10−5 |
| ErbB-signaling pathway | 2.86 × 10−5 |
| Glioma | 2.86 × 10−5 |
| Epstein-Barr virus infection | 6.38 × 10−5 |
miRNAs with >fourfold less expression in A549shRNA.Sdc1 cells compared with A549shRNA.scr cells were uploaded into miRPath software version 3,32 and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using validated targets identified in DIANA-TarBase software version 7.0.33 Only pathways with a false discovery rate (FDR) of <0.0001 were listed.
ErbB, epidermal growth factor receptor.
Table 2.
Significantly Enriched Functional Pathways Altered by Up-Regulated miRNAs
| KEGG pathway | FDR |
|---|---|
| ECM–receptor interaction | 1.79 × 10−15 |
| Prion diseases | 8.47 × 10−11 |
| Protein processing in endoplasmic reticulum | 1.06 × 10−10 |
| Fatty acid biosynthesis | 2.05 × 10−7 |
| Spliceosome | 2.05 × 10−7 |
| Viral carcinogenesis | 4.03 × 10−6 |
| Hippo signaling pathway | 4.43 × 10−6 |
| Proteoglycans in cancer | 7.59 × 10−6 |
| Central carbon metabolism in cancer | 4.02 × 10−5 |
miRNAs with more than fourfold more expression in A549shRNA.Sdc1 cells compared with A549shRNA.scr cells were uploaded into miRPath software version 3,32 and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using validated targets identified in DIANA-TarBase software version 7.0.33 Only pathways with a false discovery rate (FDR) of <0.0001 were listed.
ECM, extracellular matrix.
Syndecan-1 Alters Exosome Ability to Promote Cellular Proliferation
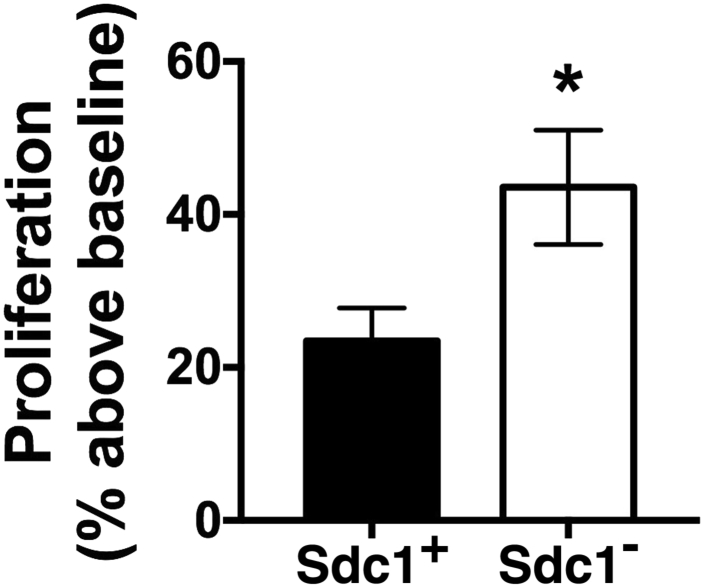
These findings indicate that the loss of syndecan-1 expression by lung cancer cells alters the miRNAs in secreted exosomes to a more tumorigenic profile, which is consistent with the in vitro and in vivo procancer phenotype linked to syndecan-1 deficiency (Figure 2). To test this concept, A549 cells were cultured with exosomes isolated from A549 cells replete with syndecan-1 (Sdc1+) and from cells deficient in syndecan-1 (Sdc1–) (Figure 4). A549 cells treated with Sdc1– exosomes had a higher proliferation rate than did cells treated with Sdc1+ exosomes. These data demonstrate that exosomes from syndecan-1–deficient cells stimulate proliferation in target cells.
Figure 4.
Proliferation is augmented by exosomes from syndecan-1 replete cells. Wild-type A549 cells were treated with exosomes from A549shRNA.scr and A549shRNA.Sdc1 cells (Sdc1+ and Sdc1−, respectively), and proliferation was measured. n = 6 per group. ∗P < 0.05 versus Sdc1+ (t-test).
Pathways Enriched in Adenocarcinoma Tumors Match the Targeted Pathways of the miRNAs Contained in the Exosomes Secreted from Lung Adenocarcinoma Cells
As a proof of concept that the change in the miRNA profile of exosomes from the loss of syndecan-1 expression in lung cancer cells promotes more tumorigenic signaling, publicly available microarray data from lung adenocarcinomas were analyzed and compared with those from normal lung (https://www.ncbi.nlm.nih.gov/geo; accession number GSE43767).34 The pathways that were affected by the significantly up- and down-regulated genes in adenocarcinoma were determined with WebGestalt (Supplemental Tables S6 and S7). As expected, the affected pathways overwhelmingly represented procancer signals. Moreover, 15 of 29 pathways (51.7%) were identical between those up-regulated in adenocarcinoma and those from down-regulated miRNAs (Table 3). Similarly, 18 of 27 pathways (66.7%) were identical between down-regulated pathways in adenocarcinoma and those from up-regulated miRNAs (Table 4). The top pathway was focal adhesion, which is related to extracellular matrix–receptor interactions, and consistent with the role of syndecan-1 in regulating focal-adhesion turnover.7
Table 3.
Functional Pathways Identified Both in the Analysis of the Predicted Targets of miRNAs in Exosomes that Were Less in Syndecan-1–Deficient Conditions and in the Analysis of Genes Up-Regulated in Adenocarcinoma Samples
| KEGG pathway | FDR |
|---|---|
| Cell cycle | 9.64 × 10−19 |
| p53 signaling pathway | 7.87 × 10−6 |
| Oocyte meiosis | 1.19 × 10−5 |
| Bladder cancer | 5.24 × 10−5 |
| RNA transport | 1 × 10−4 |
| Melanoma | 1.8 × 10−3 |
| Glioma | 2.8 × 10−3 |
| Pancreatic cancer | 5.1 × 10−3 |
| Prostate cancer | 1.2 × 10−2 |
| Chronic myeloid leukemia | 1.7 × 10−2 |
| Non–small cell lung cancer | 1.8 × 10−2 |
| ErbB-signaling pathway | 2.2 × 10−2 |
| Neurotrophin-signaling pathway | 3.2 × 10−2 |
| Endometrial cancer | 3.7 × 10−2 |
| Lysine degradation | 4.5 × 10−2 |
ErbB, epidermal growth factor receptor; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table 4.
Functional Pathways Identified Both in the Analysis of the Predicted Targets of miRNAs in Exosomes that Were More in Syndecan-1–Deficient Conditions and in the Analysis of Genes Down-Regulated in Adenocarcinoma Samples
| KEGG pathway | FDR |
|---|---|
| Focal adhesion | 1.58 × 10−16 |
| Endocytosis | 1.12 × 10−11 |
| Regulation of actin cytoskeleton | 7.85 × 10−10 |
| Small cell lung cancer | 4.70 × 10−9 |
| Gap junction | 2.09 × 10−5 |
| Ubiquitin mediated proteolysis | 2.70 × 10−5 |
| Neurotrophin signaling pathway | 7.18 × 10−5 |
| Prion diseases | 2 × 10−4 |
| Adherens junction | 2 × 10−4 |
| Chronic myeloid leukemia | 2 × 10−4 |
| Prostate cancer | 3 × 10−4 |
| ECM–receptor interaction | 7 × 10−4 |
| Glioma | 5.3 × 10−3 |
| Protein export | 5.4 × 10−3 |
| Dorsoventral axis formation | 6.6 × 10−3 |
| Spliceosome | 1.2 × 10−2 |
| Protein processing in endoplasmic reticulum | 1.5 × 10−2 |
| Cell cycle | 3.9 × 10−2 |
ECM, extracellular matrix; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Furthermore, putative miRNAs that target the differentially regulated genes in adenocarcinoma cells were identified and compared with the differentially expressed miRNAs contained in exosomes from A549 cells. On comparison of putative miRNAs from up-regulated genes to the list of down-regulated miRNAs, 27 of 43 miRNAs (62.8%) were present (Table 5). The reverse analysis between miRNAs predicted to target the down-regulated genes in adenocarcinoma and the up-regulated miRNAs in A549 exosomes identified overlap in 30 of the 91 miRNAs (33.0%). These data support the idea that the loss of syndecan-1 by lung adenocarcinoma cells alters the miRNA profile in secreted exosomes to generate a more tumorigenic milieu.
Table 5.
List of Putative miRNAs Identified by miRvestigator35 that Were Also Found to Be Differentially Expressed in Exosomes
| From down-regulated genes in adenocarcinoma and up-regulated in exosomes from A549shRNA.Sdc1 | From up-regulated genes in adenocarcinoma and down-regulated in exosomes from A549shRNA.Sdc1 |
|---|---|
| hsa-miR-485-3p | hsa-miR-30a-5p |
| hsa-miR-550b-3p | hsa-miR-361-3p |
| hsa-miR-3129-5p | hsa-miR-152-3p |
| hsa-miR-30d-3p | hsa-miR-28-3p |
| hsa-miR-136-5p | hsa-miR-24-2-5p |
| hsa-miR-409-3p | hsa-miR-452-5p |
| hsa-miR-543 | hsa-miR-340-5p |
| hsa-miR-323a-3p | hsa-miR-574-3p |
| hsa-let-7f-1-3p | hsa-miR-140-3p |
| hsa-miR-92a-1-5p | hsa-miR-27a-3p |
| hsa-let-7e-3p | hsa-miR-181a-5p |
| hsa-miR-29b-1-5p | hsa-miR-34c-3p |
| hsa-miR-493-5p | hsa-miR-664a-3p |
| hsa-let-7a-3p | hsa-miR-181d-5p |
| hsa-miR-128-3p | hsa-miR-30b-5p |
| hsa-miR-27b-5p | hsa-miR-101-3p |
| hsa-miR-495-3p | hsa-miR-203a-3p |
| hsa-miR-381-3p | hsa-miR-584-5p |
| hsa-let-7d-3p | hsa-miR-28-5p |
| hsa-miR-628-3p | hsa-miR-181c-5p |
| hsa-miR-1304-5p | hsa-miR-374b-5p |
| hsa-miR-382-5p | hsa-miR-374c-3p |
| hsa-let-7b-3p | hsa-miR-194-5p |
| hsa-miR-935 | hsa-miR-4683 |
| hsa-miR-125b-1-3p | hsa-miR-143-3p |
| hsa-miR-629-5p | hsa-miR-205-5p |
| hsa-miR-23b-3p | hsa-miR-10b-5p |
| hsa-miR-93-5p | |
| hsa-miR-16-2-3p | |
| hsa-miR-17-5p |
These are miRNAs from down-regulated genes in adenocarcinoma and up-regulated in A549shRNA.Sdc1 conditions.
Discussion
The tumor microenvironment has long been recognized to regulate cancer progression, and recent work has demonstrated that tumor-derived exosomes play a role in specifically targeting and reprogramming cells within the cancer niche.24, 25, 26, 20, 42 Certainly, exosomes released from tumor cells have been identified as important mediators that reshape the tumor microenvironment.19 Our data suggest the loss of syndecan-1 expression by high-grade lung adenocarcinomas facilitates the malignant process by transforming the tumor milieu to one that is more tumorigenic, through alterations in exosome cargo. Indeed, the miRNA profiles in exosomes from syndecan-1–deficient lung adenocarcinoma cells shifted in a way that would promote signaling tumor growth and progression. This work demonstrates that syndecan-1 (or loss thereof) in cancerous cells facilitates tumor progression by controlling the miRNA content packaged in exosomes.
Increased serum levels of syndecan-1 have been associated with worsened outcomes in lung cancer.43 To reconcile this finding with ours and others' demonstrating that tumors with lower syndecan-1 expression confer a survival disadvantage,13, 14 it can be inferred that excessive syndecan-1 shedding contributes to the lower syndecan-1 expression by the lung tumor. However, syndecan-1 surface expression is most likely not solely regulated by proteases. Indeed, there is a constitutive shedding of syndecan-1 from the cell membrane that regulates the normal turnover of this proteoglycan (Supplemental Figure S1). Moreover, cleavage of syndecan-1 is not fully removed from the lung epithelium in acute lung injury models in which protease activity has a quick and dramatic spike.4, 6, 44, 45 Finally, the data demonstrate that lower syndecan-1 mRNA levels are associated with a worsened clinical prognosis, which mirrors findings from prior survival studies that evaluated protein levels of syndecan-1,13, 14 and suggest that transcription is likely a major driver in the overall expression of syndecan-1.
Normal tissues produce innate antitumor signals that help to contain and suppress the formation of malignant cells.3 However, as the cancer progresses, it reshapes the tumor microenvironment to become more conducive to tumor growth and metastasis.2 In one aspect, loss of syndecan-1 expression diminishes the suppressive signals of various oncogenic pathways that are carried in exosomes (ie, decreased miRNA levels in exosomes from syndecan-1–deficient cells). In fact, many of the pathways targeted by the suppressed miRNAs in exosomes from syndecan-1–deficient A549 cells are directly implicated in carcinogenesis (ie, non–small cell lung cancer, viral carcinogenesis, bladder cancer, proteoglycans in cancer, central carbon metabolism in cancer, chronic myeloid leukemia, pancreatic cancer, prostate cancer, glioma, melanoma). Moreover, these miRNAs were also found to affect tumorigenic events such as fatty acid biosynthesis and epidermal growth factor receptor (EGFR) and neurotrophin signaling.46, 47, 48, 49 Cell cycle is also up-regulated, consistent with the fact that syndecan-1–deficient conditions produced larger tumors both in vitro and in vivo. Furthermore, we experimentally demonstrated that exosomes from syndecan-1–deficient conditions augment cell proliferation in comparison to exosomes from syndecan-1–expressing cells.
The loss of syndecan-1 expression by lung cancer cells may also alter miRNA content in exosomes to provide greater inhibition of various tumor-suppressive signals (ie, elevated miRNA levels in exosomes from syndecan-1–deficient cells). Interestingly, hsa-miR-485-3p, the expression of which was 184-fold higher in exosomes from syndecan-1–expressing A549 cells, has been found to suppress metastasis of breast and prostate carcinoma cells.50, 51 Congruous with this fact, extracellular matrix–receptor interaction was the functional pathway that had the highest significance in this analysis.
These studies demonstrate the ability of syndecan-1 to alter exosome contents, which in turn can affect various target cells within the malignancy niche. The mechanism by which syndecan-1 regulates exosome cargo loading has not been determined. However, syndecan-1 facilitates exosome biogenesis, and alterations to this process may affect miRNA loading into exosomes.16, 52, 53 Neutral sphingomyelinase-2 and Kirsten ras have both been shown to mediate miRNA packaging.38, 39, 54 RNA binding proteins have also been implicated in shuttling miRNAs into exosomes.55, 56, 57, 58 Moreover, certain motifs or RNA modifications made after translation preferentially sort miRNAs into exosomes.57, 59
The majority of miRNAs identified in the RNASeq profiling had only modest changes in expression. However, many of these miRNAs likely have overlapping roles in controlling various cellular pathways, either by targeting identical genes or by suppressing various adapter proteins within any given signaling cascade, thus enhancing the role of small changes to miRNA levels in controlling intracellular signaling networks. Moreover, cancer is a chronic disease that forms over an extended period, and a small divergence in cellular signals may have profound effects over time. Therefore, the power in these results may not be primarily driven by large expression changes in a small number of miRNAs but more importantly may be due to a cumulative effect of small changes in a large number of miRNAs to alter the tumor microenvironment.
TarBase was used to identify miRNA targets for functional pathway analysis.32, 33 The advantage of TarBase is that the interactions between the miRNA and mRNA are experimentally validated in comparison to programs that use predictive algorithms (eg, TargetScan).33 However, TarBase likely underestimates the number of true targets.60 In contrast, in silico methods may overestimate the number of targets and introduce false-positive genes into the functional analysis.61 Supporting the validity of our approach, the functional pathways regulated by miRNA changes in exosomes from A549 cells had a high degree of similarity to the changes in tissue samples from adenocarcinoma biopsy samples. Furthermore, putative miRNAs that would target the differentially regulated genes in adenocarcinoma also had a large proportion of matches to the results from exosome profiling.
Here, we show that syndecan-1 loss in lung cancer cells promotes tumor growth in vitro and in vivo. We demonstrate that syndecan-1 specifically changes the miRNAs packaged in exosomes released from lung cancer cells. These miRNA alterations promote a cancer pathway in the absence of syndecan-1 and are consistent with the activated pathways found in lung adenocarcinoma cells. Although the analysis was focused on the miRNA content of exosomes, it is possible that other types of cargo are altered by syndecan-1 and may also participate in reshaping the tumor microenvironment. Indeed, exosome contents are quite diverse, containing other types of small RNAs (eg, lncRNA), mRNAs, proteins, and lipids.18 Additionally, the results do not specifically pinpoint the target cells on which the exosomes are acting. However, a recent study demonstrated that several types of cells can take up tumor-derived extracellular vesicles in vivo.43 Nonetheless, this work implies that a loss of syndecan-1 by lung cancer alters the miRNA content within exosomes to promote malignant signals that mold the cancer niche. These data solidify syndecan-1 expression as a possible prognostic indicator in patients with lung cancer. Moreover, future research may include identifying specific miRNA signatures within exosomes as prognostic and/or diagnostic modalities for lung cancer.
Footnotes
Supported by NHLBI/NIH grants HL084396 (P.C.) and HL103868 (P.C.), and the Samuel Oschin Comprehensive Cancer Institute Lung Cancer Research Award (P.C.).
T.P. and R.B. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2017.12.009.
Supplemental Data
Supplemental Figure S1.
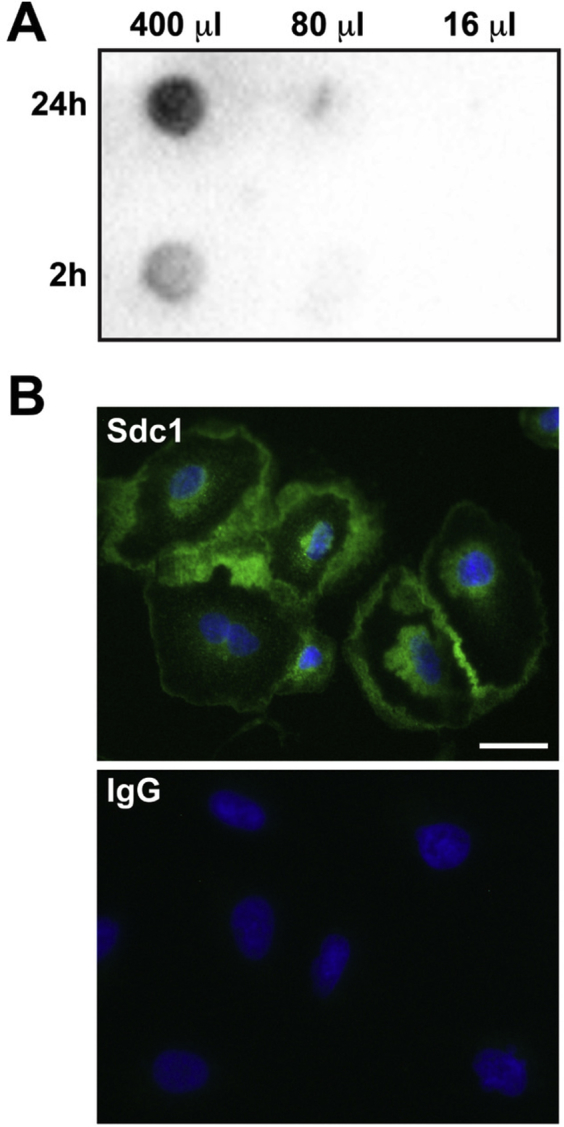
Syndecan-1 (Sdc1) is constitutively shed from A549 cells. A: Dot blot of conditioned medium from A549 cells demonstrates constitute shedding of syndecan-1. A549 cells were washed and fresh medium was added. After 2 and 24 hours, conditioned medium was collected and the respectively labeled microliters were added to the well of a dot-blot apparatus. Membranes were processed for dot blot using a mouse anti-human syndecan-1 antibody (MI15; BD Biosciences, San Jose, CA). B: Syndecan-1 immunofluorescence on A549 cells reveals persistent presence on the cell surface despite constitutive shedding. A549 cells were plated on chamber slides and processed for syndecan-1 immunostaining using a mouse anti-human syndecan-1 antibody (MI15) followed by an Alexa Fluor 488–conjugated anti-mouse IgG. Scale bar = 20 μm.
References
- 1.Herbst R.S., Heymach J.V., Lippman S.M. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Bissell M.J., Hines W.C. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brauer R., Ge L., Schlesinger S.Y., Birkland T.P., Huang Y., Parimon T., Lee V., McKinney B.L., McGuire J.K., Parks W.C., Chen P. Syndecan-1 attenuates lung injury during influenza infection by potentiating c-Met signaling to suppress epithelial apoptosis. Am J Respir Crit Care Med. 2016;194:333–344. doi: 10.1164/rccm.201509-1878OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couchman J.R. Syndecans: proteoglycan regulators of cell-surface microdomains? Nat Rev Mol Cell Biol. 2003;4:926–937. doi: 10.1038/nrm1257. [DOI] [PubMed] [Google Scholar]
- 6.Chen P., Abacherli L.E., Nadler S.T., Wang Y., Li Q., Parks W.C. MMP7 shedding of syndecan-1 facilitates re-epithelialization by affecting alpha(2)beta(1) integrin activation. PLoS One. 2009;4:e6565. doi: 10.1371/journal.pone.0006565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altemeier W.A., Schlesinger S.Y., Buell C.A., Parks W.C., Chen P. Syndecan-1 controls cell migration by activating Rap1 to regulate focal adhesion disassembly. J Cell Sci. 2012;125:5188–5195. doi: 10.1242/jcs.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altemeier W.A., Schlesinger S.Y., Buell C.A., Brauer R., Rapraeger A.C., Parks W.C., Chen P. Transmembrane and extracellular domains of syndecan-1 have distinct functions in regulating lung epithelial migration and adhesion. J Biol Chem. 2012;287:34927–34935. doi: 10.1074/jbc.M112.376814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beauvais D.M., Ell B.J., McWhorter A.R., Rapraeger A.C. Syndecan-1 regulates alphavbeta3 and alphavbeta5 integrin activation during angiogenesis and is blocked by synstatin, a novel peptide inhibitor. J Exp Med. 2009;206:691–705. doi: 10.1084/jem.20081278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato M., Saunders S., Nguyen H., Bernfield M. Loss of cell surface syndecan-1 causes epithelia to transform into anchorage-independent mesenchyme-like cells. Mol Biol Cell. 1995;6:559–576. doi: 10.1091/mbc.6.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stepp M.A., Pal-Ghosh S., Tadvalkar G., Rajjoub L., Jurjus R.A., Gerdes M., Ryscavage A., Cataisson C., Shukla A., Yuspa S.H. Loss of syndecan-1 is associated with malignant conversion in skin carcinogenesis. Mol Carcinog. 2010;49:363–373. doi: 10.1002/mc.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binder Gallimidi A., Nussbaum G., Hermano E., Weizman B., Meirovitz A., Vlodavsky I., Götte M., Elkin M. Syndecan-1 deficiency promotes tumor growth in a murine model of colitis-induced colon carcinoma. PLoS One. 2017;12:e0174343. doi: 10.1371/journal.pone.0174343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah L., Walter K.L., Borczuk A.C., Kawut S.M., Sonett J.R., Gorenstein L.A., Ginsburg M.E., Steinglass K.M., Powell C.A. Expression of syndecan-1 and expression of epidermal growth factor receptor are associated with survival in patients with nonsmall cell lung carcinoma. Cancer. 2004;101:1632–1638. doi: 10.1002/cncr.20542. [DOI] [PubMed] [Google Scholar]
- 14.Anttonen A., Heikkilä P., Kajanti M., Jalkanen M., Joensuu H. High syndecan-1 expression is associated with favourable outcome in squamous cell lung carcinoma treated with radical surgery. Lung Cancer. 2001;32:297–305. doi: 10.1016/s0169-5002(00)00230-0. [DOI] [PubMed] [Google Scholar]
- 15.Nackaerts K., Verbeken E., Deneffe G., Vanderschueren B., Demedts M., David G. Heparan sulfate proteoglycan expression in human lung-cancer cells. Int J Cancer. 1997;74:335–345. doi: 10.1002/(sici)1097-0215(19970620)74:3<335::aid-ijc18>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 16.Baietti M.F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., Zimmermann P., David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 17.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 18.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melo S.A., Sugimoto H., O'Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A., Lucci A., Ivan C., Calin G.A., Kalluri R. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J., Zanesi N., Crawford M., Ozer G.H., Wernicke D., Alder H., Caligiuri M.A., Nana-Sinkam P., Perrotti D., Croce C.M. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umezu T., Ohyashiki K., Kuroda M., Ohyashiki J.H. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2012;32:2747–2755. doi: 10.1038/onc.2012.295. [DOI] [PubMed] [Google Scholar]
- 23.Le M.T.N., Hamar P., Guo C., Basar E., Perdigão-Henriques R., Balaj L., Lieberman J. miR-200–containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124:5109–5128. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C.M., Nitadori-Hoshino A., Hoffman C., Badal K., Garcia B.A., Callahan M.K., Yuan J., Martins V.R., Skog J., Kaplan R.N., Brady M.S., Wolchok J.D., Chapman P.B., Kang Y., Bromberg J., Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W60–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlachos I.S., Paraskevopoulou M.D., Karagkouni D., Georgakilas G., Vergoulis T., Kanellos I., Anastasopoulos I.-L., Maniou S., Karathanou K., Kalfakakou D., Fevgas A., Dalamagas T., Hatzigeorgiou A.G. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43:D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng L., Wang J., Cao B., Zhang Y., Wu B., Di X., Jiang W., An N., Lu D., Gao S., Zhao Y., Chen Z., Mao Y., Gao Y., Zhou D., Jen J., Liu X., Zhang Y., Li X., Zhang K., He J., Cheng S. Gene expression profiling in human lung development: an abundant resource for lung adenocarcinoma prognosis. PLoS One. 2014;9:e105639. doi: 10.1371/journal.pone.0105639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plaisier C.L., Bare J.C., Baliga N.S. miRvestigator: web application to identify miRNAs responsible for co-regulated gene expression patterns discovered through transcriptome profiling. Nucleic Acids Res. 2011;39:W125–W131. doi: 10.1093/nar/gkr374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gazdar A.F., Girard L., Lockwood W.W., Lam W.L., Minna J.D. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1310–1321. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DuPage M., Dooley A.L., Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cha D.J., Franklin J.L., Dou Y., Liu Q., Higginbotham J.N., Demory Beckler M., Weaver A.M., Vickers K., Prasad N., Levy S., Zhang B., Coffey R.J., Patton J.G. KRAS-dependent sorting of miRNA to exosomes. eLife. 2015;4:e07197. doi: 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenzie A.J., Hoshino D., Hong N.H., Cha D.J., Franklin J.L., Coffey R.J., Patton J.G., Weaver A.M. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 2016;15:978–987. doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikediobi O.N., Davies H., Bignell G., Edkins S., Stevens C., O'Meara S. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan M.R., Humphries M.J., Bass M.D. Synergistic control of cell adhesion by integrins and syndecans. Nat Rev Mol Cell Biol. 2007;8:957–969. doi: 10.1038/nrm2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zomer A., Maynard C., Verweij F.J., Kamermans A., Schäfer R., Beerling E., Schiffelers R.M., de Wit E., Berenguer J., Ellenbroek S.I.J., Wurdinger T., Pegtel D.M., van Rheenen J. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046–1057. doi: 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joensuu H., Anttonen A., Eriksson M., Mäkitaro R., Alfthan H., Kinnula V., Leppä S. Soluble syndecan-1 and serum basic fibroblast growth factor are new prognostic factors in lung cancer. Cancer Res. 2002;62:5210–5217. [PubMed] [Google Scholar]
- 44.Li Q., Park P.W., Wilson C.L., Parks W.C. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell. 2002;111:635–646. doi: 10.1016/s0092-8674(02)01079-6. [DOI] [PubMed] [Google Scholar]
- 45.Zuo D., Chen Y., Li W., Liu R., He Y., Ren L., Zhou L., Deng T., Wang X., Ying G., Ba Y. Shed syndecan-1 is involved in chemotherapy resistance via the EGFR pathway in colorectal cancer. Br J Cancer. 2014;111:1965–1976. doi: 10.1038/bjc.2014.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciardiello F., Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 47.Sinkevicius K.W., Kriegel C., Bellaria K.J., Lee J., Lau A.N., Leeman K.T., Zhou P., Beede A.M., Fillmore C.M., Caswell D., Barrios J., Wong K.-K., Sholl L.M., Schlaeger T.M., Bronson R.T., Chirieac L.R., Winslow M.M., Haigis M.C., Kim C.F. Neurotrophin receptor TrkB promotes lung adenocarcinoma metastasis. Proc Natl Acad Sci U S A. 2014;111:10299–10304. doi: 10.1073/pnas.1404399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chopin V., Lagadec C., Toillon R.A., Le Bourhis X. Neurotrophin signaling in cancer stem cells. Cell Mol Life Sci. 2016;73:1859–1870. doi: 10.1007/s00018-016-2156-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gouw A.M., Eberlin L.S., Margulis K., Sullivan D.K., Toal G.G., Tong L., Zare R.N., Felsher D.W. Oncogene KRAS activates fatty acid synthase, resulting in specific ERK and lipid signatures associated with lung adenocarcinoma. Proc Natl Acad Sci U S A. 2017;114:4300–4305. doi: 10.1073/pnas.1617709114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lou C., Xiao M., Cheng S., Lu X., Jia S., Ren Y., Li Z. MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1α expression. Cell Death Dis. 2016;7:e2159. doi: 10.1038/cddis.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Formosa A., Markert E.K., Lena A.M., Italiano D., Finazzi-Agro E., Levine A.J., Bernardini S., Garabadgiu A.V., Melino G., Candi E. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2013;33:5173–5182. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 52.Wei J.-X., Lv L.-H., Wan Y.-L., Cao Y., Li G.-L., Lin H.-M., Zhou R., Shang C.-Z., Cao J., He H., Han Q.-F., Liu P.-Q., Zhou G., Min J. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61:1284–1294. doi: 10.1002/hep.27660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iavello A., Frech V.S.L., Gai C., Deregibus M.C., Quesenberry P.J., Camussi G. Role of Alix in miRNA packaging during extracellular vesicle biogenesis. Int J Mol Med. 2016;37:958–966. doi: 10.3892/ijmm.2016.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kosaka N., Iguchi H., Hagiwara K., Yoshioka Y., Takeshita F., Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shurtleff M.J., Yao J., Qin Y., Nottingham R.M., Temoche-Diaz M.M., Schekman R., Lambowitz A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci U S A. 2017;114:E8987–E8995. doi: 10.1073/pnas.1712108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shurtleff M.J., Temoche-Diaz M.M., Karfilis K.V., Ri S., Schekman R., Nilsen T.W. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife. 2016;5:e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Cabo F., Pérez-Hernández D., Vázquez J., Martin-Cofreces N., Martinez-Herrera D.J., Pascual-Montano A., Mittelbrunn M., Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santangelo L., Giurato G., Cicchini C., Montaldo C., Mancone C., Tarallo R., Battistelli C., Alonzi T., Weisz A., Tripodi M. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 59.Koppers-Lalic D., Hackenberg M., Bijnsdorp I.V., van Eijndhoven M.A.J., Sadek P., Sie D., Zini N., Middeldorp J.M., Ylstra B., de Menezes R.X., Wurdinger T., Meijer G.A., Pegtel D.M. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 60.Sethupathy P., Corda B., Hatzigeorgiou A.G. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Riffo-Campos Á.L., Riquelme I., Brebi-Mieville P. Tools for sequence-based miRNA target prediction: what to choose? Int J Mol Sci. 2016;17:1987. doi: 10.3390/ijms17121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.