Abstract
The subesophageal zone (SEZ) of the Drosophila brain houses the circuitry underlying feeding behavior and is involved in many other aspects of sensory processing and locomotor control. Formed by the merging of four neuromeres, the internal architecture of the SEZ can be best understood by identifying segmentally reiterated landmarks emerging in the embryo and larva, and following the gradual changes by which these landmarks become integrated into the mature SEZ during metamorphosis. In previous works, the system of longitudinal fibers (connectives) and transverse axons (commissures) has been utilized as a scaffold that provides internal landmarks for the neuromeres of the larval ventral nerve cord. We have extended the analysis of this scaffold to the SEZ and, in addition, reconstructed the tracts formed by lineages and nerves in relationship to the connectives and commissures. As a result we establish reliable criteria that define boundaries between the four neuromeres (tritocerebrum; mandibular neuromere, maxillary neuromere, labial neuromere) of the SEZ at all stages of development. Fascicles and lineage tracts also demarcate seven columnar neuropil domains (ventromedial, ventro-lateral; centromedial; central; centrolateral; dorsomedial; dorsolateral) identifiable throughout development. These anatomical subdivisions, presented in the form of an atlas including confocal sections and 3D digital models for the larval, pupal and adult stage, allowed us to describe the morphogenetic changes shaping the adult SEZ. Finally, we mapped MARCM-labeled clones of all secondary lineages of the SEZ to the newly established neuropil subdivisions. Our work will facilitate future studies of function and comparative anatomy of the SEZ.
Keywords: Drosophila, brain, subesophageal zone, tritocerebrum, gnathal ganglion, tract, neural lineage, compartment, development, RRID:BDSC_9313, RRID:BDSC_32185, RRID:AB_528402, RRID:AB_2314331, RRID:AB_528404
1 Introduction
Processing of gustatory information and the control of feeding behavior in insects is executed by neural circuits located in the subesophageal ganglion (SEG) of the brain (Bullock and Horridge, 1965; Dethier, 1976; Wang et al., 2004; Scott, 2005; Cobb et al., 2009; Freeman and Dahanukar, 2015; Schwarz et al., 2017). In addition, command centers for a variety of other behaviors are also situated in the SEG (larval wandering and ecdysis behavior: Dominick and Truman, 1986a/b; Zitnan and Adams, 2000; stridulation: Lins and Lakes-Harlan, 1994; respiration: Ramirez, 1998; locomotor behavioral choice: Matsuura et al., 2002; reproductive behavior: Certel et al., 2007; Sakurai et al., 2013; aggression: Zhou et al., 2008; Andrews et al., 2014; courtship behavior: Auer and Benton, 2016). The SEG consists of the three fused segmental ganglia that innervate the structures surrounding the mouth of the animal. In primitive insects, three separate segments (called gnathal segments) with movable appendages (mandible, maxilla, labium) form the mouthparts of the animal. In flies, the gnathal segments are strongly reduced and structurally modified. Mandibles are essentially absent; of the maxillary appendages, only the small maxillary palps remain; the labial appendages form the fused proboscis (Ferris, 1950; Bryant, 1978). A similar reduction has taken place in the internal structure of the suboesophageal ganglion. For example, the number of neural lineages, which for the thorax amounts to 30 pairs per neuromere (i.e., 90 per three hemineuromeres), is reduced to a total of 14 for all three gnathal neuromeres together (Kuert et al., 2014). This reduction is mostly brought about by programmed cell death, which sets in shortly after gnathal neuroblasts have delaminated (Kuert et al., 2014; Urbach et al., 2016). Furthermore, commissural axon tracts and overall volume of the subesophageal neuropil are decreased. Furthermore, as a result of the pronounced condensation that affects the fly nervous system as a whole, the subesophageal ganglion is fused with the basal part of the supraesophageal ganglion into a composite domain called the subesophageal zone (SEZ; Ito et al., 2014). Anteriorly, the tritocerebrum (the neuromere innervating the mouth cavity and the gut) has become fully incorporated into the subesophageal zone (Fig. 1a). The tritocerebrum contains neural circuits that sense food stuffs in the mouth and integrate this information with input from the viscera (parameters like extension of the gut or nutrient levels in the tissues, which reflect the need for food intake), and then generate a motor output controlling feeding behavior (Dethier, 1976; Rajashekhar and Singh, 1994).
Figure 1.
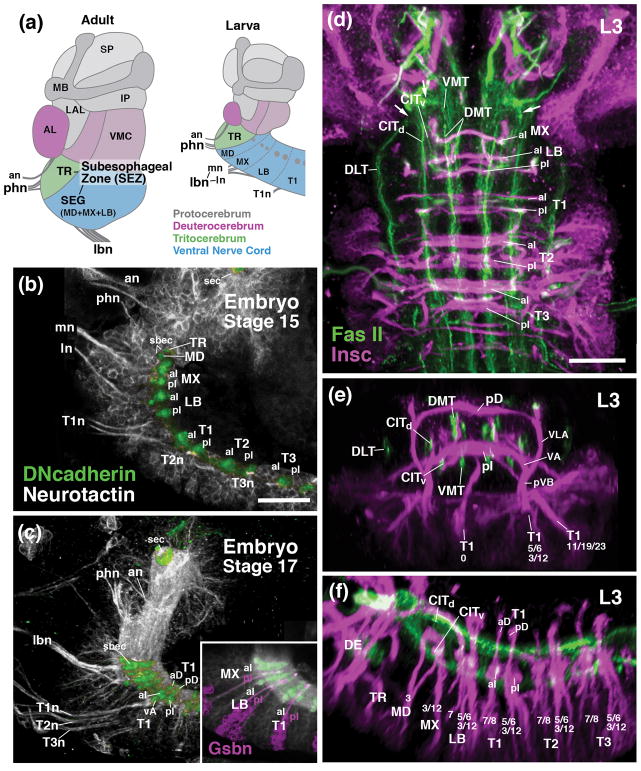
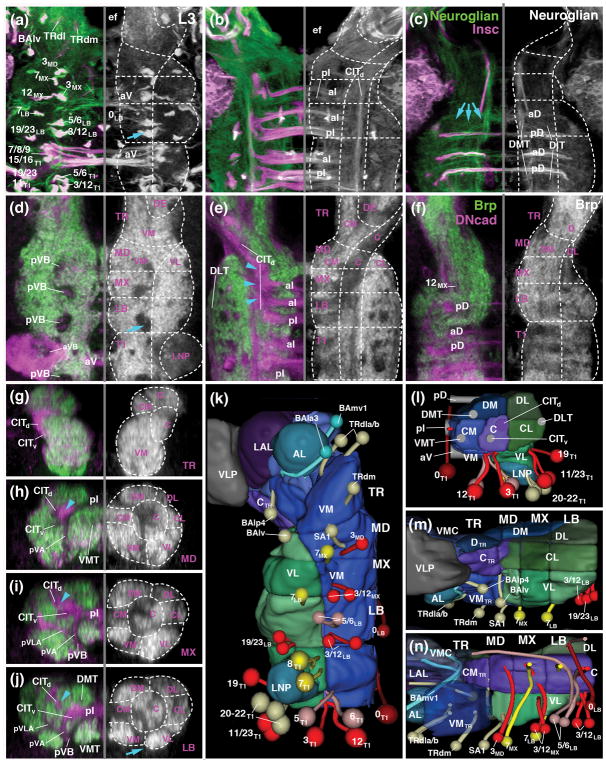
Metameric organization of the subesophageal zone. (a) Schematic parasagittal section of adult brain (left) and larval brain showing metameric brain architecture, composed of protocerebrum (gray; SLP superior protocerebrum; IP inferior protocerebrum, MB mushroom body, LAL lateral accessory lobe), deuterocerebrum (magenta; AL antennal lobe; VMC ventromedial cerebrum), tritocerebrum (TR; green), and subesophageal ganglia (SEG) (blue; MD mandibula; MX maxilla; LB labium). (b, c) Parasagittal confocal sections of embryonic brain (B: stage 15; C: stage 17; anterior to the left, dorsal up). Labeling of primary neurons with anti-Neurotactin (BP106; white) and neuropil with anti-DN-cadherin (green). Note metameric arrangement of peripheral nerves (an antennal nerve; phn pharyngeal nerve; mn maxillary segmental nerve; ln labial segmental nerve; lbn combined maxillary-labial nerve; T1n-T3n nerves of thoracic segments 1–3) and commissures (aD anterior dorsal commissure; pD posterior dorsal commissure; aI anterior intermediate commissure; pI posterior intermediate commissure; sbec subesophageal commissure, formed by crossing fibers of mandibula and tritocerebrum; sec supraesophageal commissure). (d–f) Z-projections of confocal sections of third instar larval nerve cord (d: horizontal plane; E: frontal plane at level of T1 neuromere; f: parasagittal plane). Labeling with Insc-Gal4>UAS-mcd8-GFP (magenta; secondary neurons with their lineage-associated tracts; lineages numbered according to nomenclature of Truman et al., 2004) and anti-Fasciclin II (green; longitudinal long axon tracts; CITd dorsal central intermediate tract; CITv ventral central intermediate tract; DLT dorsal lateral tract; DMT dorsal medial tract; VMT ventral medial tract). For other abbreviations: see List of Abbreviations. Bars: 25μm (a–c; d–h)
Sensory and motor axons connect to the SEZ via two composite peripheral nerves called labial nerve, and pharyngeal nerve (Fig. 1a). The labial nerve, formed by axon bundles of both labial and maxillary origin, enters the central part of the SEZ and carries chemosensory and mechanosensory afferents from the maxillary palps, proboscis and head capsule, and motor axons that move the proboscis. The pharyngeal nerve, which conducts sensory afferents from the mouth cavity and the foregut, enters the tritocerebrum, located at the anterior tip of the SEZ (Rajashekhar and Singh, 1994). Motor axons of the pharyngeal nerve control the pharyngeal dilator muscles (effect suction during feeding), movement of the proboscis, as well as motility of the foregut.
Very little is known about the anatomy and function of the large number of interneurons, which constitute the majority of neurons in the SEZ by far, and whose arborizations form the “association neuropil” compartments surrounding the sensory centers. A multitude of Gal4 and LexA markers is now becoming available (e.g., Jenett et al., 2012), many of them expressed in specific subsets of central interneurons. These lines are used as tools for anatomical mapping and functional manipulation to address the circuitry in the SEZ. As a prerequisite to integrate the great number of disparate patterns of projection and branching revealed by the reporter lines expressed in the SEZ, we undertake in this and the accompanying paper (Kendroud et al., 2017) an analysis of the neural lineages and compartments, developmental and structural units which allow one to construct a detailed neuroanatomical map grounded in development, similar to the one existing for the brain (Pereanu and Hartenstein, 2006; Cardona et al., 2009; Pereanu et al., 2010; Lovick et al., 2013; Wong et al., 2013; Ito et al., 2014; Hartenstein et al., 2015).
Lineages of the Drosophila central nervous system are clusters of neurons, typically in the range of 100–150 cells, that all descend from a common progenitor cell, called neuroblast. Neuroblasts represent a fixed population of uniquely identifiable cells that delaminate from the neurectoderm of the embryo and divide asymmetrically in a stem cell mode. Each division produces one daughter cell that keeps dividing as a neuroblast, and a second daughter cell, called ganglion mother cell (GMC), which undergoes one further mitosis. The mitosis of the GMC is also asymmetric and gives rise to an “A” daughter neuron and a “B” daughter neuron (Truman et al., 2010). “A” neurons and “B” neurons form the so called “A” hemilineage and “B” hemilineage, respectively. Most neuroblasts produce lineages of 10–20 primary neurons during the embryonic period. A few neuroblast generate as few as 4 neurons (e.g., NB7-3), or more than 35 neurons (e.g., NB7-1; Bossing et al., 1996; Schmidt et al., 1997; Schmid et al., 1999). Primary neurons differentiate into the larval brain and ventral nerve cord (Hartenstein et al., 2008). After a period of mitotic quiescence that lasts from mid-embryogenesis to early larval development (Lovick and Hartenstein, 2015), neuroblasts resume their activity and produce lineages of 100–150 secondary neurons (Bello et al., 2008). Primary neurons and secondary neurons together constitute the adult brain.
Neuroblasts and the lineages they produce represent genetic modules. Distinct combinations of transcription factors become active in a neuroblast and shape the morphology and function of the lineage of neurons produced by this neuroblast (Pearson and Doe, 2004; Brody and Odenwald, 2005; Kohwi and Doe, 2013). Aside from their relevance as genetic modules, lineages and hemilineages also form structural units. Neurons forming a hemilineage typically remain clustered together throughout development, and axons emitted by neurons of one hemilineage form a coherent fascicle, the primary and secondary axon tracts (PAT, SAT; for review see Hartenstein et al., 2008; Spindler and Hartenstein, 2010). In many cases, one hemilineage undergoes programmed cell death, resulting in lineages that consist of a single cluster/axon tract (Truman et al., 2010; Kumar et al., 2009; Lovick et al., 2016). Once neurons differentiate, the axonal and dendritic arborizations of a given (hemi)lineage are spatially confined to an individual neuropil compartment, or parts thereof; examples are the calyx of the mushroom body or the antennal lobe, formed by dendrites of four, or five, neighboring lineages, respectively (Ito et al., 1997; Lai et al., 2008; Ito et al., 2013; Wong et al., 2013; Yu et al., 2013). In other words, lineages and their tracts represent a scaffold of connections that determines the “macrocircuitry” of the insect nervous system.
In previous studies, the axon tracts of secondary lineages of the ventral nerve cord (VNC) and the SEZ were mapped at the late larval stage, when these structures are most obvious (Truman et al., 2004; Kuert et al., 2012; 2014). The lineages of the thoracic segments were also presented as clones for the adult (Harris et al., 2015; Shepherd et al., 2016). In this paper we reconstruct secondary lineages of the SEZ from the larval to adult stage, and establish the spatial relationship between lineage-associated tracts and the systems of longitudinal fibers (connectives) and transverse axons (commissures) that were defined in previous works (e.g., Landgraf et al., 2003; Nassif et al., 2003; Truman et al., 2004) as internal landmarks for the neuromeres of the larval ventral nerve cord. Our analysis allows us to define boundaries between the four neuromeres of the SEZ [tritocerebrum; mandibular neuromere (more simply called “mandibula” in the following), maxillary neuromere (“maxilla”), labial neuromere (“labium”)] at all stages of development. Fascicles and lineage tracts also demarcate discrete columnar neuropil domains along the dorso-ventral and medio-lateral axis of the neuropil. Based on the totality of these landmarks we establish an anatomical atlas of the larval and adult SEZ, and analyze the morphogenetic changes that take place during the transition between these two stages.
2 Material and Methods
2.1. Drosophila Stocks
Flies were reared at 25°C using standard fly media unless otherwise noted. The Drosophila stocks utilized in this study include,10xUAS-mCD8::GFP (Bloomington Drosophila Stock Center (BDSC) #32185; RRID:BDSC_32185),, Tdc2-Gal4 (Selcho et al., 2012; 2014; Bloomington Drosophila Stock Center (BDSC) #9313; RRID:BDSC_9313), Oregon RT, gsbn-Gal4 (He and Noll, 2013; ).
2.2. Immunohistochemistry
The following primary antibodies were used: mouse anti-Neurotactin (Nrt, BP106; RRID:AB_528404), mouse anti-Neuroglian (Nrg, BP104; RRID:AB_528402), and rat anti-DN-cadherin (DN-Ex #8; RRID:AB_2314331) antibodies from Developmental Studies Hybridoma Bank (DSHB, University of Iowa, Iowa City, Iowa; each diluted 1:10). For antibody labeling standard procedures were followed (e.g., Ashburner, 1989). For fluorescent staining, the following secondary antibodies were used: Alexa Fluor 546 goat anti-Mouse IgG (H+L) (#A11030; Invitrogen, Carlsbad, CA; used at 1:500) and Cy5 goat anti-Rat IgG (H+L) (112-175-143; Jackson Immunoresearch, West Grove, PA; used at 1:400).
All larvae, pupae and adults were grown at 25°C on standard food media. Adults were aged 3 to 5 days post-eclosion before dissection. For antibody labeling, standard procedures were followed (Ashburner, 1989). Briefly, dissected brains were fixed with phosphate buffered saline (PBS), pH 7.4, containing 4% paraformaldehyde for 25 – 30 min’s. They were then washed with 1× PBS, pH 7.4, containing 0.1 % Triton X-100 for 3 X 10 min’s. Samples were then incubated in blocking buffer (2% bovine serum albumin (BSA) in 1X PBS, pH 7.4, containing 0.1 % Triton X-100) for 1 hour at RT. They were then incubated with primary antibody diluted in blocking buffer overnight at 4C. They were then washed 3 X 15 min in 1X PBS, pH 7.4, containing 0.1 % Triton X-100 @ RT. The samples were washed in blocking buffer 1 X 20 min’s. They were then incubated with secondary antibody diluted in blocking buffer overnight at 4C. Samples were washed in 1X PBS, pH 7.4, containing 0.1 % Triton X-100 for 3 X 15 min’s and mounted in Vectashield (Vector Laboratories). Drosophila brains labeled with antibody markers were viewed as whole-mounts by confocal microscopy [LSM 700 Imager M2 using Zen 2009 (Carl Zeiss Inc.); lenses: 40× oil (numerical aperture 1.3)]. Complete series of optical sections were taken from preparations between 1.2 and 2-μM intervals.
2.3. Markers
The DN-cadherin antibody (DSHB DN-EX #8), a marker for neuropil, is a mouse monoclonal antibody raised against a peptide encoded by exon 8, amino acid residues 1210–1272 of the Drosophila CadN gene. The antibody detected two major bands, 300 kDa and 200 kDa molecular weights on Western blot of S2 cells only after transfection with a cDNA encoding the DN-cadherin protein (Iwai et al., 1997). In addition, the specificity of this antibody was tested with immunostaining of Drosophila embryos. Signal was hardly detectable in homozygous mutant, l(2)36DaM19 with nonsense mutation causes premature termination of protein translation. In contrast, this antibody gave a signal in mutant embryos expressing a DN-cadherin transgene.
The Neurotactin antibody (DSHB BP106) labels secondary neurons and their axons. It is a mouse monoclonal antibody raised against the first 280 aminoterminal amino acid residues (Hortsch et al., 1990) of the Drosophila Nrt gene. The monoclonal antibody detected the same Drosophila embryonic pattern to that of a polyclonal antisera raised against a fusion protein using part of the Neurotactin cDNA (Hortsch et al., 1990). In addition, another monoclonal antibody, MAb E1C, against Neurotactin gave a similar expression pattern in Drosophila embryos to that of BP106 (Piovant and Léna, 1988).
The Neuroglian antibody (DSHB BP104) labels secondary neurons and axons in the adult brain. It is a mouse monoclonal antibody from a library generated against isolated Drosophila embryonic nerve cords (Bieber et al., 1989). The Nrg antibody was used to purify protein from whole embryo extracts by immunoaffinity chromatography. Protein microsequencing of the purified protein was performed to determine that the 18 N-terminal amino acids that is identical to the sequence determined for the N-terminus of the protein based on a full-length cDNA clone (Bieber et al., 1989).
2.4. Generation and documentation of clones
Clones were generated by Flp-mediated mitotic recombination at homologous FRT sites (Lee and Luo, 2001). Mitotic clones were induced during the late first instar/early second instar stages by heat-shocking at 38°C for 30 minutes to 1 hour (approximately 12h-44h after larval hatching). GFP-labeled MARCM clones contain the following genotype:
hsflp/+; FRTG13, UAS-mCD8GFP/FRTG13, tub-GAL80; tub-Gal4/+ or
FRT19A GAL80, hsflp, UAS-mCD8GFP/elavC155-Gal4, FRT19A; UAS-CD8GFP/+
To generate the figure panels illustrating clones (Fig. 8, 9), z-projections of the individual MARCM clones were registered digitally with z-projections of a standard brain (“2D registration”). To this end, the standard brain was subdivided along the antero-posterior axis into six slices of approximately 20 μm thickness. These slices, each one characterized by one or more easily recognized landmark structures (antennal lobe, optic tubercle, ellipsoid body, fan-shaped body, lateral bend of antennal lobe tract, calyx), are introduced and utilized in previous papers (Pereanu et al., 2010; Lovick et al., 2013; Wong et al., 2013). The process of 2D registration involved the following steps:
Figure 8.
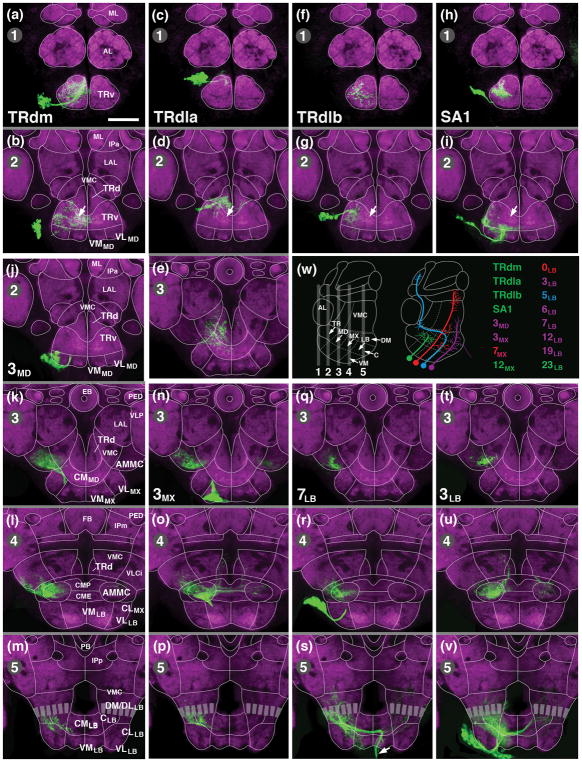
Clones representing secondary lineages of SEZ. Panels (a–v) show z-projections of frontal (according to body axis) confocal sections of adult brain labeled with anti-DN-cadherin (neuropil; magenta). Z-projections illustrate five different planes, numbered 1–5 in upper left corner of panels, along the anterior-posterior axis. Planes are indicated as numbered gray bars on schematic sagittal section shown in panel (w; left). Thin white lines in (a–v) show boundaries between neuropil domains; domains are annotated on panels of the left column (a, b, k, l, m); for abbreviations see List of Abbreviations. MARCM clones are labeled by GFP (green; see Material and Methods). Upper rows of panels (a–i) represent the four lineages associated with the anterior SEZ (a, b: TRdm; c–e: TRdla; f, g: TRdlb; h, i: SA1). Arrows in (b, c, g, i) point at DN-cadherin rich domain within ventromedial tritocerebrum that corresponds to gustatory center (see accompanying paper by Kendroud et al., 2017). Panels of bottom rows (j–v) present lineages targeting the central neuropil column of the SEZ (j–m: 3MD; n–p: 3MX; q–s: 7LB; t–v: 3LB). Arrow in (s) points at descending tract formed by lineage 7LB. Each clone is represented by two to four contiguous panels; panels belonging to the same clone are separated by gray lines; the entire clone is framed by thick white boundaries. Name of lineage represented by clone is given in first panel representing that clone. For example, clone of lineage 3MD (j, k, l, m) is visible on the four z-projections corresponding to levels 2 [j; level of medial mushroom body lobes (ML)], 3 [k; level of ellipsoid body (EB)], 4 [l; level of fan-shaped body (FB)], and 5 [m; level of middle part of protocerebral bridge (PB)]. (w) shows schematic sagittal section of brain and lists SEZ lineages shown in Figures 8 and 9. Lineages with mainly local arborizations are colored green; those with wide projections in adjoining domains along the anterior-posterior or dorso-ventral axis are in purple. Lineages 7MX and 0LB, shown in red, and 5LB, in blue, connect distant, non-contiguous regions of the brain. Bar: 50μm.
Figure 9.
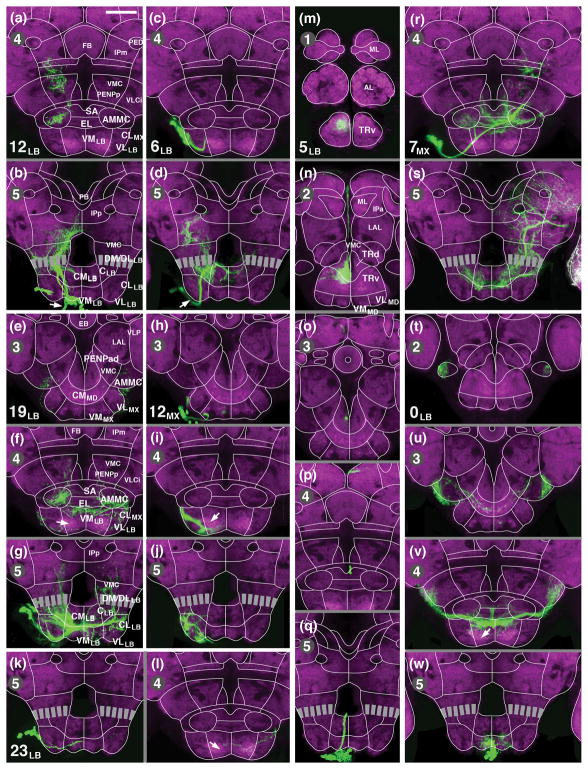
Clones representing secondary lineages of SEZ. Panels (a–w) show z-projections of frontal confocal sections of adult brain labeled with anti-DN-cadherin (neuropil; magenta). Panels are constructed and are arranged as described in legend of Figure 8. Panels of two columns to the left (a–l) illustrate lineages with arborizations restricted to the posterior SEZ (a, b: 12LB; c, d: 6LB; e–g: 19LB; h–j: 12MX; k, l: 23LB). Panels of the third column (m–q) show lineage 5LB, which projects from the labium through the tritocerebrum to the superior protocerebrum. The right column depicts lineage 7MX, which links the SEZ with the SPG (r, s), and 0LB, with bilaterally symmetric projections in the SEZ and VLCi (t–w). Arrows in (f, i, l) point at DN-cadherin rich domain within ventromedial maxilla/labium that corresponds to central sensory endings of labial nerve (see accompanying paper by Kendroud et al., 2017). Arrows in (b, d) points at descending tracts formed by lineages 6LB and 12LB. For abbreviations, see List of Abbreviations. Bar: 50μm
The confocal stack depicting a given clone was imported into the FIJI program (Schindelin et al., 2012; National Institutes of Health, http://rsbweb.nih.gov/ij/ and http://fiji.sc/) and digitally oriented such that the peduncle was aligned with the z-axis of the stack;
Z-projections of the 20μm slices of a clone corresponding to the standard levels described above were generated.
Z-projections of slices of clones and corresponding z-projections of standard brain were imported as two layers into a file generated by the Adobe Photoshop program. Using few standard landmarks (location of dorsal and ventral midline of SEZ neuropil, entry portals of SEZ lineages), the layer containing the clone (rendered temporarily semitransparent) was optimally fitted to the underlying layer representing the standard brain.
The optimally-fitted layer containing the clone was re-opened in FIJI, and then merged with the red channel (BP104 or DN-cadherin) of the standard brain.
2.5. Generation of Three-Dimensional Models
Digitized images of confocal sections were imported into FIJI (Schindelin et al., 2012; http://fiji.sc/). Complete series of optical sections were taken at 2μm intervals. Since sections were taken from focal planes of one and the same preparation, there was no need for alignment of different sections. Models were generated using the 3-dimensional viewer as part of the FIJI software package. Digitized images of confocal sections were imported using TrakEM2 plugin in FIJI software (Cardona et al., 2012). Digital atlas models of cell body clusters and SATs were created by manually labeling each lineage and its approximate cell body cluster location in TrakEM2.
3 Results
3.1. Lists of abbreviations used throughout text and figures
0, 2, 3, 5, 6, 7, 8, 11, 12, 19, 20–22, 23 identifiers for lineages of the ventral nerve cord, including gnathal ganglia (as defined in Truman et al., 2004; Kuert et al., 2014); aD anterior dorsal commissure; aI anterior intermediate commissure; AL antennal lobe; an antennal nerve; AMMC antenno-mechanosensory and motor center; aVA anterior ventral arch; aVB anterior vertical bundle; BAla1–4, BAmv1/2 deuterocerebral lineages (as defined in Pereanu and Hartenstein, 2006); BAlv tritocerebral secondary lineage (as defined in Pereanu and Hartenstein, 2006); C central neuropil domain; CITd dorsal central intermediate tract; CITv ventral central intermediate tract; CL centrolateral neuropil domain; CM centromedial neuropil domain; CME centromedian ellipsoid; CMP centromedian plate; DE deuterocerebrum; DL dorsolateral neuropil domain; DLT dorsal lateral tract; DM dorsomedial neuropil domain; DMT dorsal medial tract; EB ellipsoid body; FB fan-shaped body; IP inferior protocerebrum; IPa anterior inferior protocerebrum; IPm medial inferior protocerebrum; IPp posterior inferior protocerebrum; LAL lateral accessory lobe; LB labium; lbn combined labial nerve; ln labial segmental nerve; LNP leg neuropil; MB mushroom body; MD mandibula; ML medial lobe of mushroom body; MX maxilla; mn maxillary segmental nerve; PB protocerebral bridge; pD posterior dorsal commissure; PED peduncle of mushroom body; PENPa anterior periesophageal neuropil; pI posterior intermediate commissure; phn combined pharyngeal nerve; pVB posterior vertical bundle; SA1 mandibular secondary lineage with unclear homology in other neuromeres; sbec subesophageal commissure; sec supraesophageal commissure; SEG subesophageal ganglion; SEZ subesophageal zone; SP superior protocerebrum; T1–T3 thoracic neuromeres 1–3; T1n–T3n nerves of thoracic segments 1–3; TR tritocerebrum; TRd dorsal tritocerebrum; TRdla, TRdlb, TRdm: tritocerebral secondary lineages with unclear homologies in other neuromeres; TRv ventral tritocerebrum; VA ventral arch; VL ventrolateral neuropil domain; VLA ventro-lateral arch; VLCi inferior ventrolateral cerebrum; VLP ventrolateral protocerebrum; VM ventromedial neuropil domain; VMC ventromedial cerebrum; VMT ventral medial tract
3.2. Segmental ganglia form the embryonic SEZ
Whereas the segmental organization of the SEZ is obscured in the adult brain (Fig. 1a) by the merging and morphogenetic distortion of the gnathal neuromeres that takes place during metamorphosis, it is revealed clearly in the pattern of lineages and axon fascicles formed in the embryonic and early larval brain. Fascicles of the VNC, labeled by anti-Neuroglian (BP104), form two main commissural bundles, the anterior and posterior intermediate commissure, in each neuromere of the stage 15 embryo (Fig. 1a, b). In addition, a thinner set of dorsal and ventral commissures makes its appearance in the late embryo (stage 17; Fig. 1c). Each neuromere emits a segmental nerve with motor axons to the musculature and sensory axons from the sensilla of the corresponding segment (Fig. 1b, c). Along the anterior-posterior axis (neuraxis), longitudinal tracts (connectives) divide the neuropil into longitudinal columns from late embryonic stages onward (Nassif et al., 1998; Landgraf et al. 2003; Truman et al., 2004; Cardona et al., 2010; Fig. 1d–f). The dorsomedial tract (DMT) and dorso-intermediate tract (DIT) extend close to the roof of the neuropil, right underneath the dorsal set of commissures (Fig. 1e, f). The central tracts (CITd, CITv), as well as the ventromedial tract (VMT) are in contact with the intermediate commissures (Fig. 1e, f); the dorsolateral tract extends along the lateral surface of the neuropil (Fig. 1e, f).
The pattern of commissures and connectives continues forward into the SEZ. The posterior two gnathal neuromeres (maxilla and labium) each possesses both intermediate commissures, although somewhat reduced in size (Fig. 1b). Likewise, both maxilla and labium emit a segmental nerve (Fig. 1b). From late embryonic stages onward, these two nerves travel close to each other over much of their peripheral trajectory (Fig. 1c), forming the compound “labial nerve” described for the larval and adult brain. We will in the following use the term “labial segmental nerve” (ln) and “maxillary segmental nerve (mn) when referring to the individual fiber bundles connected to their respective neuromeres, and “compound labial nerve” (or simply “labial nerve”; lbn), when talking about the compound nerve carrying both bundles. The anterior two neuromeres, tritocerebrum and mandibula, form but one small commissure each, as observed by Page (2004); both of these commissures are closely spaced right underneath the esophageal foramen and form the subesophageal commissure (Fig. 1b, c). A compound nerve with axons to and from the pharynx and foregut connects to the tritocerebral neuromere (Fig. 1b, c); no separate mandibular nerve exists. The complicated nomenclature that is in use for the system of peripheral fiber bundles associated with the tritocerebrum requires clarification. In the embryo, three different components were distinguished (Campos-Ortega and Hartenstein, 1997). They include the frontal connective, with axons to and from the stomatogastric nervous system; the labral nerve, from the dorsal pharyngeal (=labral) sensory organs; and the hypopharyngeal nerve, a thin bundle formed by sensory axons of the hypopharyngeal sense organ (later renamed “posterior pharyngeal sense organ”; Gendre et al., 2004). In the adult, the hypopharyngeal nerve, together with the adjoined frontal connective (“stomodeal nerve”) is called “pharyngeal nerve”; the labral nerve is now the “accessory pharyngeal nerve” (Rajashekhar and Singh, 1994; Singh, 1997). For the larva, Stocker and colleagues (Gendre et al., 2004; Colomb et al., 2007) used the term “labral nerve” for all sensory axons entering the tritocerebrum. Finally, also for the larva, Spiess and colleagues (Schoofs and Spiess, 2007; Spiess et al., 2008), followed by Pankratz and colleagues (Schoofs et al., 2014; Hückesfeld et al., 2015), combined the proximal portion of the labral nerve, antennal nerve, and frontal connective into one entity, the “antennal nerve”, based on the fact that all three (in the fly larva, not the adult) are surrounded by a common glial sheath. We will in the following use for all stages of development, from embryo to adult, the term approximating the adult nomenclature, “compound pharyngeal nerve”, or simply “pharyngeal nerve” (phn), when referring to the entire system of peripheral nerve fibres connected to the tritocerebrum.
The longitudinal tracts of the ventral nerve cord continue forward into the SEZ (Fig. 1d). At the level of the mandibula/tritocerebrum, tracts turn laterally, following the outline of the esophageal foramen that penetrates the SEZ. The tracts converge and form a complex system of anastomoses, before continuing into the supraesophageal ganglion. The dorsolateral, central, and ventromedial tracts approach each other near the lateral surface of the tritocerebral neuropil (arrows in Fig. 1d); from there, fibers sort out and continue forward as the posterior and lateral cervical tract (Nassif et al., 1998; 2003). The dorsomedial tract remains at a more medial level, flanking the esophageal foramen; it continues as the medial cervical connective (=median bundle) into the protocerebrum (Nassif et al., 1998; 2003).
3.3. Lineage associated tracts as landmarks of ganglionic anatomy: the canonical pattern
Lineages whose axons enter the neuropil in vertically oriented bundles are distributed in a stereotyped, segmental pattern. These bundles can be recognized in large part already in the late embryo and early larva (Fig. 2a–c). During later larval stages, neuroblasts generate clusters of secondary neurons whose axon tracts follow the primary bundles, forming a pattern described for the late larval VNC by Truman et al. (2004) and for the SEZ by Kuert et al. (2012, 2014). The pattern is illustrated in Fig. 2d–i and represented as a digital 3D model in Fig. 2j–l. Lineages of the VNC have been related to their respective neuroblasts in two recent publications (Birkholz et al., 2015; Lacin and Truman, 2016). A look-up table that shows the association between embryonic neuroblasts and the lineages they produce is shown in Fig. 2m.
Figure 2.
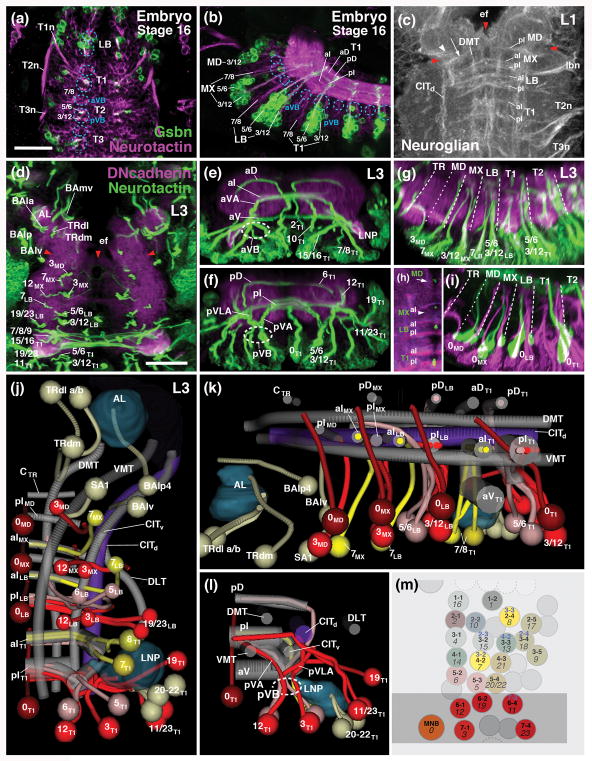
Lineage architecture of the subesophageal zone. (a, b) Z-projections of confocal sections of stage 16 embryo (a: horizontal plane, anterior to the top; b: parasagittal plane, anterior to the left) labeled with anti-Neurotactin (magenta; labels all primary neurons) and gsbn-Gal4>UAS-mcd8-GFP (green; labels subset of posterior lineages in each metamere; Urbach et al., 2016). (c) Z-projection of horizontal confocal sections of first larval instar SEZ, labeled with anti-Neuroglian (white; labels primary neurons). (d–g) Z-projections of confocal sections of third instar larval nerve cord (d: horizontal plane; e: frontal plane at anterior level of T1 neuromere; f: frontal plane at posterior level of T1 neuromere; g: parasagittal plane). Labeling anti-Neurotactin (green; secondary neurons with their lineage-associated tracts; lineages numbered according to nomenclature of Truman et al., 2004 and Kuert et al., 2014) and anti-DN-cadherin (magenta; neuropil). (h, i) Z-projections of confocal sections of third instar larval nerve cord (h: horizontal plane; i: parasagittal plane) labeled with Tdc2-Gal4>UAS-mcd8-GFP (green, labels primary lineage 0 of each metamere) and anti-Neurotactin (magenta; labels secondary lineages and their tracts). Hatched lines in (g, h) demarcate neuromere boundaries. (j–l) Digital 3D models of long axon tracts and lineages of SEZ and thoracic neuromere T1. Panels show ventral (j), medial (k) and posterior (l) view of right half of nerve cord. Long axon tracts are rendered in gray, with the exception of the central intermediate tract (CITd) which is shown in purple. Subset of lineages forming bundles of tracts that form useful landmarks in defining the neuroanatomy of the segmental ganglia SEZ are shown in different colors (red: posterior, engrailed-positive lineages; bright yellow: anterior lineages; light yellow: other lineages of the T1 and SEZ neuromeres). The antennal lobe (AL) and leg neuropil (LNP) are rendered turquois. (m) Schematic map of neuroblasts of right half of thoracic neuromere. Neuroblasts forming secondary lineages are identified by nomenclature coined for embryo (e.g., 1-1; Doe, 1992) and third instar larva (e.g., 16; Truman et al., 2004). Posterior neuroblasts are colored red; two anterior neuroblasts giving rise to lineages 7 and 8 are colored yellow. The association of neuroblasts and their respective lineages is based upon Birkholz et al. (2015) and Lacin and Truman (2016). For several neuroblasts, different lineage associations were established by these two groups; in these cases, blue numbers at the top of neuroblast indicate the results of Lacin and Truman (2016). Gray shading indicates expression of engrailed. For other abbreviations see List of Abbreviations. Bar: 25μm.
In a “canonical” neuromere of the VNC (e.g., T1) of the late larva, one can distinguish two conspicuous vertical bundles of lineage tracts. At the level of the anterior commissures, a group of lineages (7–9, 15, 16) defines the anterior vertical bundle (aVB; Fig. 2a, b, d, e; termed “lateral cylinder” in the late larva; Truman et al., 2004). From among this bundle, axon tracts of lineages 7 and 8 turn medially, forming the anterior ventral arch (aVA) and cross in the anterior intermediate commissure (Fig. 2e). Posteriorly, at the level of the posterior intermediate commissure, lineages 3, 5, 6, and 12 form the posterior vertical bundle (pVB; Fig. 2a, b, d, f, l). Within the neuropil, this massive vertical bundle splits into a branch that turns medially (posterior ventral arch (pVA) of Truman et al., 2004) towards the posterior intermediate commissure (Fig. 2f), and a second branch (posterior ventrolateral arch (pVLA) of Kuert et al., 2014) that projects laterally, and then dorsally (Fig. 2f). A second group of posterior lineages (11, 19, 23) forms the posterior-lateral bundle that also projects towards the posterior intermediate commissure (Fig. 2f, l).
The posterior-lateral and posterior-medial vertical bundles, as well as the entry point of the median lineage 0, represent landmarks defining the boundary between adjacent neuromeres. Thus, lineages forming these bundles are derived from the engrailed-positive row of neuroblasts that flanks the posterior boundary of a neuromere in the embryonic ectoderm (Birkholz et al., 2015; Lacin and Truman, 2016; Fig. 2m). The neuron clusters generated by these neuroblasts, and the tracts they emit, remain stationary, occupying the same position as the neuroblasts themselves. Tracts of lineages 3, 12, 19, and 23 project straight vertically into the intermediate posterior commissure, which is thereby defined as the most posterior element of the neuromere (Fig. 2g, j, k). The unpaired median lineage 0, which can be visualized by expression of Tdc2-Gal4 (Cole et al., 2005), demarcates neuromere boundaries in the mid-sagittal plane (Fig. 2h, i). Lineage tract 0 extends diagonally, entering ventrally at the level of the posterior commissure; passing in between the anterior and posterior commissure in the center of the neuromere, and terminating dorsally at the level of the anterior commissure (Fig. 2h, i).
3.4. Lineage associated tracts as landmarks of ganglionic anatomy in the larval SEZ
The pattern of secondary lineage tracts is reduced in the SEZ (Kuert et al., 2014), but members of the posterior (engrailed-positive) sets of lineages are retained, so that neuromere boundaries can be defined. In the labium, 8 secondary lineages can be detected. These include most lineages forming the posterior vertical bundles and posterior commissure (lineages 0, 3, 5, 6, 12, 19, 23), and one anterior lineage projecting along the anterior commissure (7LB; Figs. 2d, g, j). In the maxilla, one finds two secondary lineages forming a reduced posterior vertical bundle (lineages 3MX and 12 MX; called SA4 and SA5, respectively, in Kuert et al., 2014) and one anterior lineage (7 MX; SA3 in Kuert et al., 2014; Figs. 2d, g, j). The maxilla also possesses an anterior and postero-medial vertical bundle of primary lineage-associated tracts which turn into the anterior and posterior commissure, respectively (Fig. 2b). In the larva, only one lineage (7MX) forms secondary neurons, whose tract crosses in the anterior intermediate commissure (Fig. 2j, k). In the mandibula, primary lineage tracts converge from laterally and medially onto the one existing commissure, corresponding to the pattern formed by the posterior lineages 19/23 and 3/12 in the other neuromeres (Fig. 2c, arrow). This and the fact that it flanks mandibular lineage 0 posteriorly (Fig. 2K) indicates that this commissure represents the posterior commissure of the mandibular neuromere. The mandibula possesses only two secondary lineages, one that can be homologized to posterior lineage 3 (3MD; called SA2 in Kuert et al., 2014), and one lineage (SA1) of unclear serial homology (Fig. 2d, j, k).
In the anterior domain of the SEZ, lineages were defined as belonging to the tritocerebrum based on their expression of the Hox gene labial (Kuert et al., 2012). The pattern of tritocerebral lineage bundles deviates too much from the canonical VNC pattern to identify them with their serial homologs in the VNC. There exist five lineages in all (TRdm, TRdla, TRdlb, BAlv, BAlp4; Kuert et al., 2012), which are all engrailed-negative, indicating that none of their axon tracts demarcates the posterior boundary of the tritocerebrum. We here tentatively use the anterior lower edge of the esophageal foramen (medially) and a noticeable furrow in the neuropil surface (ventro-laterally) as the boundary between tritocerebrum and mandibula (red arrowheads in Fig. 2c, d). Anteriorly, the border between tritocerebrum and deuterocerebrum is well demarcated by axon bundles of the posterior (engrailed-positive) deuterocerebral lineage BAla3 (Fig. 2d; Kumar et al., 2009) and other deuterocerebral lineages (BAmv1/2; Fig. 2d), as well as discontinuities in the neuropil surface.
3.5. Neuropil domains of the larval VNC and SEZ
The lineages and neuropil tracts introduced above allow one to formulate a segmentally organized domain structure of the late larval VNC and SEZ. This domain structure will be outlined in detail, because it serves as the basis to generate a developmentally meaningful topology of the adult SEZ. Global neuronal markers distinguish between three major categories of structural elements within the late larval neuropil. (1)The bulk of the neuropil volume consists of primary neurons with their branched axons and dendrites, inter connected by (primary) long axon tracts. (2) Secondary neurons form discrete lineage tracts, which “tunnel” through the existing neuropil, following primary long axon tracts. Secondary axons emit tufts of filopodia along their length, which later, during pupal development, give rise to definitive axonal and dendritic branches (Omoto et al., 2017). Primary and secondary neurons are both positive for anti-Neuroglian, whereby stronger labeling occurs in long axon tracts (Hartenstein et al., 2015; Lovick et al., 2016; Fig. 3a–c). Anti-Neurotactin or insc-Gal4>UAS-mCD8-GFP label secondary axon tracts (Fig. 3a–c). DN-cadherin is expressed globally in the entire neuropil, but occurs at higher level in long axon tracts formed by both primary and secondary neurons (i.e., the connectives and commissures; Fig. 3d–j). DN-cadherin also strongly labels the filopodial tufts flanking secondary axon tracts (Omoto et al., 2017). (3) Synapses, formed by differentiated primary neurons, are distributed all over the neuropil, but are absent from the domains where secondary elements have invaded. Synaptic markers such as anti-Bruchpilot (Brp) are strongly expressed in primary neurons, but are excluded from undifferentiated secondary neurons (Fig. 3d–j). The combination of these markers reveals a columnar neuropil structure: cylindrical volumes, formed by connectives and commissures, thickened by filopodia-studded axons of secondary lineages, are strongly positive for DN-cadherin, but lack expression of Brp (Fig. 3d–f), or show a striated appearance, with multiple Brp-negative parallel strands formed by DN-cadherin-positive long axons, embedded in a matrix of Brp-positive material (Fig. 3f). We will call these domains “tract neuropils” (rather than simply “tracts”) in the following.
Figure 3.
Neuropil domains of the ventral nerve cord and SEZ. (a–c) Z-projections of horizontal confocal sections of third instar larva nerve cord labeled with anti-Neuroglian (green on left halves, white on right halves; labels primary neurons), and Insc-Gal4>UAS-mcd8-GFP (magenta; secondary lineages and their tracts). The three panels show a ventral plane (a; approximately 10–15μm above ventral neuropil surface), central plane (b; 20–25μm above ventral surface) and dorsal plane (c; 30–35μm above ventral surface). (d–f) Z-projections of confocal sections of another larval specimen cropped and oriented like the one shown in (a–c). The horizontal planes in (d–f) correspond to the ones of the panels (a–c) above. Labeling with anti-Brp (green on left halves, white on right halves; labels differentiated synaptic neuropil) and anti-DN-cadherin (magenta; labels all neurons, with high levels of expression in secondary neurons). (g–j) show z-projections of digitally rotated frontal sections of third instar VNC at the levels of tritocerebrum (g), mandibula (h), maxilla (i) and labium (j). Labeling with anti-Brp (green, white) and anti-DN-cadherin (magenta). In all panels, thick grey line demarcates midline. Hatched lines indicate boundaries between columnar neuropil domains, as described in the text (C central domain; CL centrolateral domain; CM centromedial domain; DL dorsolateral domain; DM dorsomedial domain; VL ventrolateral domain; VM ventromedial domain). (k–n) Digital 3D models of SEZ and T1 neuromere. Panels show ventral (k), posterior (l), lateral (m) and medial (n) view of one side of nerve cord. Surfaces of columnar neuropil domains are rendered in different colors. Subset of lineages useful as landmarks are shown as pipes (lineage-associated tract) and spheres (cell body cluster). Coloring as in models shown in Fig.2j–l. In (n), medial neuropil domains are left out, to allow view onto lineage tracts. For other abbreviations see List of Abbreviations. Bar: 25μm
Tract neuropils subdivide each neuromere into seven longitudinal domains which we will term ventromedial, ventrolateral, centromedial, central, centrolateral, dorsomedial, and dorsolateral domain (Fig. 3a–l). The ventromedial and ventrolateral domains are located ventrally of the VMT and CITv (Fig. 3a, d, j, k, l). The boundary between the two ventral domains is defined by sequence of portals through which the vertical bundles of lineage tracts (pVB in Fig. 3d; see above) enter the neuropil. The portals lie at the same medio-lateral level as the central longitudinal tracts (Fig. 3e, j; CITd, CITv). The three thoracic segments of the late larva possess enlarged, laterally protruding ventrolateral domains, which will give rise to the massive leg neuropils (LNP) of the adult VNC (Fig. 3d, k, l).
The central and ventromedial tract neuropils and commissural neuropils allow one to distinguish a central domain that surrounds CITd and CITv and the volume in between; a centrolateral domain located laterally of CITd/v and DIT (Fig. 3b, e, j, l); and a centromedial domain that includes the VMT at its ventral boundary and the massive commissural tract neuropils, formed by the anterior and posterior intermediate commissures of each neuromere (Fig. 3e, j, l). The dorsal “roof” of the larval VNC and SEZ comprises the dorsomedial and dorsolateral domain (Fig. 3c, f, j, l), which can be distinguished by the fact that dorsal commissural bundles and superficial longitudinal fiber strands of the DIT are present in the former, and absent in the latter (blue arrows in Fig. 3c).
Neuropil domains in the posterior three neuromeres of the SEZ essentially resemble their thoracic and abdominal counterparts; however, the reduction in volume of the SEZ neuromeres, consisting mainly in a shortening along the antero-posterior axis, that was already noticeable in the early larva, has become more pronounced at late larval stages (Fig. 3a–f, k). This further reduction is caused in part by the greatly decreased number of SEZ lineages that enter a secondary phase of proliferation (Kuert et al., 2014; see previous section). The labial neuromere is approximately one third shorter than any of the thoracic neuromeres; the maxilla, mandibula and tritocerebrum have approximately half of the length of a thoracic segment (Fig. 3f, k). Aside from overall changes in neuromere length, the size and shape of the columnar neuropil domains of the larval labium and maxilla resemble those of a canonical thoracic neuromere described above. This is different in the anteriorly adjacent mandibula and, even more so, the tritocerebrum. Thus, in the anterior part of the SEZ, the central tract neuropils (CITd and CITv) turn towards the lateral surface of the neuropil and merge with each other; as a result, the ventrolateral and centrolateral domain is very small in the mandibula, and absent in the tritocerebrum (Fig. 3e, g, h). Conversely, the VMT widens, resulting in a greatly increased ventromedial domain (Fig. 3d, g, k) which borders the conspicuous antennal lobe (=the ventromedial domain of the deuterocerebrum) anterolaterally (Fig. 3k). The centromedial domain of the tritocerebrum is also enlarged; it forms an elongated, pyramidal domain easily recognizeable by its high DN-cadherin signal, flanking the esophageal foramen (Fig. 3e, g, n). Likewise, the central tritocerebral domain is enlarged, bordering the deuterocerebrum (antero-medially) and protocerebrum (antero-laterally; Fig. 3g, k, m). At a dorsal level, due to the changed topology of longitudinal tracts and commissures, it is difficult to clearly discern a boundary between the dorsomedial and dorsolateral domain within the tritocerebrum; we designated the neuropil located dorsal of the converging CITd/v/VMT tract neuropils as “dorsal tritocerebral domain” (D in Fig. 3f, g, m, n).
The central domain of the SEZ, which surrounds the CIT tract neuropils, deserves special mentioning for its prominent role in reconstructing the metamorphic changes shaping the SEZ, as described in the following section. The vertically oriented axon tracts of the lineages 3LB, 3MX and 3MD project from ventromedially into this domain, (blue arrowheads in Fig. 3e, h–j). This results in a high level of DN-cadherin signal in the center of the central domain, which can be easily followed throughout metamorphosis into the adult brain.
3.6. Lineage associated tracts persist throughout metamorphosis and constitute landmarks that define neuromeres of the adult SEZ
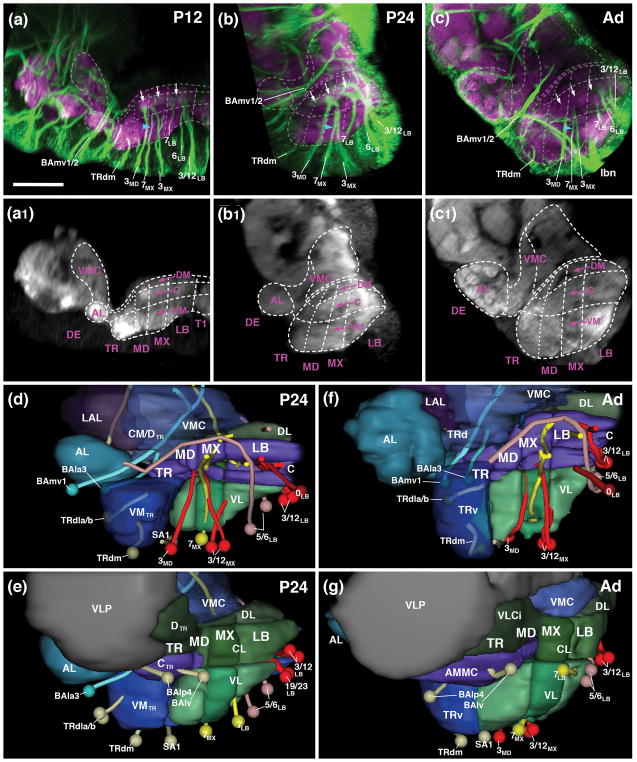
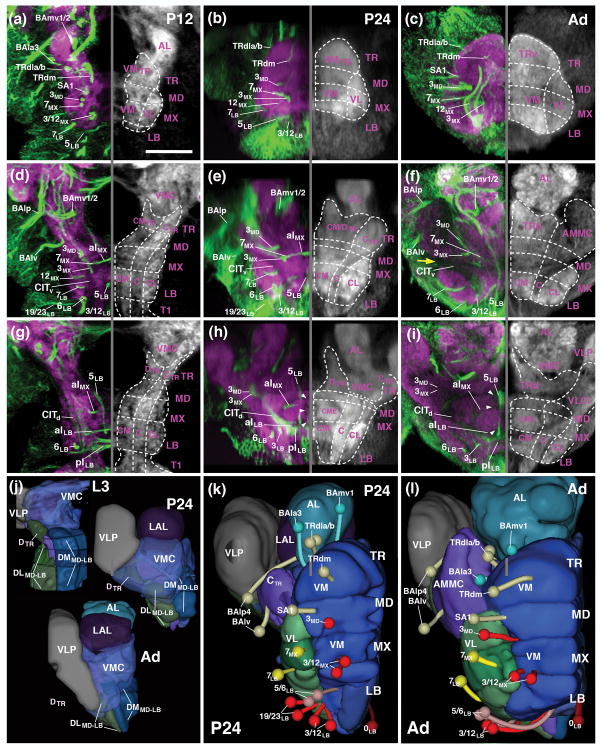
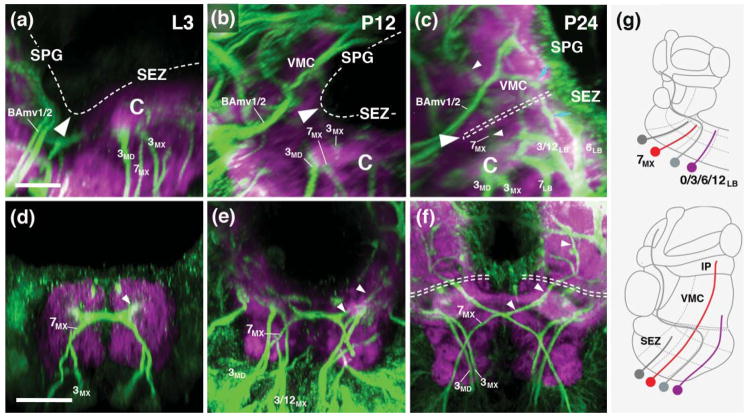
Neuropil fascicles and lineage associated tracts described in the previous section can be readily identified at all pupal stages and in the adult, which makes it possible to establish the relationship between neuropil domains of the adult and larval SEZ. As discussed in more detail below, one of the most significant metamorphic changes in the SEZ is an unequal increase in size of ventral versus dorsal neuropil domains. As a result, vertical bundles of lineage-associated tracts, extending more or less parallel to each other in the larva and the early pupa (P12)(see Figs. 2g, k; 4a), become tilted, with more anterior vertical bundles tilted backward, and posterior ones forward (Fig. 4b, c, d, f). In the following figures that feature sagittal sections, frontal sections and horizontal sections of the developing SEZ (Figs. 4–6), we digitally oriented the confocal stack in such a way that the neuraxis near the middle of the SEZ (maxillary domain) is roughly parallel to the horizontal plane. In this orientation, the anterior vertical bundle of the maxilla (linage 7MX) extends roughly parallel to the vertical plane (blue arrowheads in Fig. 4a–c). The chiasmatic crossing of 7MX in the centromedial domain of the maxilla forms a characteristic hallmark of cross sections of the maxillary neuromere of the SEZ visible throughout metamorphosis and in the adult (blue arrowheads in Figs. 5g–i). 7MX is flanked anteriorly and posteriorly by the tracts of 3MD and 3MX. Extending parallel to each other from the ventral surface towards the central domain of the SEZ, these tracts define the boundaries of the maxillary neuromere (Figs. 4a–c1).
Figure 4.
Metamorphosis of the SEZ. (a–c1) Z-projections of parasagittal confocal sections of pupal brain (a/a1: P12; b/b1: P24) and adult brain (c/c1) labeled with anti-Neuroglian (green; labels secondary axon tracts), and anti-DN-cadherin (neuropil; magenta in a–c; white in a1–c1, which represent the same z-projections as panels a–c). Z-projections include contiguous sections approximately 15–25μm distant from the midline, illustrating the metamerically organized vertical bundles of secondary lineages. Hatched lines indicate boundaries between metameres and columnar neuropil domains, as described in the text. White arrows in (a–c) point at DN-cadherin-rich core of central neuropil column, targeted by lineages 3 and 7 of all neuromeres. Blue arrowhead in (a–c) shows characteristic trajectory of lineage 7MX, which converges with 3MX towards dorsally. (d–g) Digital 3D models of SEZ at 24hr pupal stage (d, e) and adult (f, g). Panels show medial (d, f) and lateral view (e, g) of one side of brain. Surfaces of columnar neuropil domains are rendered in different colors. In (d, f), medial neuropil domains are left out, or rendered semi-transparent to allow view onto lineage tracts. Subset of lineages useful as landmarks are shown as pipes (lineage-associated tract) and spheres (cell body cluster). Coloring as in models shown in Figs. 2 and 3. For abbreviations see List of Abbreviations. Bar: 25μm
Figure 6.
Metamorphosis of the SEZ. Panels (a–i) show Z-projections of horizontal confocal sections of pupal brain at 12hrs APF (P12; left column; a, d, g), 24hrs (P24; middle column; b, e, h) and adult brain (Ad; right column; c, f, i). Preparations are labeled with anti-Neuroglian (green on left halves of panels; labels secondary axon tracts), and anti-DN-cadherin (neuropil; magenta on left halves of panels; white on right halves). Z-projections represent different levels along the dorso-ventral axis [(a–c) ventral level, approximately 10–15μm above ventral neuropil surface), (d–f) central plane, 20–25μm above ventral surface; (g–i) dorsal plane; 30–35μm above ventral surface]. Hatched lines indicate boundaries between columnar neuropil domains, as described in the text. White arrowheads in (h, i) point at DN-cadherin-poor poor commissural fiber systems delineating centromedial ellipsoid (CME). Yellow arrow in (f) indicates location where DN-cadherin-poor CITd and CITv converge at the later neuropil surface. (j–l) Digital 3D models of SEZ at late larval stage [L3; upper left of (j)], 24hr pupal stage [P24; upper right of (j), and (k)], and adult [Ad; bottom of (j), and (l)]. Panels show dorsal view (j) and ventral view (k, l) of one side of the brain. Surfaces of columnar neuropil domains are rendered in different colors. In (j), ventromedial cerebrum (VMC) is rendered semi-transparent to allow view onto dorsal SEZ domains which become covered by the VMC at P24 and adult. In (k, l), secondary lineages are shown as pipes (lineage-associated tract) and spheres (cell body cluster). Coloring as in models shown in Figs. 2–4. For abbreviations see List of Abbreviations. Bar: 25μm
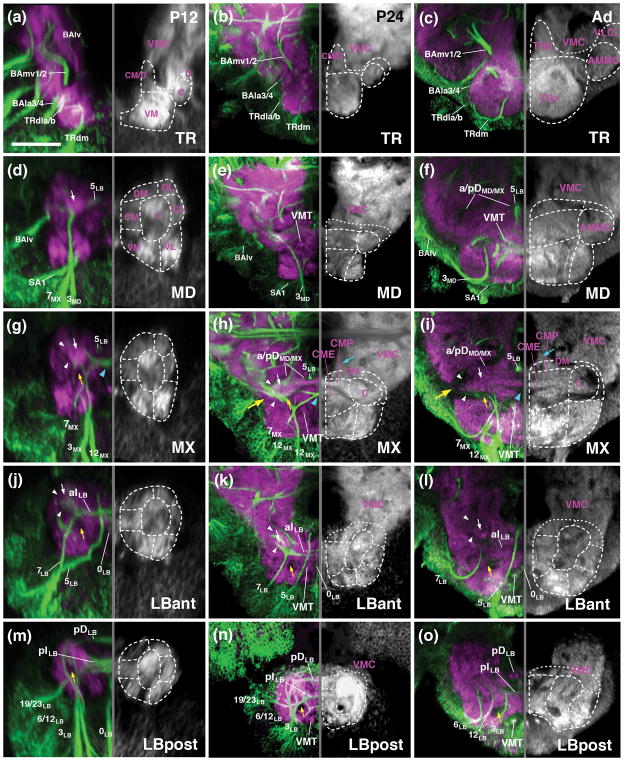
Figure 5.
Metamorphosis of the SEZ. All panels show Z-projections of frontal confocal sections of pupal brain at 12hrs APF (P12; left column; a, d, g, j, m), 24hrs (P24; middle column; b, e, h, k, n) and adult brain (Ad; right column; c, f, i, l, o). Preparations are labeled with anti-Neuroglian (green on left halves of panels; labels secondary axon tracts), and anti-DN-cadherin (neuropil; magenta on left halves of panels; white on right halves). Z-projections represent different levels along the antero-posterior axis [(a–c) tritocerebrum (TR); (d–e) mandibula (MD); (g–i) maxilla (MX); (j–l) anterior labium (LBant); (m–o) posterior labium (LBpost)]. Hatched lines indicate boundaries between columnar neuropil domains, as described in the text. White arrows point at DN-cadherin-rich core of central neuropil column, targeted by lineages 3 and 7 of all neuromeres. White arrowheads point out DN-cadherin-poor poor periphery of central neuropil column, which corresponds to bundles of long axons (CITd and CITv tracts). Blue arrowhead in (g, h) shows chiasmatic midline crossing of lineage 7MX. Large yellow arrow in (h, i) indicates location where DN-cadherin-poor CITd and CITv converge at the later neuropil surface. Small yellow arrows point at DN-cadherin-poor CITv bundle. For abbreviations see List of Abbreviations. Bar: 25μm
In the posterior SEZ, the anterior vertical bundle (tract of 7LB) and posterior vertical bundle (3LB, 12 LB) define anterior and posterior levels, respectively, of the labial neuromere throughout all developmental stages (P12, P24, Adult; Figs. 4a–c1, d, f; 5j–o). The commissure formed by 7LB (aI commissure of the labial neuromere) is strongly Neuroglian-immunoreactive during pupal stages (Fig. 5j, k), but is only faintly visible in the adult brain (Fig. 5l). In contrast, labeling of the 5LB tract, which extends vertically right posteriorly of the 7LB commissure and then arches anteriorly, is strong in the pupa and adult (Fig. 5j–l). Likewise, the pI commissure, formed by crossing branches of the 3LB lineage, represents a conspicuous landmark structure of the posterior labium from larva to adult (Fig. 5m–o). Tracts of lineages 6LB and 12LB, forming part of the posterior vertical bundle along with 3LB, extend vertically into the dorsal labial neuromere, with 6LB located slightly more anteriorly and medially than 12LB (Figs. 4a–c; 5m–o). At a dorsal level, 6LB turns medially into the posterior dorsal commissure of the labium (Fig. 5n). Neurglian expression of this structure declines in the late pupa and adult (Fig. 5o). Similarly, labeling of the 19/23LB tracts, which enter the posterior labial neuropil at a lateral level and then proceed medially to form a posterior-ventral bundle within the pI commissure (Fig. 5m, n), becomes faint in the adult.
Linage-associated landmarks in the anterior SEZ are the conspicuous vertical tract of 3MD which defines the posterior boundary of the mandibula (Figs. 4a–c, d, f; 5d–f). Right in front of 3MD, the thin tract of lineage SA1, follows a medially directed trajectory and crosses right in front of 3MD (Figs. 4d, f; 5d–f). More anteriorly, tracts of TRdl and TRdm enter the ventromedial tritocerebral domain from laterally and medially, respectively (Figs. 4a–c, d, f; 5a–c). The border between the tritocerebrum and the deuterocerebrum is demarcated by the engrailed-positive lineages BAla3/4 (Kumar et al., 2009; Figs. 4a–c, d–g; 5a–c). As discussed in a previous section for the late larva, the boundary between tritocerebrum and mandibula is ill defined, since no engrailed-positive tritocerebral secondary lineage tracts exist.
3.7. Metamorphic changes in the pattern of neuropil domains of the SEZ
Lineage-related tracts, in addition to the longitudinal fiber systems that also remain visible from larva to adult, allow one to follow the columnar neuropil domains defined above for the larva throughout all pupal stages (P12, P24, P48) into the adult brain. Conspicuosly, the central domain formed around the cylindrical, DN-cadherin-rich CITd tract neuropil, rests like a beam on the series of pillar-shaped vertical bundles formed by 3MD, 3MX, and 3LB, respectively (small arrows in Figs. 4a–c; 5d–l). Longitudinal fiber masses low in DN-cadherin flank the CITd tract neuropil laterally (white arrowheads in Figs. 5g–o) and form the boundary between the central and centrolateral column. The CITv is also characterized as a DN-cadherin-negative bundle that extends ventromedially of the CITd (yellow arrows in Fig. 5g–o). The lateral turn and convergence of the DN-cadherin-negative CITd and CITv fiber masses, constitutes a characteristic, highly visible feature of the metamorphosing and adult SEZ at the anterior maxillary/mandibular level (large yellow arrow in Figs. 5h, i; 6f). Anterior to this level, the central domain increases in volume throughout pupal development, possibly as a result of the differentiation of secondary neurons (3MD, 3MX, and 3LB) and massive sensory afferents (Johnston’s organ of the antenna and other mechanoreceptors) projecting into it (see below and accompanying paper by Kendroud et al., 2017). This enlarged central columnar domain of the mandibular and tritocerebral neuromere represents the antenno-mechanosensory and motor center (AMMC) of the adult brain (Figs. 5c, f; 6f).
The centromedial and ventromedial columnar domains also undergo strong growth and differentiation during metamorphosis. The centromedial domain features masses of crossing fiber systems forming the intermediate commissures, some of them positively labeled by anti-Neuroglian (crossing tracts of 7MX, 7LB, 3LB, forming the aI MX, aI LB, and pI LB, respectively; Figs. 5g–o; 6d–i), others appearing as DN-cadherin-poor volumes (white arrowheads in Fig. 6h, i). The VMT, similar in diameter as the CITv and defined by a central DN-cadherin positive component encircled by DN-cadherin-poor fibers, is a noticeable structure extending along the boundary between centromedial and ventromedial column (Fig. 5h–o). In the late pupa and adult, the centromedial domain of the maxilla and mandibula differentiates into two distinct compartments. At a more dorsal level, the neuropil forms a strongly DN-cadherin-immunoreactive plate (“centromedial plate”; CMP) that interconnects the laterally adjacent central columns (Fig. 5h, i). Ventro-posteriorly of the plate, the neuropil surrounding the chiasm formed by the lineage 7MX tracts compacts into an elliptical domain (“centromedial ellipsoid”; CME; Figs. 5h, i, 6h, i).
The ventromedial domain of the SEZ also increases in size. This growth is particularly prominent in the tritocerebrum and mandibula (Figs. 4a–c1, d, f; 6a–c, k, l; ), whose ventromedial domains are innervated by differentiating neurons of multiple lineages (TRdla/b, TRdm, SA1; see below), as well as the large number of sensory afferents from the mouthparts and pharynx (see accompanying paper by Kendroud et al., 2017). In the posterior labium, the ventromedial domain is small or absent: the VMT, defining the floor of the centromedial domain (see above) extends along the ventral surface of the posterior labial neuropil (Fig. 5n, o) on its way to the cervical connective (the connection between SEZ and thoracic ganglia).
Longitudinal fiber systems (DMT, DIT) and dorsal commissures (DTR/MD, aDMX, pDMX, aDLB, pDLB) define the narrow dorsal domain of the SEZ. Most of these fiber systems downregulate Neuroglian expression during pupal stages, but remain detectable as DN-cadherin-low domains all the way towards the adult. The pDLB commissure, represented by the tract of lineage 6LB, demarcates the posterior-dorsal surface of the SEZ (Fig. 5n, o). Dorsal commissural bundles of the maxilla and mandibula develop into a thick, DN-cadherin-negative “plate” covering the CITd tract neuropil and dorsomedial neuropil at this level (Fig. 5f, h, i). The DMT, which extends within the dorsomedial domain, is represented by the axon bundle of lineage 5LB; the forward directed 5LB tract joins the DMT at a mid-labial level and continues forward with the DMT into the tritocerebrum (Figs. 4d, f; 5d–l).
The boundary between the SEZ and supraesophageal neuropil (SPG) of the pupal and adult brain is difficult to define, in part because fiber masses originating in the two domains coalesce. This process can be followed by reconstructing secondary lineages, in particular 7MX and some of the posterior labial lineages (6LB, 12LB). During the early stages of metamorphosis (P0–P24), the supraesophageal ganglion tilts sharply backwards, such that its posterior-ventral surface attaches itself to the dorsal surface of the labium and maxilla (large white arrowheads in Figs. 4a, b; 7a–c). As described above, the DN-cadherin-poor longitudinal and transverse fiber systems demarcate where SEZ and SPG meet. By P24, lineage tracts of 7MX and 6LB have extended further dorsally, “breaking through” the boundary between SEZ and SPG. The (contralateral) 7MX tract reaches all the way into the inferior protocerebrum (small white arrowheads in Fig. 7a–f). Similarly, dorsally directed tracts of the posterior labial lineages pass the line of demarcation between SEZ and SPG and reach into the posterior VMC, part of the deuterocerebrum (blue arrows in Fig. 7c).
Figure 7.
Coalescence of supraesophageal (SPG) and subesophageal neuropils (SEZ). (a–f) Z-projections of parasagittal (a–c) and frontal (d–f) confocal sections of third instar larval brain (a, d), 12hr pupal brain (b, e) and 24hr pupal brain (c, f) labeled with anti-Neuroglian (green; labels secondary axon tracts), and anti-DN-cadherin (neuropil; magenta). Hatched line in (a–c) and (f) demarcates dorsal surface of neuropil. Large white arrowheads point at boundary between SPG and SEZ. Note posterior tilt of SPG between larval and 24hr pupal stage. From 24hr pupal development onward, the dorsal deuterocerebrum (called the ventromedial cerebrum (VMC) in the standard nomenclature for the adult brain), which forms the most ventral part of the SPG, covers the dorsal surface of the SEZ [hatched double line in (c, f)]. Also note growth of tract of lineage 7MX [small white arrowheads in (c–f)]) into SPG. Blue arrows in (c) point at tract of lineage 12LB, which also reaches into the SPG. (g) Schematic sagittal section of late larval brain (top) and pupal brain (bottom), depicting extension of SEZ lineages 7MX and 0/3/6/12LB into SPG. For other abbrveviations, see List of Abbreviations. Bar: 10μm (a–c); 25μm (d–f).
More anteriorly, at the level of the AMMC, secondary lineages of the SEZ, including 3MD, 3MX, 3LB, 7LB, and 0LB, also extend “through the roof” of the SEZ into the SPG. This process can be best appreciated in GFP-labeled MARCM clones of individual lineages (see below). Near the midline of the brain, a discrete neuropil domain of the ventromedial cerebrum, called posterior periesophageal neuropil (PENPp) or ‘cantle’ (Ito et al., 2014), continues in the shape of a triangular process out of the dorsal surface of the AMMC (Fig. 5). This domain receives terminal arborizations of 3MX and 3LB (Fig. 8o, u). Laterally, terminal branches of these lineages, in addition to fibers of 3MD and 7LB, reach beyond the AMMC into a SPG compartment called inferior ventrolateral cerebrum (VLCi), or “wedge” (Ito et al., 2014; Fig. 8l, o, r, u). Even more prominently, dense terminal arbors of 0LB reach bilaterally into the VLCi (Fig. 9v)
3.8. Projection of secondary lineages within the neuropil domains of the SEZ
Using the MARCM technique, GFP-labeled clones of secondary lineages were induced by a heat pulse applied to early larvae, and analyzed in preparations of adult brains. Clones could be unambiguously assigned to lineages based on the projection of their axon tracts. Lineages of the tritocerebrum (TRdm, TRdla, TRdlb) and lineage SA1, which is located at the boundary between tritocerebrum and mandibula and could belong to either of these neuromeres, innervate the ventral and centromedial neuropil domain of the tritocerebrum (Fig. 8a–i). A note in regard to nomenclature: the tritocerebrum of the Drosophila brain, as delineated on the basis of projections form the pharyngeal nerves, stomatogastric nervous system, and pars intercerebralis by Rajashkhar and Singh (1994), was termed “anterior periesophageal neuropil (PENPa) in the anatomical description by Ito et al. (2014). Two major domains, a ventral and a dorsal one, can be distinguished. The ventral domain (“ventral tritocerebrum”; TRv in Figs. 5c, 6c, 7, 8), which receives the compound pharyngeal nerve, evolves from the ventromedial neuropil column discussed throughout this paper. In the adult nomenclature, this domain is also called “prow” (Ito et al., 2014). The dorsal domain (“dorsal tritocerebrum”; TRd in Figs. 5c, 6f, 7, 8) corresponds to the neuropil that receives input from stomatogastric afferents, but mainly from central fiber systems that descend from the pars intercerebralis (Rajashekhar and Sing, 1994). This domain evolves from the centromedial and dorsal neuropil columns of the tritocerebrum described here. In the classical fly literature (e.g., Strausfeld, 1976), but also the recent compilation of anatomical terms (Ito et al., 2014), the dorsal tritocerebrum received the epithet “flange”.
Terminal arborizations of TRdm more or less evenly fill the ventral tritocerebrum, but are excluded from the dorsal tritocerebrum (Fig. 8a, b). By contrast, TRdla projects to a restricted dorsolateral domain of the TRv; its main terminal arborizations are in the TRd and the posteriorly adjacent centromedial mandibula (Fig. 8c–e). TRdlb is excluded from the TRd, like TRdm; in the TRv, innervates the ventro-lateral domain (Fig. 8f, g). SA1 overlaps with TRdla/b in the TRv. In addition, it covers the ventral domain of the mandibula (Fig. 8h, i). Significantly, with the exception of TRdm, none of these lineages projects to the DN-cadherin-rich medial part of the TRv which constitutes the gustatory center of the SEZ (white arrows in Fig. 8b, d, g, i; see accompanying paper by Kendroud et al., 2017).
The projection of two additional secondary lineages assigned to the tritocerebrum based on their expression of the Hox gene labial, BAlp4 and BAlv (Kuert et al., 2012), have been described previously (Wong et al., 2013) and are not depicted in Fig. 8. Briefly, BAlv, whose tract targets the central column of the neuropil at the boundary of tritocerebrum and mandibula (see Fig. 5d–f), includes exclusively locally branching neurons that arborize in the AMMC and the posterior-laterally neighboring ventro-lateral cerebrum (VLCi). BAlp4 is part of a quartet of closely apposed lineages, BAlp1–4, whose tracts enter the neuropil at the boundary between lateral tritocerebrum and deuterocerebrum (Lovick et al., 2013; Wong et al., 2013; see Fig. 5d–f). BAlp4 has proximal branches in the antennal lobe and medially adjacent TRd (part of the tritocerebrum); its axons project along the antennal lobe tract to the superior protocerebrum (Wong et al., 2013).
Lineages whose tracts form part of the vertical bundles (3MD, 3MX, 3LB, 7LB) innervate the central columnar domain that extends throughout the SEZ (Fig. 8j–v). Anterior members of this groups (3MD, 3MX) have their densest terminal arbors in more anterior levels of this column (the AMMC; Fig. 8k, l, n, o), posterior members (3LB, 7LB) have a more posterior focus (posterior AMMC and central labial domain; Fig. 8r, s, u, v). However, there is a wide overlap, and each of these lineages reaches all three neuromeres. Towards dorsolaterally, arborizations of 3MD and 3MX “spill over” into the adjacent VLCi of the supraesophageal ganglion (Fig. 8l, o). Towards medially and dorsomedially, 3MX and 3LB have projections into the centromedial maxilla (Fig. 8o, u), as well as the PENPp which forms part of the ventral cerebrum of the SPG (Fig. 8o, u). At a posterior level, in the labial neuromere, 3LB and 7LB also reach up into the posterior VMC, as well as the centrolateral labium, and the ventromedial labium (Fig. 8s, v). Both of these lineages cross the midline and form strong connections with the posterior central domain and the posterior VMC of the contralateral SEZ (Fig. 8s, u, v). Lineage 7LB forms a tract that descends via the contralateral cervical connective towards the thoracic ganglia (Fig. 8Ss).
Most of the remaining lineages, including 12LB, 6LB, 19LB, 23LB, and 12MX restrict their arborization to the labial neuromere (Fig. 9a–l). Lineages 12LB and 6LB are focused on the central domain, but have also major branches to the centromedial and centrolateral domain, and the dorsally adjacent VMC (Fig. 9a, b, d). 12LB and 6LB extend strong descending tracts in the ipsilateral and contralateral cervical connective, respectively (arrows in Fig. 9b, d). 19 LB forms a thick commissural tract and has most terminal arbors bilaterally in the centromedial and ventromedial labial domain (Fig. 9g), but also sends branches dorsally into the VMC (Fig. 9f, g) and anteriorly into the central domain of the maxilla and mandibula (Fig. 9f). Lineage 23LB comprises only a small number of neuronal cell bodies, with a thin axon bundle that follows the trajectory of 19LB and crosses via the posterior labial commissure, but forms only sparse projection in the contralateral ventral domains (Fig. 9k, l). Projections of 12MX are restricted to the ventral domains of the labium and maxilla. They occupy several dispersed, narrow foci near the lateral and ventral surface of this neuropil (Fig. 9h–j). As stated above for most of the anterior SEZ lineages, the DN-cadherin-rich neuropil domain of the ventral maxilla and labium, which is occupied by terminal afferents of sensory receptors (see accompanying paper by Kendroud et al., 2017) is virtually devoid of projections from 12MX, and receives only very sparse innervation by any of the other labial lineages (arrow in Fig. 9f, i, l, v).
Three SEZ lineages, 5LB, 7MX, and 0LB project to neuropil domains that are far remote from the location of their cell bodies. The tract of 5LB extends forward along the roof of the SEZ all the way to the tritocerebrum, where it forms dense arborizations in the TRd (Fig. 9n) and a small region of the anteriorly adjacent TRv (Fig. 9m, n). The axon tract continues via the median bundle into the superior medial protocerebrum (SMP) where it terminates in a restricted medial neuropil area (Fig. 9n, o). Lineage 7MX crosses in the intermediate commissure of the maxillary neuromere and projects dorsally all the way to contralateral inferior protocerebrum (see previous section; Fig. 9r, s). Terminal arborizations are found along the entire projection, locally in the central and centromedial domains of the maxilla and labium (Fig. 9r, s), and distally in the ventromedial cerebrum, posterior lateral protocerebrum, as well as the posterior, medial and lateral inferior protocerebrum (Fig. 9s). 0LB is the only unpaired secondary lineage, derived from the median neuroblast (MNB) of the labial segment. The lineage forms bilaterally symmetric projections; proximal terminal arbors fill the part of the ventromedial domain and the dorsally adjacent centromedial domain of the labium/maxilla (Fig. 9v). The bilateral axon tract continues along the lateral surface of the SEZ and sends dense terminal arbors to lateral domains of the AMMC and adjacent VLCi (Fig. 9t–v).
4 Discussion
4.1. Pattern elements of the canonical ganglion
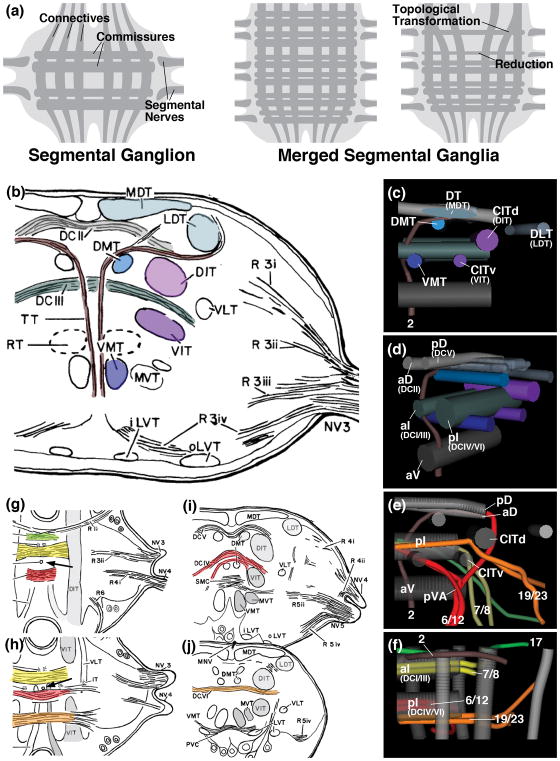
We provide in this paper a detailed account of the developing neuroanatomy of the Drosophila subesophageal zone (SEZ). Our work is guided by the fact that the SEZ is formed by a union of four reduced and structurally modified segmental ganglia. The canonical structure of an insect ganglion has been described in detail for a number of insects, notably cockroach, locust and honey bee (Gregory, 1974; Tyrer and Gregory, 1982; Rehder, 1988). Landmark structures that define the anatomy of the ganglion are long axon tracts which form longitudinal connections within and between ganglia (connectives), as well as transverse connections between left and right hemiganglia (commissures). These long axon tracts, distinct from the surrounding synaptic neuropil, can be recognized histologically or with the help of specific markers. Longitudinal tracts form a system of medial, intermediate and lateral elements. For some of these, Drosophila homologs can be readily proposed, based on their position within the neuromere relative to each other and other landmarks (Fig. 10). For example, the dorsomedial tract (DMT) and dorso-intermediate longitudinal tract (DIT; CITd in Drosophila) of the locust ventral ganglia flank an intermediate set of commissures dorsally; the ventromedial tract (VMT) and ventro-intermediate tract (VIT; CITv in Drosophila) extend ventrally of these commissures (Fig. 10b, c, d). These and other likely correspondences between canonical longitudinal tracts (as well as commissural tracts discussed in the following) and their Drosophila counterparts need to be confirmed by comparing in detail what types of neurons or lineages set up the tracts in different species.
Figure 10.
(a) Schematic representation of canonical segmental ganglion (left), fused ganglia (center), fused ganglia with reduction and topological changes of pattern elements (right). (b) Drawing of frontal section of locust mesothoracic ganglion, anterior level. (c–f) Digital 3D models of Drosophila larval thoracic neuromere (right half); posterior view (c, e; midline at left of panel); posterior-medial view (d); dorsal view (f). In (c–f), commissures are shown in different shades of gray [light: anterior and posterior dorsal commissure (aD, pD; corresponding to locust commissures DCII and DCV, respectively); blue-gray: anterior and posterior intermediate commissure (aI, pI; corresponding to locust commissures DCI/III and IV/VI, respectively); dark gray: anterior ventral commissure (aV)]. In (c, d), several longitudinal fascicles are rendered in the same colors as fascicles of locust ganglion shown in (b) proposed to be homologous to the Drosophila elements. Dark blue: ventromedial tract (VMT); light blue: dorsomedial tract (DMT); silver-blue: dorso-intermediate tract (DIT), corresponding to medio-dorsal tract (MDT) in locust; purple: ventral bundle of central-intermediate tract (CITv), corresponding to ventro-intermediate tract (VIT) in locust; magenta: dorsal bundle of central-intermediate tract (CITd), corresponding to dorso-intermediate tract (DIT) in locust. In (e, f), longitudinal fascicles and commissures are in gray; tracts of selected lineages are rendered in different colors (red: lineages 6/12; yellow: lineages 7/8; orange: lineages 19/23; green: lineage 17; maroon: lineage 2). (g, h) Horizontal sections of locust mesothoracic ganglion at dorsal level (g) and mid-dorsal level (h). Arrow points at median vertical trachea separating anterior from posterior neuromere. (i, j) Frontal sections of locust mesothoracic ganglion at mid-posterior level (i) and posterior level (j). Selected commissures and tracts with trajectories similar to the Drosophila lineage tracts shown in (e, f) are rendered in corresponding colors. Panels (b, g–j) from Tyrer and Gregory (1982) (with permission).
Among commissural tracts, a system of dorsal, intermediate, and ventral elements can be distinguished. In the Drosophila larva, Truman et al. (2004) named the anterior and posterior dorsal commissure (aD, pD), anterior and posterior intermediate commissure (aI, pI), and anterior ventral commissure (aV). Distinct lineage tracts subdivide these commissures into smaller components. For example, lineages 19 and 23 project straight medially and cross the midline at a posterior position within the pI commissure (Fig. 10e, f). By contrast, lineages 6 and 12, which also cross in the pI, approach the midine from a more medial position, forming the conspicuous ventral arch (pVA), and crossing anterior of the axons of 19/23 (Fig. 10e, f). In locusts, Tyrer and Gregory (1982) distinguish only between a dorsal and a ventral set of commissures (Fig. 10g–j); in both, there is an anterior and posterior subset, set apart by a median, vertical trachea that penetrates the center of each neuromere (arrow in Fig. 10g, h; this trachea is not present in the Drosophila larva; Pereanu et al., 2007; V.H., unpublished). On the basis of position and trajectory, we propose that the two components of the Drosophila pI commissure formed by lineages 6/12 (located more anterior and forming an arch; Fig. 10e, h) correspond to the locust DCIV; commissural tract. Tracts of lineages 19/23 (located more posterior, forming a horizontal bar; Fig. 10e, j) corresponds to commissure DCVI in locust. Likewise, the Drosophila aI commissure is comprised of an anterior and posterior bundle, formed by lineage 17 and 7/8, respectively (Fig. 10f). Lineage 2 forms a conspicuous tract that projects vertically close to the midline, penetrates the aI commissure in between the bundles formed by 17 and 7/8, and turns laterally near the dorsal surface of the neuropil, joining the aD commissure (Truman et al., 2004; Fig. 10e, f). In locusts, a tract, called T-tract, with exactly the same attributes has been described (Tyrer and Gregory, 1982); extending upward, it passes in between the DCI and DCIII commissures, which thereby are likely to correspond to the Drosophila lineage 17 and lineage 7/8 bundle of the aI (Fig. 10b, g). At a dorsal level, the T-tract turns laterally, joining the DCII commissure, the probable counterpart of the Drosophila aD (compare Fig. 10b, e). Commissure DCV, located in the posterior half of the neuromere, is likely to correspond to Drosophila pD, defined by the tract of lineage 6, which describes a shallow arch dorsal of the steeper ventral arch/pI commissure formed by lineages 6/12 (Fig. 10e, i)
4.2. Reduced pattern elements in the fused subesophageal ganglion
In many cases, segmental ganglia undergo a process of condensation and fusion. This happens for example for ganglia T3 and A1–3 in locust, which fuse together into the metathoracic ganglion. In this case, all internal pattern elements described for the canonical ganglion still appear to persist, although individual tracts maybe thinner and compressed into a smaller volume (Tyrer and Gregory, 1982; Fig. 10a, center). Another common fusion, observed in all insect orders, is that of the three ganglia innervating the mouthparts into a compound subesophageal ganglion. Here, aside from the compression of pattern elements, neuromeres lose individual elements (Fig. 10a, right). For example, in locust, commissures of the three neuromeres of the subesophageal ganglion are reduced. Similar to what we describe in this paper for Drosophila, the degree to which reduction occurs increases from posterior to anterior: in the labium, all dorsal commissures identified for the canonical thoracic segments can still be distinguished; in the maxilla, some commissures (e.g., DCI and III) have fused into a single bundle; in the mandibula, only the posterior commissures (DCIV and DCV) are clearly discernible (Tyrer and Gregory, 1982). A similar reduction that is strongest in the anterior SEG is observed in the honey bee (Rehder, 1988). This reduction matches the pattern described for the SEZ of Drosophila. The labium still possesses anterior and posterior commissural bundles, both represented by primary and secondary lineages. In the maxilla, anterior and posterior commissures are reduced; the anterior commissure has a single secondary lineage tract (7MX), the posterior commissure has none. In the mandibula, a single commissure, formed exclusively by primary axons, can be distinguished. As argued in the Results section, this thin mandibular commissure most likely corresponds to the posterior commissure, as confirmed for locusts and bees. It is worth noting that the reduction in internal neuropil pattern elements (e.g., commissures), which shows the same trend of increasing severity from posterior to anterior in flies, locusts, bees (and, probably, by inference, in other insect clades as well), does not match other differences in the subesophageal ganglion in these clades. For example, the number and distribution of subesophageal motor neurons, which largely mirrors the size of the external mouthparts, is very different in bees and flies. Drosophila has strongly reduced mandibular and maxillary appendages, and possesses no muscles or motor neurons in the corresponding segments. By contrast, in the honey bee, maxillary and mandibular appendages are large, movable structures, involved in feeding, and are innervated by sizeable populations of motor neurons located in the maxillary and mandibular neuromere (Rehder, 1989). It appears that different evolutionary pressures can act independently on different aspects of ganglionic structure: some lineages, like those setting up commissural connections in the mandibula, can be strongly reduced, whilst other lineages, including the ones producing motor neurons, are retained.
The recognition that many of the neuropil elements of the Drosophila nervous system are associated with specific lineages is helpful in analyzing neuroanatomy, in particular in cases where, like the SEZ, several neuromeres have become reduced and are fused into a single composite structure. Traditionally, neuromere boundaries in fused ganglia were only loosely defined by surface landmarks, such as the point of entry of the segmental nerves, and the prominent bulges of the ventral neuropil associated with these nerves (Tyrer and Gregory, 1982; Rehder, 1988; 1989). As described in this work, the vertical tracts and commissures formed by certain lineages provide additional landmarks that are firmly grounded in development. In the embryo, the neurectoderm, representing the primordium of the nervous system, forms a simple monolayer from which neuroblasts delaminate. Systems of genes expressed in transverse and longitudinal stripes provide positional information for the neuroectoderm and the neuroblasts; they define the neuroblasts located posteriorly or anteriorly, medially or laterally (Doe, 1992; Bossing et al., 1996; Schmidt et al., 1997). The cluster of neurons derived from each neuroblast, and the location where the axons of this cluster enter the neuropil, still largely reflects the location of that neuroblast. Thus, the axon tracts of lineages 3 and 12, derived from the engrailed (en)-positive, posterior-medial neuroblasts 7-1 and 6-1 (Birkholz et al., 2015; Lacin and Truman, 2016; Shepherd et al., 2016), enter at a posterior-medial position and form the posterior commissure of each neuromere. Lineages 19, 11 and 23, produced by posterior-lateral neuroblasts 6-2, 6-4 and 7-4, enter posterior-laterally and project into the posterior commssure as well. All (secondary) lineages contributing to the anterior commissure are derived from neuroblasts of the anterior four rows, including lineage 7 (neuroblast 4-2), 8 (2–4) and 17 (2–5; Birkholz et al., 2015; Lacin and Truman, 2016; Shepherd et al., 2016). It remains to be seen in how far the medio-lateral architecture of the neuropil, at least in regard to long axon tracts, also reflects the original positioning of neuroblasts in the neurectoderm.
As described previously, the number of lineages in the SEZ is reduced. The amount of lineage reduction roughly parallels the reduction of the overall neuropil volume and neuropil pattern elements, with the mandibular neuromere affected most strongly, followed by maxilla and labium. Interestingly, the reduction of lineages is developmentally progressive. In the early embryo, the labial neuromere has only 3 lineages less than the thoracic neuromeres; the maxilla has 5 less, and the mandibula has 12 less (Urbach et al., 2016). Subsequently, many of these neuroblasts in the mandibula and maxilla undergo fewer rounds of mitosis and/or die. When the phase of secondary neuroblast proliferation starts, there are only two neuroblasts left in the mandibula, 3 in the maxilla, and 10 in the labium (Kuert et al., 2014). Despite this numerical reduction, lineages are of great help in defining neuromere boundaries within the SEZ neuropil. Thus, the posterior vertical bundle, formed by lineages 3 and 12, can be identified in the labium, maxilla and mandibula throughout development. Commissural tracts that are easily defined in the embryo and larva become (in the absence of specific markers) obscured during metamorphosis; however, both anterior and posterior commissures of the labium, defined by lineage tracts of 3LB and 7LB are visible, as is the anterior commissure of the maxilla (7MX). Of equal importance is the oblique bundle formed by lineage 0, which is labeled in all gnathal neuromeres by the expression of Tdc2-Gal4/octopamine (Selcho et al., 2012; 2014). This bundle enters at a posterior position, at the same level as adjacent 3 and 12, but then projects diagonally in between the anterior and posterior commissure, and reach the anterior neuromere boundary where it bifurcates. Interestingly, the “midline tract” in the SEG of honey bee shows the same trajectory (Rehder, 1988); it probably corresponds to the tract of the lineage 0 homologs in this species.
The vertical and commissural tracts associated with specific lineages will likely be of essential help also when comparing the neuroanatomy of different species. Neuroblast maps have been established for two other species, the locust (Bate, 1976; Zacharias et al., 1993) and flour beetle (Biffar and Stollewerk, 2014). In both, the orthogonal embryonic pattern of 7 rows and 4–5 columns per hemineuromere is conserved. In both locust and flour beetle, reductions in neuroblast number occur in the gnathal neuromeres, and are strongest in the mandibula. It remains to be seen whether the projections of neurons derived from these neuroblasts form a pattern that resembles the one described for Drosophila in this and numerous other studies. This information is not yet available for other species, largely due to the absence of specific markers or clonal marking techniques. The possible exception is one lineage, the unpaired lineage 0, which includes a subset of neurons that are octopaminergic and can be labeled by reporter constructs or antibodies against this transmitter. As shown in this study, the unpaired lineages 0 of the mandibular, maxilla and labium possess neurons expressing the octopamine reporter Tdc2 (Selcho et al., 2012; 2014). Axons of octopminergic, so called ventral unpaired medial (VUM), neurons of the mandibular and maxillary cluster form two converging median bundles that extend dorso-anteriorly towards the dorsal surface of the SOG. The tracts bifurcate around the esophageal foramen, where neurons give off collateral terminal branches that innervate the dorsal neuropil domains of the anterior SEZ, as well as the adjoining VMC of the supraesophageal ganglion. The bifurcated distal axons continue into supraesophageal ganglion where they innervate many neuropil compartments, including the antennal lobe, lateral horn and mushroom body in a bilaterally symmetric pattern. The labial VUM cluster of octopaminergic neurons produces mostly bilaterally descending axons towards the thoracic ganglia (Selcho et al., 2014). In other taxa, including honey bee (Schröter et al., 2007), wasp (Haverkamp and Smid, 2014), hawkmoth (Dacks et al., 2005), and cockroach (Sinakevitch et al., 2005), octopaminergic VUM neurons with similar morphologies have been described, which are likely also representative of the lineage 0 in these species. In the wasp and bee, it was confirmed that subesophageal neurons form three metameric clusters belonging to the three SEG neuromeres. In the moth, it is apparently only the labial neuromere that gives rise to octopaminergic VUM neurons. It should be noted that, aside from the unpaired lineage 0, other (so far unspecified) lineages contribute to the diverse pool of octopminergic neurons; conversely, among the lineage 0 derivatives, only a small subset (in the order of 3–10 per neuromere) expresses octopamine, whereas most neurons do not. For example, in Drosophila, none of the labial secondary neurons of lineage 0 express Tdc2-Gal4 (this study); secondary lineage 0 neurons also show a different projection pattern than their primary Tdc2-Gal4-positive siblings. Thus, whereas a tight association between lineage 0 and octopaminergic phenotype is evolutionarily ancient, the exact number and projection pattern of lineage 0 neurons adopting this phenotype is subject to change.
4.3. The domain structure of the SEZ neuropil
The supraesophageal ganglion (SPG) of the fly and other insects is divided into many compartments. Some of these, the so called “structured” compartments, which include the optic lobe, antennal lobe, and individual components of the mushroom body and central complex, are set apart from each other and the surrounding “unstructured” neuropil by thick glial sheaths, and can be easily distinguished in histological sections without the help of specific markers. For the unstructured neuropil of the Drosophila brain, compartment boundaries were established on the basis of glial densities, accompanying prominent systems of long fiber tracts. For example, the boundaries between the compartments of the superior and inferior protocerebrum, or the ventromedial cerebrum and ventrolateral protocerebrum, are demarcated by such tracts, which appear as domains with reduced synaptic density in preparations labeled with (synaptic) markers such as anti-DN-cadherin, or anti-Bruchpilot (Pereanu et al., 2010; Ito et al., 2014). Boundaries between these compartments are more “permeable” than those between structured compartments, allowing nerve fibers to cross. Nevertheless, in many cases, the projection pattern of neuronal lineages respect these boundaries to a considerable degree (Ito et al., 2013; Wong et al., 2013; Yu et al., 2013), indicating that there exist systems of adhesive and repulsive cues that set up the boundaries during development.
The same conclusions can be drawn for the neuropil of the SEZ, where long axon tracts set up an orthogonal system of longitudinal, transverse and vertical tracts which allow one to define columnar neuropil domains, similar to the unstructured compartments of the supraesophageal ganglion. Tracts, which, as argued above, are likely evolutionarily conserved, were used in previous studies to compartmentalize the neuropil of the embryonic/larval VNC. For example, based on the systems of longitudinal FasII-positive tracts and sensory innervation, Zlatic et al. (2009) distinguished a medial, intermediate and lateral column, each of which further subdivided into a dorsal slice (#1) in their nomenclature, ventral slice (#4), and two central slices (#2, #3). We essentially followed the same subdivision, slightly modifying the boundaries in the central part of the neuropil. Here, based on the labeling with anti-DN-cadherin and anti-Bruchpilot which let one distinguish (1) domains rich in synapses from (2) others that lack synapses, but are filled with tracts of through-fibers and (in the larva) filopodial tufts of these fibers, we defined a single central column that surrounds the CITd and CITv fiber tracts. The central column is set apart from a centromedial domain, defined by the systems of transverse fibers forming the intermediate commissures, and a centrolateral domain, which lies outside the “cage” formed by the vertical fiber bundles formed by lineages 3, 12 and 6. Ventrally, the entry portals and vertical bundles of numerous lineages (3, 12: posterior vertical bundle; 7: anterior vertical bundle) demarcate a dividing line separating a ventromedial from a ventro-lateral column.
Most aspects of the domain structure defined for the larva remain intact during metamorphosis and serve as a scaffold to compartmentalize the SEZ of the adult. Importantly, the domain structure provides an adequate tool to describe projection patterns of neuron populations, such as those of the secondary lineages that develop in the SEZ. Many boundaries, such as those between central and ventral neuropil domains, demarcate dividing lines between territories innervated by secondary lineages, as shown in this paper. For example, the ventromedial tritocerebrum is densely innervated by several lineages, whereas ventral domains of the mandibula, maxilla and anterior labium are virtually devoid of secondary terminal arborizations. Sharp lines demarcate the projection of several lineages to the tritocerebrum (e.g., 5LB, high in the centromedial tritocerebrum (TRd); TRdm, excluded from this domain). Also, boundaries surrounding the central column, which extends throughout the SEZ, define a territory that is innervated strongly by some lineages (3MD, 3MX, 3LB, 7LB) and avoided by others (e.g., 12MX, 5LB, TRdm, TRdla/b, SA1). The centromedial domain of the mandibula and maxilla (“centromedial plate” and ventrally adjoining “centromedial ellipsoid”) are innervated by only two secondary subesophageal lineages, 7MX and 0LB. The domain structure also represents a useful tool to analyze the projection of sensory afferents to the SEZ, as described in detail in the accompanying paper by Kendroud et al. (2017). It remains to be seen in how far primary neurons, which in general show a more widespread projection than secondary neurons, fit into the domain structure of the SEZ.
It is worth pointing out that boundaries between neuropil domains of the SEZ appear to be very permeable for neuronal arbors along the anterior-posterior axis (i.e., between neuromeres) and towards dorsally (boundary between SEZ and SPG). Thus, for example, most secondary lineages innervating the central column spread throughout the entire length of the SEZ, rather than being restricted to the neuromere from which the lineage originates. This has been also shown for most of the individually labeled SEG interneurons described in the literature, whose dendritic and/or axonal arbors spread widely throughout the SOG (e.g., Kien et al., 1990; Lins and Lakes-Harlan, 1994). It is likely that this type of projection, which transcends neuromere boundaries, parallels the pattern of projection of sensory afferents, and the resulting distribution of sensory centers.
Acknowledgments
This work was funded by Sinergia Grant CRSII3_136307/1 to Dr. Simon Sprecher (Principal Investigator) and Volker Hartenstein (Co-Investigator), and by NIH Grant R01 NS054814 to V.H. K.T.N. was supported by the NSF Graduate Research Fellowship Program (No. DGE-0707424). J.J.O. was supported by the Ruth L. Kirschstein National Research Service Award (No.GM007185).
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: V.H., P.A.K. Acquisition of data: J.J.O., K.T.N., D.W., P.A.K., J.K.L., A.Y.H. Analysis and interpretation of data: V.H., P.A.K., H.R. Drafting of the manuscript: V.H.
References
- Andrews JC, Fernández MP, Yu Q, Leary GP, Leung AK, Kavanaugh MP, Kravitz EA, Certel SJ. Octopamine neuromodulation regulates Gr32a-linked aggression and courtship pathways in Drosophila males. PLoS Genetics. 2014;10(5):e1004356. doi: 10.1371/journal.pgen.1004356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Drosophila; pp. 214–217. [Google Scholar]
- Auer TO, Benton R. Sexual circuitry in Drosophila. Current Opinion in Neurobiology. 2016;38:18–26. doi: 10.1016/j.conb.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Bate CM. Embryogenesis of an insect nervous system. I. A map of the thoracic and abdominal neuroblasts in Locusta migratoria. Journal of Embryology and Experimental Morphology. 1976;35:107–23. [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Development. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber AJ, Snow PM, Hortsch M, Patel NH, Jacobs JR, Traquina ZR, Schilling J, Goodman CS. Drosophila neuroglian: a member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 1989;59:447–460. doi: 10.1016/0092-8674(89)90029-9. [DOI] [PubMed] [Google Scholar]
- Biffar L, Stollewerk A. Conservation and evolutionary modifications of neuroblast expression patterns in insects. Developmental Biology. 2014;388:103–16. doi: 10.1016/j.ydbio.2014.01.028. [DOI] [PubMed] [Google Scholar]
- Birkholz O, Rickert C, Nowak J, Coban IC, Technau GM. Bridging the gap between postembryonic cell lineages and identified embryonic neuroblasts in the ventral nerve cord of Drosophila melanogaster. Biology Open. 2015;4:420–34. doi: 10.1242/bio.201411072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Developmental Biology. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Regulation of temporal identities during Drosophila neuroblast lineage development. Current Opinion in Cell Biology. 2005;17:672–5. doi: 10.1016/j.ceb.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Bryant PJ. Pattern formation in imaginal discs. In: Ashburner M, Wright TRF, editors. The Genetics and Biology of Drosophila. 2c. Academic Press; London, New York & San Francisco: 1978. pp. 229–335. [Google Scholar]
- Bullock TH, Horridge GA. Structure and function in the nervous systems of invertebrates. Vol. 1. New York, NY: W. H. Freeman; 1965. [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. Springer Verlag; Berlin Heidelberg New York: 1997. [Google Scholar]
- Cardona A, Saalfeld S, Arganda I, Pereanu W, Schindelin J, Hartenstein V. Identifying neuronal lineages of Drosophila by sequence analysis of axon tracts. Journal of Neuroscience. 2010;30:7538–53. doi: 10.1523/JNEUROSCI.0186-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, Douglas RJ. TrakEM2 software for neural circuit reconstruction. PLoS One. 2012;7(6):e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certel SJ, Savella MG, Schlegel DC, Kravitz EA. Modulation of Drosophila male behavioral choice. Proceedings of the National Academy of Science U S A. 2007;104:4706–11. doi: 10.1073/pnas.0700328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb M, Scott K, Pankratz M. Gustation in Drosophila melanogaster. SEB Experimental Biology Series. 2009;63:1–38. [PubMed] [Google Scholar]
- Cole SH, Carney GE, McClung CA, Willard SS, Taylor BJ, Hirsh J. Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertility. Journal of Biological Chemistry. 2005;280:14948–55. doi: 10.1074/jbc.M414197200. [DOI] [PubMed] [Google Scholar]
- Colomb J, Grillenzoni N, Ramaekers A, Stocker RF. Architecture of the primary taste center of Drosophila melanogaster larvae. Journal of Comparative Neurology. 2007;502:834–47. doi: 10.1002/cne.21312. [DOI] [PubMed] [Google Scholar]
- Dacks AM, Christensen TA, Agricola HJ, Wollweber L, Hildebrand JG. Octopamine-immunoreactive neurons in the brain and subesophageal ganglion of the hawkmoth Manduca sexta. Journal of Comparative Neurology. 2005;488:255–68. doi: 10.1002/cne.20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethier VG. The Hungry Fly: A Physiological Study of the Behavior Associated with Feeding. Harvard University Press; 1976. [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–63. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Dominick OS, Truman JW. The physiology of wandering behaviour in Manduca sexta. III. Organization of wandering behaviour in the larval nervous system. Journal of Experimental Biology. 1986;121:115–32. doi: 10.1242/jeb.121.1.115. [DOI] [PubMed] [Google Scholar]
- Ferris GF. External Morphology of the Adult. In: Demerec M, editor. Biology of Drosophila. Wiley; New York: 1950. pp. 368–419. [Google Scholar]
- Gendre N, Lüer K, Friche S, Grillenzoni N, Ramaekers A, Technau GM, Stocker RF. Integration of complex larval chemosensory organs into the adult nervous system of Drosophila. Development. 2004;131:83–92. doi: 10.1242/dev.00879. [DOI] [PubMed] [Google Scholar]
- Gregory GE. Neuroanatomy of the mesothoracic ganglion of the cockroach Periplaneta americana (L.). I. The roots of the peripheral nerves. Philosophical Transactions of the Royal Society London Series B Biological Sciences. 1974;267:421–65. doi: 10.1098/rstb.1974.0005. [DOI] [PubMed] [Google Scholar]
- Harris RM, Pfeiffer BD, Rubin GM, Truman JW. Neuron hemilineages provide the functional ground plan for the Drosophila ventral nervous system. Elife. 2015 Jul;20:4. doi: 10.7554/eLife.04493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Spindler S, Pereanu W, Fung S. The development of the Drosophila larval brain. Advances in Experimental Medicine and Biology. 2008;628:1–31. doi: 10.1007/978-0-387-78261-4_1. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Younossi-Hartenstein A, Lovick JK, Kong A, Omoto JJ, Ngo KT, Viktorin G. Lineage-associated tracts defining the anatomy of the Drosophila first instar larval brain. Developmental Biology. 2015;406:14–39. doi: 10.1016/j.ydbio.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp A, Smid HM. Octopamine-like immunoreactive neurons in the brain and subesophageal ganglion of the parasitic wasps Nasonia vitripennis and N. giraulti. Cell and Tissue Research. 2014;358:313–29. doi: 10.1007/s00441-014-1960-3. [DOI] [PubMed] [Google Scholar]
- He H, Noll M. Differential and redundant functions of gooseberry and gooseberry neuro in the central nervous system and segmentation of the Drosophila embryo. Developmental Biology. 2013;382:209–23. doi: 10.1016/j.ydbio.2013.05.017. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Patel NH, Bieber AJ, Traguina ZR, Goodman CS. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases, is dynamically expressed during embryogenesis. Development. 1990;110:1327–1340. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Hückesfeld S, Schoofs A, Schlegel P, Miroschnikow A, Pankratz MJ. Localization of Motor Neurons and Central Pattern Generators for Motor Patterns Underlying Feeding Behavior in Drosophila Larvae. PLoS One. 2015;10(8):e0135011. doi: 10.1371/journal.pone.0135011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–71. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Ito M, Masuda N, Shinomiya K, Endo K, Ito K. Systematic analysis of neural projections reveals clonal composition of the Drosophila brain. Current Biology. 2013;23:644–655. doi: 10.1016/j.cub.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Ito K, Shinomiya K, Ito M, Armstrong JD, Boyan G, Hartenstein V, Harzsch S, Heisenberg M, Homberg U, Jenett A, Keshishian H, Restifo LL, Rössler W, Simpson JH, Strausfeld NJ, Strauss R, Vosshall LB Insect Brain Name Working Group. A systematic nomenclature for the insect brain. Neuron. 2014;81:755– 765. doi: 10.1016/j.neuron.2013.12.017. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, Iyer N, Fetter D, Hausenfluck JH, Peng H, Trautman ET, Svirskas RR, Myers EW, Iwinski ZR, Aso Y, DePasquale GM, Enos A, Hulamm P, Lam SC, Li HH, Laverty TR, Long F, Qu L, Murphy SD, Rokicki K, Safford T, Shaw K, Simpson JH, Sowell A, Tae S, Yu Y, Zugates CT. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendroud S, Bohra AA, Kuert P, Nguyen B, Guillermin O, Sprecher SG, Reichert H, VijayRaghavan K, Hartenstein V. Structure and Development of the Subesophageal Zone of the Drosophila Brain. II. Sensory Compartments. Journal of Comparative Neurology. 2017 doi: 10.1002/cne.24316. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kien J, Fletcher WA, Altman JS, Ramirez JM, Roth U. Organisation of intersegmental interneurons in the suboesophageal ganglion of Schistocerca gregaria (Forksal) and Locusta migratoria migratoides (Reiche and Fairmaire)(Acrididae, Orthoptera) International Journal of Insect Morphology and Embyology. 1990;19:35–60. [Google Scholar]
- Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nature Reviews Neuroscience. 2013;14:823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuert PA, Bello BC, Reichert H. The labial gene is required to terminate proliferation of identified neuroblasts in postembryonic development of the Drosophila lineage identification and lineage-specific Hox gene action during postembryonic development of the subesophageal ganglion in the Drosophila central brain. Developmental Biology. 2012;390:102–115. [Google Scholar]
- Kuert PA, Hartenstein V, Bello BC, Lovick JK, Reichert H. Neuroblast lineage identification and lineage-specific Hox gene action during postembryonic development of the subesophageal ganglion in the Drosophila central brain. Developmental Biology. 2014;390:102–15. doi: 10.1016/j.ydbio.2014.03.021. [DOI] [PubMed] [Google Scholar]
- Kumar A, Fung S, Lichtneckert R, Reichert H, Hartenstein V. Arborization pattern of engrailed-positive neural lineages reveal neuromere boundaries in the Drosophila brain neuropil. Journal of Comparative Neurology. 2009;517:87–104. doi: 10.1002/cne.22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacin H, Truman JW. Lineage mapping identifies molecular and architectural similarities between the larval and adult Drosophila central nervous system. Elife. 2016;15:5, e13399. doi: 10.7554/eLife.13399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Sánchez-Soriano N, Technau GM, Urban J, Prokop A. Charting the Drosophila neuropil: a strategy for the standardised characterisation of genetically amenable neurites. Developmental Biology. 2003;260:207–25. doi: 10.1016/s0012-1606(03)00215-x. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends in Neuroscience. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Lins F, Lakes-Harlan R. Interneurons with inhibitory effects on stridulation in grasshoppers exhibit GABA-like immunoreactivity. Brain Research. 1994;635:103–12. doi: 10.1016/0006-8993(94)91428-1. [DOI] [PubMed] [Google Scholar]
- Lovick JK, Ngo KT, Omoto JJ, Wong DC, Nguyen JD, Hartenstein V. Postembryonic lineages of the Drosophila brain: I. Development of the lineage-associated fiber tracts. Developmental Biology. 2013;384:228–257. doi: 10.1016/j.ydbio.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovick JK, Hartenstein V. Hydroxyurea-mediated neuroblast ablation establishes birth dates of secondary lineages and addresses neuronal interactions in the developing Drosophila brain. Developmental Biology. 2015;402:32–47. doi: 10.1016/j.ydbio.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovick JK, Kong A, Omoto JJ, Ngo KT, Younossi-Hartenstein A, Hartenstein V. Patterns of growth and tract formation during the early development of secondary lineages in the Drosophila larval brain. Developmental Neurobiology. 2016;76:434–451. doi: 10.1002/dneu.22325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Kanou M, Yamaguchi T. Motor program initiation and selection in crickets, with special reference to swimming and flying behavior. Journal of Comparative Physiology Series A Neuroethology Sensory Neural Behavioural Physiology. 2002;187:987–95. doi: 10.1007/s00359-001-0269-3. [DOI] [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Embryonic development of the Drosophila brain. I. Pattern of pioneer tracts. Journal of Comparative Neurology. 1998;402:10–31. [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Early development of the Drosophila brain: III. The pattern of neuropil founder tracts during the larval period. Journal of Comparative Neurology. 2003;455:417–34. doi: 10.1002/cne.10482. [DOI] [PubMed] [Google Scholar]
- Page DT. A mode of arthropod brain evolution suggested by Drosophila commissure development. Evolution and Development. 2004;6:25–31. doi: 10.1111/j.1525-142x.2004.04003.x. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annual Review of Cell and Developmental Biology. 2004;20:619–47. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. Journal of Neuroscience. 2006;26:5534–53. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Spindler S, Cruz L, Hartenstein V. Tracheal development in the Drosophila brain is constrained by glial cells. Developmental Biology. 2007;302:169–80. doi: 10.1016/j.ydbio.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Kumar A, Jenett A, Reichert H, Hartenstein V. Development-based compartmentalization of the Drosophila central brain. Journal of Comparative Neurology. 2010;518:2996–3023. doi: 10.1002/cne.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovant M, Léna P. Membrane glycoproteins immunologically related to the human insulin receptor are associated with presumptive neuronal territories and developing neurones in Drosophila melanogaster. Development. 1988;103:145–156. doi: 10.1242/dev.103.1.145. [DOI] [PubMed] [Google Scholar]
- Rajashekhar KP, Singh RN. Neuroarchitecture of the tritocerebrum of Drosophila melanogaster. Journal of Comparative Neurology. 1994;349:633–45. doi: 10.1002/cne.903490410. [DOI] [PubMed] [Google Scholar]
- Ramirez JM. Reconfiguration of the respiratory network at the onset of locust flight. Journal of Neurophysiology. 1998;80:3137–47. doi: 10.1152/jn.1998.80.6.3137. [DOI] [PubMed] [Google Scholar]
- Rehder V. A neuroanatomical map of the suboesophageal and prothoracic ganglia of the honey bee (Apis mellifera) Proceedings of the Royal Society London [Biology] 1988;235:179–202. [Google Scholar]
- Rehder V. Sensory pathways and motoneurons of the proboscis reflex in the suboesophageal ganglion of the honey bee. Journal of Comparative Neurology. 1989;279:499–513. doi: 10.1002/cne.902790313. [DOI] [PubMed] [Google Scholar]
- Sakurai A, Koganezawa M, Yasunaga K, Emoto K, Yamamoto D. Select interneuron clusters determine female sexual receptivity in Drosophila. Nature Communications. 2013;4:1825. doi: 10.1038/ncomms2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–89. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Developmental Biology. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Schoofs A, Spiess R. Anatomical and functional characterisation of the stomatogastric nervous system of blowfly (Calliphora vicina) larvae. Journal of Insect Physiology. 2007;53:349–360. doi: 10.1016/j.jinsphys.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Schoofs A, Hückesfeld S, Schlegel P, Miroschnikow A, Peters M, Zeymer M, Spiess R, Chiang AS, Pankratz MJ. Selection of motor programs for suppressing food intake and inducing locomotion in the Drosophila brain. PLoS Biology. 2014;12(6):e1001893. doi: 10.1371/journal.pbio.1001893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter U, Malun D, Menzel R. Innervation pattern of suboesophageal ventral unpaired median neurones in the honeybee brain. Cell and Tissue Research. 2007;327:647–667. doi: 10.1007/s00441-006-0197-1. [DOI] [PubMed] [Google Scholar]
- Schwarz O, Bohra AA, Liu X, Reichert H, VijayRaghavan K, Pielage J. Motor control of Drosophila feeding behavior. Elife. 2017;17:6. doi: 10.7554/eLife.19892. pii: e19892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. Taste recognition: food for thought. Neuron. 2005;48:455–64. doi: 10.1016/j.neuron.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Selcho M, Pauls D, El Jundi B, Stocker RF, Thum AS. The role of octopamine and tyramine in Drosophila larval locomotion. Journal of Comparative Neurology. 2012;520:3764–3785. doi: 10.1002/cne.23152. [DOI] [PubMed] [Google Scholar]
- Selcho M, Pauls D, Huser A, Stocker RF, Thum AS. Characterization of the octopaminergic and tyraminergic neurons in the central brain of Drosophila larvae. Journal of Comparative Neurology. 2014;522:3485–500. doi: 10.1002/cne.23616. [DOI] [PubMed] [Google Scholar]
- Shepherd D, Harris R, Williams DW, Truman JW. Postembryonic lineages of the Drosophila ventral nervous system: Neuroglian expression reveals the adult hemilineage associated fiber tracts in the adult thoracic neuromeres. Journal of Comparative Neurology. 2016;524:2677–95. doi: 10.1002/cne.23988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinakevitch I, Niwa M, Strausfeld NJ. Octopamine-like immunoreactivity in the honey bee and cockroach: comparable organization in the brain and subesophageal ganglion. Journal of Comparative Neurology. 2005;488:233–54. doi: 10.1002/cne.20572. [DOI] [PubMed] [Google Scholar]
- Singh RN. Neurobiology of the gustatory systems of Drosophila and some terrestrial insects. Microscopy Research and Technique. 1997;39:547–563. doi: 10.1002/(SICI)1097-0029(19971215)39:6<547::AID-JEMT7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Spiess R, Schoofs A, Heinzel HG. Anatomy of the stomatogastric nervous system associated with the foregut in Drosophila melanogaster and Calliphora vicina third instar larvae. Journal of Morphology. 2008;269:272–82. doi: 10.1002/jmor.10581. [DOI] [PubMed] [Google Scholar]
- Spindler SR, Hartenstein V. Bazooka mediates secondary axon morphology in Drosophila brain lineages. Neural Development. 2011;6:16. doi: 10.1186/1749-8104-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld NJ. Atlas of an Insect Brain. Springer-Verlag; Berlin: 1976. pp. 1–214. [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- Tyrer NM, Gregory GE. A guide to the neuroanatomy of locust suoesophageal and thoracic ganglia. Philosophical Transactions of the Royal Society London Series B. 1982;297:91–123. [Google Scholar]
- Urbach R, Jussen D, Technau GM. Gene expression profiles uncover individual identities of gnathal neuroblasts and serial homologies in the embryonic CNS of Drosophila. Development. 2016;143:1290–301. doi: 10.1242/dev.133546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117:981–91. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Wong DC, Lovick JK, Ngo KT, Borisuthirattana W, Omoto JJ, Hartenstein V. Postembryonic lineages of the Drosophila brain: II. Identification of lineage projection patterns based on MARCM clones. Developmental Biology. 2013;384:258–289. doi: 10.1016/j.ydbio.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GA. To feed or not to feed: circuits involved in the control of feeding in insects. Current Opinion in Neurobiology. 2016;41:87–91. doi: 10.1016/j.conb.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Yu HH, Awasaki T, Schroeder MD, Long F, Yang JS, He Y, Ding P, Kao JC, Wu GY, Peng H, Myers G, Lee T. Clonal development and organization of the adult Drosophila central brain. Current Biology. 2013;23:633–643. doi: 10.1016/j.cub.2013.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias D, Williams JLD, Meier T, Reichert H. Neurogenesis in the insect brain: cellular identification and molecular characterization of brain neuroblasts in the grasshopper embryo. Development. 1993;118:941–955. [Google Scholar]
- Zitnan D, Adams ME. Excitatory and inhibitory roles of central ganglia in initiation of the insect ecdysis behavioural sequence. Journal of Experimental Biology. 2000;203:1329–40. doi: 10.1242/jeb.203.8.1329. [DOI] [PubMed] [Google Scholar]
- Zhou C, Rao Y, Rao Y. A subset of octopaminergic neurons are important for Drosophila aggression. Nature Neuroscience. 2008;11:1059–67. doi: 10.1038/nn.2164. [DOI] [PubMed] [Google Scholar]
- Zlatic M, Li F, Strigini M, Grueber W, Bate M. Positional cues in the Drosophila nerve cord: semaphorins pattern the dorso-ventral axis. PLoS Biology. 2009;7(6):e1000135. doi: 10.1371/journal.pbio.1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]