Abstract
An understudied symptom of the genetic disorder Neurofibromatosis type 1 (NF1) is chronic idiopathic pain. We used targeted editing of Nf1 in rats to provide direct evidence of a causal relationship between neurofibromin, the protein product of the Nf1 gene, and pain responses. Our study data identified a protein-interaction network with collapsin response meditator protein 2 (CRMP2) as a node and neurofibromin, syntaxin 1A, and the N-type voltage-gated calcium (CaV2.2) channel as interaction edges. Neurofibromin uncouples CRMP2 from syntaxin 1A. Upon loss/mutation of neurofibromin, as seen in patients with NF1, the CRMP2/Neurofibromin interaction is uncoupled, which frees CRMP2 to interact with both syntaxin 1A and CaV2.2, culminating in increased release of the pro-nociceptive neurotransmitter calcitonin gene related peptide (CGRP). Our work also identified the CRMP2-derived peptide CNRP1, which uncoupled CRMP2’s interactions with neurofibromin, syntaxin 1A, as well as CaV2.2. Here, we tested if CRISPR/Cas9-mediated editing of the Nf1 gene, which leads to functional remodeling of peripheral nociceptors through effects on the tetrodotoxin-sensitive (TTX-S) Na+ voltage-gated sodium channel (NaV1.7) and CaV2.2, could be affected by using CNRP1, a peptide designed to target the CRMP2-neurofibromin interface. The data presented here shows that disrupting the CRMP2 neurofibromin interface is sufficient to reverse the dysregulations of voltage-gated ion channels and neurotransmitter release elicited by Nf1 gene editing. As a consequence of these effects, the CNRP1 peptide reversed hyperalgesia to thermal stimulation of the hindpaw observed in Nf1-edited rats. Our findings support future pharmacological targeting of the CRMP2/neurofibromin interface for NF1-related pain relief.
Keywords: CRMP2, Neurofibromin, Pain, CaV2.2, NaV1.7, Neurofibromatosis type 1
Introduction
Chronic idiopathic pain is a major symptom and comorbidity factor in the neurological disorder Neurofibromatosis type 1 (NF1) (Creange et al., 1999; Drouet et al., 2004). The prevalence of pain in NF1 patients is unknown but qualities of life-based questionnaires consistently identify both the intensity and quality of pain as having a major impact on NF1 patients (Bicudo et al., 2016; Crawford et al., 2015; Ferner, 2007; Ferner et al., 2017; Kodra et al., 2009; Riklin et al., 2017; Wolkenstein et al., 2001; Wolters et al., 2015). The first direct evidence of a causal relationship between neurofibromin, a protein encoded by the Nf1 gene, and pain responses was demonstrated by using a genome editing approach in vivo (Moutal et al., 2017a; Moutal et al., 2017f). The truncation of neurofibromin by targeting exon 39 on the Nf1 gene in sensory neurons, with a specific guide RNA (gRNA) conjugated to clustered regularly interspaced short palindromic repeats (CRISPR) associated protein-9 nuclease (Cas9), resulted in hyperalgesia (Moutal, et al., 2017a; Moutal, et al., 2017f). The underlying mechanism for the observed painful behaviors was hypothesized to via remodeling of small-diameter nociceptive sensory neurons (Moutal et al., 2017b). Data from recent studies of rat neurons from Cas9-edited Nf1 were in agreement with observations in haploinsufficient (Nf1+/−) mouse sensory neurons: upregulation of voltage-gated ion channels (Duan et al., 2014; Moutal, et al., 2017a; Moutal, et al., 2017b; Moutal, et al., 2017f; Wang et al., 2010a; Wang et al., 2010b). Specifically, the nociceptive synapse in these rodent models of NF1 is characterized by increases in N-type voltage-gated calcium (CaV2.2) as well as tetrodotoxin-sensitive (TTX-S) voltage-gated sodium (NaV1.7) currents (Duan, et al., 2014; Moutal, et al., 2017f; Wang, et al., 2010a; Wang, et al., 2010b). A consequence of this nociceptor remodeling was a reduction of rheobase (i.e., the current required to elicit the first action potential (AP)) and a concomitant increase in excitability (Moutal, et al., 2017f; Wang, et al., 2010b; Wang et al., 2005). It follows then, that release of excitatory transmitter – calcitonin gene-related peptide (CGRP) – to the spinal dorsal horn was also increased in both Nf1 edited rats and Nf1+/− mice (Hingtgen et al., 2006; Moutal, et al., 2017a). Thus, in NF1, the sensory neurons are primed at the synaptic level for facilitated nociceptive signal transmission. Further studies identified the dysregulation of the collapsin response mediator protein 2 (CRMP2) (Patrakitkomjorn et al., 2008) to be responsible for NF1-related pain.
CRMP2 was initially described as an axonal protein involved in axon guidance and growth (Fukata et al., 2002; Goshima et al., 1995; Kamata et al., 1998). The functional association of neurofibromin and CRMP2 was reported to be essential for neuronal cell differentiation; lack of expression or abnormal regulation of neurofibromin resulted in impaired function of neuronal cells (Lin et al., 2008; Patrakitkomjorn, et al., 2008). Suppression of neurofibromin using neurofibromin small interfering RNA significantly inhibited neurite outgrowth and upregulated CRMP2 phosphorylations by kinases identified as Cdk5, GSK-3β, and Rho kinase (Patrakitkomjorn, et al., 2008). Truncation of the C-terminus of neurofibromin, where CRMP2 binds (Patrakitkomjorn, et al., 2008), lead to upregulation of CRMP2 functions in ion channel trafficking (Moutal, et al., 2017a; Moutal, et al., 2017f). CRMP2 binds to and regulate the membrane localization of both CaV2.2 (Brittain et al., 2011; Brittain et al., 2009; Moutal et al., 2016a; Moutal et al., 2016b; Moutal et al., 2016c) and NaV1.7 (Dustrude et al., 2016; Dustrude et al., 2017a; Dustrude et al., 2013). CRMP2 also participates in neurotransmitter release (Chi et al., 2009; Moutal, et al., 2016b; Moutal et al., 2017e). Recent proteomic analysis identified syntaxin 1A as a novel CRMP2-binding protein whose interaction with CRMP2 was strengthened in neurofibromin-depleted cells (Moutal, et al., 2017e). These mutually exclusive interactions appear to form a ‘core complex’ that can facilitate nociceptive transmission in NF1. A peptide targeting CRMP2’s interaction domain with neurofibromin, designated CRMP2-neurofibromin regulating peptide 1 (CNRP1,) inhibited CGRP release and acute and neuropathic nociceptive behaviors (Moutal, et al., 2017e).
Although an obligatory role for CRMP2 has been established for manifestation of NF1-related pain (Moutal, et al., 2017a), the precise manner by which neurofibromin’s interaction with CRMP2 leads to alterations in ion channel function is unclear. Here, we tested if targeting the CRMP2/neurofibromin interaction in Nf1 edited in sensory neurons could reverse ion channel dysregulation, sensory neuron excitability, and hyperalgesia. Our data show that the CNRP1 peptide from CRMP2 is sufficient to reverse alterations of sensory neurons and nociceptive behaviors in NF1without producing motor impairment or anxiety.
Experimental procedures
Animals
Pathogen-free, adult male and female Sprague–Dawley rats (150–200 g; Harlan Laboratories) were housed in temperature (23±3 °C) and light (12-h light/12-h dark cycle; lights on 07:00–19:00) controlled rooms with standard rodent chow and water available ad libitum. The Institutional Animal Care and Use Committee of the College of Medicine at the University of Arizona approved all experiments. All procedures were conducted in accordance with the Guide for Care and Use of Laboratory Animals published by the National Institutes of Health and the ethical guidelines of the International Association for the Study of Pain. Animals were randomly assigned to treatment or control groups for the behavioral experiments. Animals were initially housed three per cage but individually housed after the intrathecal cannulation on a 12 h light-dark cycle with food and water ad libitum. All behavioral experiments were performed by experimenters who were blinded to the experimental groups and treatments.
Materials
t-CNRP1 peptide (tat cell penetrating peptide sequence: YGRKKRRQRRR fused to CNRP1: HVTEGSGRYIPRKPF) (Moutal, et al., 2017e) was synthesized and HPLC-purified (>95% purity) by Genscript Inc. (Piscataway, NJ, USA). Scramble and random sequence based peptides conjugated to various cargoes as controls have been previously studied as controls in molecular, biochemical and behavioral assays and demonstrated to have no effects (Brittain, et al., 2011; Francois-Moutal et al., 2015; Ju et al., 2012; Piekarz et al., 2012). All chemicals, unless noted were purchased from Sigma (St. Louis, MO). Antibodies were purchased as follows: anti-CaV2.2 polyclonal antibody (Cat# TA308673, Origene Technologies, Inc, Rockville, MD), anti-NaV1.7 (Cat# 75-103, NeuroMab, Davis, CA) or anti-neurofibromin C-terminal (Abcam Cat# ab17963).
gRNA strategy for Nf1 gene targeting
Our strategy to truncate neurofibromin focused on targeting exon 39 of the Nf1 gene using a guide RNA (gRNA) as described previously (Moutal, et al., 2017a; Moutal, et al., 2017f). We targeted this exon to express a C-terminally truncated neurofibromin protein, since 80% of NF1 patients express a C-terminally truncated neurofibromin (Esposito et al., 2015). The gRNA sequence (GGCAGTAACCCTTTGTCGTT) was inserted into the BbsI restriction site of the pSpCas9(BB)-2A-GFP plasmid (PX458, Cat#48138, Addgene, Cambridge, MA) (Ran et al., 2013), a plasmid that allows for simultaneous expression of (i) the Cas9 enzyme; (ii) the gRNA; and (iii) a green fluorescent protein (GFP) – to control for transfection efficiency. All plasmids were verified by Sanger sequencing (Eurofins, Louisville, KY).
Culturing and transfection of rat primary Dorsal Root Ganglia (DRG) Neurons
Rat DRG neurons were isolated from 150–174g Sprague-Dawley rats and then transfected exactly as previously described (Brittain, et al., 2011). In brief, removing dorsal skin and muscle and cutting the vertebral bone processes parallel to the dissection stage exposed DRGs. DRGs were then collected, trimmed at their roots, and digested in 3 ml bicarbonate free, serum free, sterile Dulbecco’s Modified Eagle’s medium (DMEM; Cat# 11965, Thermo Fisher Scientific, Waltham, MA) solution containing neutral protease (3.125 mg.ml−1, Cat#LS02104, Worthington, Lakewood, NJ) and collagenase Type I (5 mg.ml−1, Cat# LS004194, Worthington, Lakewood, NJ) and incubated for 45 min at 37°C under gentle agitation. Dissociated DRG neurons (~1.5 × 106) were then gently centrifuged to collect cells and washed with DRG media: DMEM containing 1% penicillin/streptomycin sulfate from 10 000 μg/ml stock, 30 ng.ml−1 nerve growth factor, and 10% fetal bovine serum (Hyclone). The pelleted cells were re-suspended in Nucleofector transfection reagent containing plasmids. Then, cells were subjected to electroporation protocol O-003 in an Amaxa Biosystem (Lonza, Basel, Switzerland) and plated onto poly-D-lysine- and laminin-coated glass (12- or 15-mm) coverslips. Transfection efficiencies were routinely between 20% and 30% with about ~10% cell death. Small diameter (~ <30 μm) neurons were selected to target Aδ- and C-fiber nociceptive neurons. All cultures were used within 48 hours after transfection.
Patch clamp electrophysiology
Recordings were obtained from acutely dissociated DRG neurons as described (Dustrude, et al., 2016; Moutal, et al., 2017e; Moutal, et al., 2017f). To isolate calcium currents, Na+ and K+ currents were blocked with 500 nM TTX (Alomone Labs) and 30 mM tetraethylammonium chloride (Sigma). Extracellular recording solution (at ~310 mOsm) consisted of the following (in millimolar): 110 N-methyl-D-glucamine, 10 BaCl2, 30 tetraethylammonium chloride, 10 HEPES, 10 glucose, pH at 7.4, 0.001 TTX, and 0.01 nifedipine. The intracellular recording solution (at ~310 mOsm) consisted of the following (in millimolar): 150 CsCl2, 10 HEPES, 5 Mg-ATP, 5 BAPTA, pH at 7.4. To isolate the contributions of N-type (CaV2.2) channels, we used the following subunit-selective blockers (all purchased from Alomone Labs, Jerusalem): Nifedipine (10 μM, L-type); ω-agatoxin GIVA (200 nM, P/Q-type) (Mintz et al., 1992); SNX-482 (200 nM, R-type) (Newcomb et al., 1998); and 3,5-dichloro-N-[1-(2,2-dimethyl-tetrahydro-pyran-4-ylmethyl)-4-fluoro-piperidin-4-ylmethyl]-benzamide (TTA-P2, 1 μM, T-type) (Choe et al., 2011).
Whole cell voltage clamp and current clamp recordings were performed at room temperature using an EPC 10 Amplifier-HEKA as previously described (Dustrude, et al., 2013). The internal solution for voltage clamp sodium current recordings contained (in mM): 140 CsF, 1.1Cs-EGTA, 10 NaCl, and 15 HEPES (pH 7.3, 290–310 mOsm/L) and external solution contained (in mM): 140 NaCl, 3 KCl, 30 tetraethylammonium chloride, 1 CaCl2, 0.5 CdCl2, 1 MgCl2, 10 D-glucose, 10 HEPES (pH 7.3, 310–315 mosM/L). For DRG current clamp experiments the internal solution contained (in mM): 140 KCl, 10 NaCl, 1 MgCl2, 1 EGTA, 10 HEPES (pH 7.2), and 1 ATP-Mg (pH 7.3, 285-295 mOsm/L) and external solution contained (in mM): 154 NaCl, 5.6 KCl, 2 CaCl2, 2.0 MgCl2, 1.0 Glucose, and 10 HEPES (pH 7.4, 305-315 mOsm/L).
DRG neurons were subjected to current-density (I–V) and fast-inactivation voltage protocols. In the I–V protocol, cells were held at a −80 mV holding potential prior to depolarization by 20 ms voltage steps from −70 mV to +60 mV in 5 mV increments. This allowed for collection of current density data to analyze activation of sodium channels as a function of current versus voltage and also peak current density which was typically observed near ~ 0–10 mV and normalized to cell capacitance (pF). In the fast-inactivation protocol, cells were held at a −80 mV holding potential prior to hyperpolarizing and repolarizing pulses for 500 ms between −120 mV to −10 mV in 5 mV increments. This step conditioned various percentages of channels into fast-inactivated states so that a 0 mV test pulse for 20 ms could reveal relative fast inactivation normalized to maximum current. DRGs were subjected to current-density (I–V) protocol and H-infinity (pre-pulse inactivation protocol). To estimate TTX-R contributions, I-V protocol was run after 10 min incubation with 500 nM TTX. Following holding at −100 mV, 200 ms voltage steps from −70 mV to +60 mV in 5 mV increments allowed for analysis of peak currents. Small and medium diameter DRG neurons which homogenously present TTX-S, TTX-R Na+ currents (Roy et al., 1992) and N-type Ca2+ currents (Scroggs et al., 1992), were selected for all recordings.
Fire-polished recording pipettes with 2 to 5 megaohms (mΩ) resistances were used for all recordings. Whole-cell recordings were obtained using a HEKA EPC-10 USB (HEKA Instruments Inc, Bellmore, NY); data were acquired using a Patchmaster (HEKA) and analyzed with a Fitmaster (HEKA). Capacitive artifacts were fully compensated, and series resistance was compensated by ~70%. Recordings made from cells with greater than a 5 mV shift in series resistance compensation error were excluded from analysis. All experiments were performed at room temperature (~23°C).
Confocal imaging
Immunocytochemistry was performed on DRGs that were transfected as described before (Moutal, et al., 2017f). Briefly, cells were fixed using 4% paraformaldehyde, 4% sucrose for 20 minutes at room temperature. Permeabilization was achieved with a 30-min incubation in phosphate-buffered saline (PBS) containing 0.1 % Triton X-100, following which nonspecific binding sites were saturated by PBS containing 3 % BSA for 30 minutes. Cell staining was performed with primary antibodies in PBS with 3 % BSA overnight at 4°C. Cells were then washed three times in PBS and incubated with PBS containing 3 % BSA and secondary antibodies (Alexa 488 goat anti-mouse and Alexa 594 goat anti-rabbit from Life Technologies) for 1 hour at room temperature. Immunofluorescent micrographs were acquired on a Nikon C1si scanning confocal microscope using CFI Plan APO VC ×60 oil immersion objective with 1.4 numerical aperture. Camera gain and other relevant settings were kept constant. Membrane to cytosol ratio was determined by defining regions of interest on the cytosol and the membrane of each cell using Image J. Total fluorescence intensity was normalized to the area of the region of interest before calculating the membrane to cytosol ratio. Raw images were used for quantification while representative pictures were background subtracted and contrast enhanced for better visualization of the reader.
Implantation of intrathecal catheter
For intrathecal drug administration, rats were chronically implanted with catheters as described by Yaksh and Rudy (Yaksh et al., 1976). Rats were anesthetized with halothane and placed in a stereotactic head holder. The occipital muscles were separated from their occipital insertion and retracted caudally to expose the cisternal membrane at the base of the skull. Polyethylene tubing was passed caudally from the cisterna magna to the level of the lumbar enlargement. Animals were allowed to recover and were examined for evidence of neurologic injury. Animals with evidence of neuromuscular deficits were excluded.
In vivo transfection of CRISPR plasmids
For in vivo transfection, the plasmids pSpCas9(BB)-2A-GFP-Nf1-gRNA or pSpCas9(BB)-2A-GFP-Nf1-control-gRNA were diluted to 0.5 μg/μl in 5% sterile glucose solution. Then, Turbofect in vivo transfection reagent (Cat#R0541, Thermo Fisher Scientific, Waltham, MA) was added following manufacturer’s instructions. Finally, 20 μL of the plasmid complexes were injected intrathecally in Sprague Dawley rats.
Immunohistofluorescence and epifluorescence imaging
L5 DRGs were dissected from adult rats and then fixed using 4% paraformaldehyde for 4 hrs at room temperature (RT). DRGs were next transferred into a 30% sucrose solution and left at 4°C until sinking of the tissues could be observed (~3 days). Tissues were cut at 10 μm thickness using the Bright OTF 5000 Microtome Cryostat (Hacker Instruments and Industries, Inc., Winnsboro, SC), and fixed onto gelatin coated glass slides and kept at −20°C until use. DRG slices were permeabilized and saturated using PBS containing 3% BSA, 0.1% triton X-100 solution for 30 min at RT, and then anti-neurofibromin C-terminal (Abcam Cat# ab17963) was added overnight. The slices were then washed 3X in PBS, and incubated with PBS containing 3% BSA, 0.1% triton X-100 containing secondary antibody (Alexa 594 goat anti-rabbit (Life Technologies)) for at least 3 hrs at RT. After 3 washes (PBS, 10 min, RT), either DAPI was used to stain the nuclei of cells. Slides were mounted and stored at 4°C until analysis. Immunofluorescent micrographs were acquired on an Olympus BX51 microscope with a Hamamatsu C8484 digital camera using a 20X UplanS Apo 0.75 numerical aperture objective. For a given experiment, all images were taken using identical acquisition parameters by individuals blinded to the treatment groups.
Measurement of thermal withdrawal latency
The method of Hargreaves et al. (Hargreaves et al., 1988) was used. Rats were acclimated within Plexiglas enclosures on a clear glass plate maintained at 30°C. A radiant heat source (high-intensity projector lamp) was focused onto the plantar surface of the hind paw. When the paw was withdrawn, a motion detector halted the stimulus and a timer. A maximal cutoff of 33.5 sec was used to prevent tissue damage.
Elevated plus maze (EPM)
The EPM consists of four elevated (50cm) arms (50cm long and 10cm wide) with two opposing arms containing 30cm high opaque walls. EPM testing occurred in a quiet testing room with ambient lighting at ~500 lux. On day of testing, rats were allowed to acclimate to the testing room for 20 minutes. Each rat was placed in a closed arm, facing the enter platform and cage mates started in the same closed arm. Each rat was allowed 5 minute to explore the EPM and then returned to its home cage. Between animals the EPM was cleaned thoroughly with Versa-Clean (Fisher Scientific). EPM performance was recorded using an overhead video camera (MHD Sport 2.0 WiFi Action Camera, Walmart.com) for later quantification. Open and closed arm entries were defined as the front two paws entering the arm, and open arm time began the moment the front paws entered the open arm and ended upon exit. An anxiety index was also calculated; the index combines EPM parameters into one unified ratio with values ranging from 0 to 1, with a higher value indicating increased anxiety (Huynh et al., 2011). The following equation was used for calculation of the anxiety index:
Rotarod
Following placement of the intrathecal catheters, the rats were trained to walk on a rotating rod (8 rev/min; Rotamex 4/8 device) with a maximal cutoff time of 180 seconds. Training was initiated by placing the rats on a rotating rod and allowing them to walk on the rotating rod until they either fell off or 180 seconds was reached. This process was repeated 6 times and the rats were allowed to recover for 24 hours before beginning the treatment session. Prior to treatment, the rats were run once on a moving rod in order to establish a baseline value. Saline or t-CNRP1 was administered spinally via the intrathecal catheter. Assessment consisted of placing the rats on the moving rod and timing until either they fell off or reached a maximum of 180 seconds. This was repeated 30 min after injection and then every hour for a total time course of 5 hours.
Statistical analyses
All data was first tested for a Gaussian distribution using a Shapiro-Wilk test (Graphpad Prism 7 Software). The statistical significance of differences between means was determined by either Student’s t test, non-parametric or parametric analysis of variance (ANOVA) followed by post hoc comparisons (Tukey) using GraphPad Prism 7 Software. All behavioral data were analyzed by non-parametric two-way ANOVA (post hoc: Student-Neuman–Kuels) in FlashCalc (Dr. Michael H. Ossipov, University of Arizona, Tucson, AZ, USA). Differences were considered to be significant if p≤ 0.05. All data were plotted in GraphPad Prism 7. No outlier data were removed.
Results
t-CNRP1 reverts increased CaV2.2 currents in Nf1-edited sensory neurons
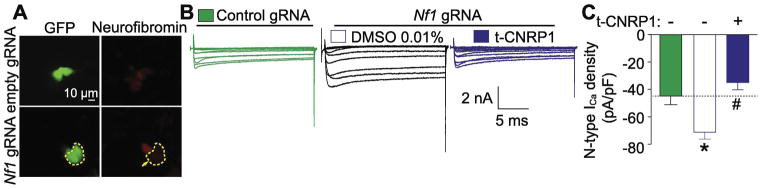
We first generated Nf1 edited DRG sensory neurons (Figure 1A) using a CRISPR/Cas9 expressing plasmid together with a gRNA targeting exon 39 of the rat Nf1 gene. As described previously (Moutal, et al., 2017f), Nf1 gene editing resulted in increased CaV2.2 currents compared to unedited DRG neurons (Figure 1). We then asked whether t-CNRP1, a peptide targeting the CRMP2/neurofibromin interaction, could normalize this upregulated CaV2.2 activity. Sensory neurons were treated overnight 10μM of t-CNRP1 (where t is the trans-acting activator of transcription (TAT) domain of the HIV-1 protein (Schwarze et al., 1999) that facilitates penetration into cells) and then measured Ca2+ currents (Figure 1B). CaV2.2 currents were recorded in the presence of blockers of all other voltage-gated calcium channels in DRGS (see Methods). Under these conditions, t-CNRP1, reversed the increase of Ca2+ currents induced by Nf1 gene editing (Figure 1C) to the level of non-edited DRG neurons (dotted line, Figure 1C). These results suggest a role for the CRMP2 neurofibromin interface in increasing CaV2.2 functions in NF1.
Figure 1. Increased Ca2+ currents in Nf1 edited sensory neurons are normalized by t-CNRP1.
(A) Representative micrograph showing neurofibromin levels in DRG neurons transfected with either pSpCas9(BB)-2A-GFP (control gRNA) or pSpCas9(BB)-2A-GFP-Nf1 gRNA plasmids. GFP fluorescence identifies transfected neurons. In this experiment, neuron without GFP has robust expression of neurofibromin, whereas the adjacent neuron with GFP fluorescence (circled) demonstrates significantly decreased neurofibromin expression (marked by an arrow). Scale bar is 10 μm. (B) Representative family of current traces are illustrated for neurons transfected with control or Nf1 gRNA and treated with 10uM t-CNRP1 or DMSO 0.01%. (C) Peak CaV2.2 current density, at +10 mV, in DRG neurons unedited or transfected by Nf1 gRNA containing plasmid and treated with 10 μM t-CNRP1 or DMSO 0.01%. Line shows peak CaV.2.2 current level in Nf1 unedited DRG neurons. Mean ± s.e.m., asterisks denote statistical significance compared with 0.01% DMSO treated cells (p<0.05, non-parametric one-way ANOVA). Recordings are from small and medium DRG neurons.
t-CNRP1 reverts increased TTX-S Na+ currents in Nf1-edited sensory neurons
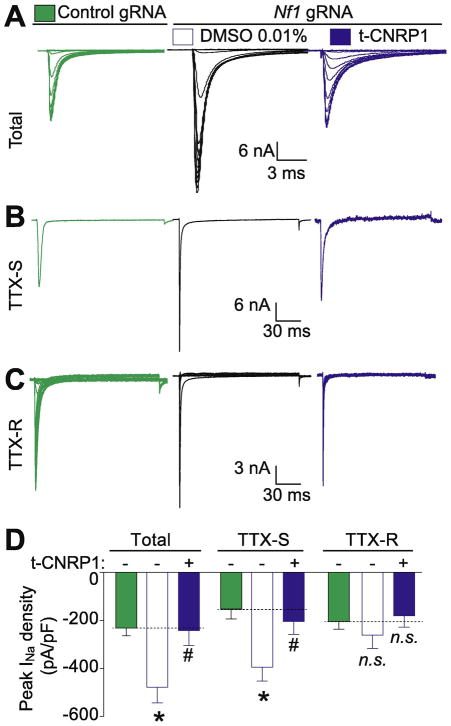
Our recent report demonstrated an increase of total Na+ currents, mostly TTX-S voltage gated Na+ channels in sensory neurons isolated from mice following intrathecal editing with CRISPR/Cas9 with a gRNA targeting exon 39 of Nf1 (Moutal, et al., 2017f), thereby implicating neurofibromin as a possible regulator of Na+ channels. Our earlier work had shown that CRMP2 plays a role in regulating TTX-S current density through NaV1.7 channels (Dustrude, et al., 2016; Dustrude, et al., 2017a; Moutal et al., 2017c). Consequently, here we tested if perturbing the CRMP2 neurofibromin interface could affect Na+ channel dysregulation. Total (Figure 2A), TTX-S (Figure 2B) and TTX-R (Figure 2C) Na+ currents were increased in Nf1 edited sensory neurons compared to unedited neurons. Targeting the CRMP2/neurofibromin interface with t-CNRP1 normalized the increased total Na+ current density to the level of unedited DRG neurons (Figure 2D). This effect was due to t-CNRP1 mediated normalization of TTX-S, but not TTX-R, Na+ currents (Figure 2D). These results show that the CRMP2/neurofibromin interface regulates Na+ current density.
Figure 2. Increased total and TTX-S Na+ currents in Nf1 edited sensory neurons are normalized by t-CNRP1.
Representative family of total (A), tetrodotoxin-sensitive (TTX-S) (B), and TTX-resistant (TTX-R) (C) Na+ current traces are illustrated for Nf1-edited sensory neurons and treated with 10 μM t-CNRP1 or DMSO 0.01% compared to unedited neurons. (D) Peak current density for Total, TTX-S or TTX-R Na+ current density in unedited DRG neurons or transfected by Nf1 gRNA containing plasmid and treated with either 10 μM t-CNRP1 or DMSO 0.01%. Line shows peak current level in DRG neurons transfected with the empty plasmid. Mean ± s.e.m., asterisks denote statistical significance compared with 0.01% DMSO treated cells (p<0.05, non-parametric one-way ANOVA). n.s., not-significant; P>0.05; non-parametric one way ANOVA. Recordings are from small and medium DRG neurons.
The activation V50, the half-inactivated potential for activation of TTX-S currents was −27.2 ± 0.7 mV in control neurons, 30.0 ± 0.3 mV in cells with Nf1 gRNA, and 24.7 ± 2.1 mV in cells with Nf1 gRNA treated with 10 μM t-CNRP1 (Table 1). Although the activation V50 in cells treated with t-CNRP1 represented a statistically significant difference compared to Nf1 edited neurons (n = 11 cells, p < 0.05, One-way ANOVA with Bonferroni’s multiple comparison test), the ~ 6mV shift in hyperpolarization reflected a normalization to the activation V50 of control cells. For inactivation of TTX-S currents, the trend of a depolarization shift in V50 of ~12.6 mV observed in Nf1 gRNA versus control neurons was restored by t-CNRP1 treatment to V50 values observed in control cells (Table 1). For TTX-R currents, the ~ 7 mV shift in hyperpolarization caused by Nf1 editing was negated by t-CNRP1 treatment which brought the activation V50 to levels observed in control cells (p < 0.05, One-way ANOVA with Bonferroni’s multiple comparison test) (Table 1). No significant difference for the inactivation of TTX-R currents was detected (p < 0.05, One-way ANOVA with Bonferroni’s multiple comparison test). This indicates that t-CNRP1 normalizes the biophysical properties of TTX-S and TTX-R currents after Nf1 editing in DRG neurons.
Table 1.
Boltzmann parameters for the activation and inactivation of tetrodotoxin-sensitive (TTX-S) and resistant (TTX-R) INa in control and Nf1 sgRNA-transfected dorsal root ganglion neurons.
| V0.5 (mV) | k | |
|---|---|---|
| TTX-S | ||
| INa activation | ||
| Control (n=9) | −27.2 ± 0.7 | 5.0 ± 0.3 |
| Nf1 gRNA (n=11) | −30.0 ± 0.3 | 4.3 ± 0.3 |
| Nf1 gRNA + t-CNRP1 (n=11) | −24.7 ± 2.1a | 7.0 ± 1.3 |
| INa inactivation | ||
| Control (n=9) | −64.0 ± 3.2 | 7.7 ± 1.5 |
| Nf1 gRNA (n=11) | −51.4 ± 7.0 | 17.2 ± 1.8b |
| Nf1 gRNA + t-CNRP1 (n=11) | −68.3 ± 1.3a | 13.5 ± 0.7 |
| TTX-R | ||
| INa activation | ||
| Control (n=9) | −20.0 ± 0.7 | 7.8 ± 0.7 |
| Nf1 gRNA (n=11) | −26.9 ± 0.7b | 5.8 ± 0.2 |
| Nf1 gRNA + t-CNRP1 (n=11) | −19.9 ± 1.4a | 5.8 ± 0.4 |
| INa inactivation | ||
| Control (n=9) | −30.0 ± 2.4 | 5.6 ± 1.6 |
| Nf1 gRNA (n=11) | −36.3 ± 5.1 | 8.2 ± 2.6 |
| Nf1 gRNA + t-CNRP1 (n=11) | −43.9 ± 7.1 | 15.7 ± 3.5 a |
p < 0.05 vs Nf1 sgRNA (One-way ANOVA with Bonferroni’s multiple comparison test);
p < 0.05 vs control (One-way ANOVA with Bonferroni’s multiple comparison test);
t-CNRP1 reduces CaV2.2 and NaV1.7 surface localization in Nf1-edited sensory neurons
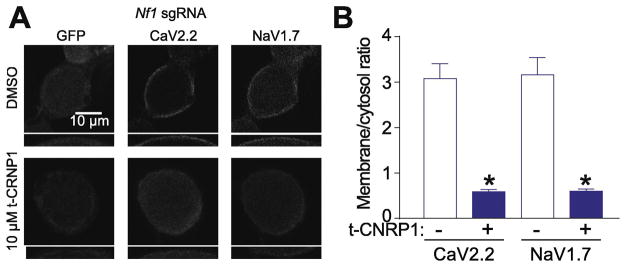
CRMP2 promotes surface expression of CaV2.2 (Brittain, et al., 2011; Brittain, et al., 2009; Brittain et al., 2012; Francois-Moutal, et al., 2015) and NaV1.7 (Dustrude, et al., 2016; Dustrude, et al., 2017a; Dustrude, et al., 2013). Since t-CNRP1 decreased both Ca2+ and Na+ current densities, we hypothesized that treating Nf1-edited sensory neurons with this peptide might reduce the surface expression of CaV2.2 and NaV1.7. After overnight treatment of Nf1-edited sensory neurons with 10μM of t-CNRP1, CaV2.2 and NaV1.7 were immunostained. We used confocal imaging to analyze the localization in close vicinity of the membrane for CaV2.2 and NaV1.7 in Nf1-edited DRG neurons (identified by GFP expression) (Figure 3A). We found that t-CNRP1 decreased the surface localization of both CaV2.2 and NaV1.7 (Figure 3) compared to control (0.01% DMSO) neurons. Thus, the decreased Ca2+ and Na+ current densities induced by t-CNRP1 are related to a decreased surface expression of these channels.
Figure 3. CaV2.2 and NaV1.7 surface localization is decreased by t-CNRP1 in Nf1 edited sensory neurons.
(A) Representative micrographs of CaV2.2 and NaV1.7 localization in Nf1-edited DRG neurons (identified by GFP expression) treated with 10 μM t-CNRP1 or DMSO 0.01% (n=12 per condition). Scale bar is 10μm (B) quantification of the relative membrane localization of CaV2.2 and NaV1.7 (compared to cytosol) in Nf1 edited DRG neurons treated with 10 μM t-CNRP1 or DMSO 0.01% (n=11 per condition). Insets in panels A and B represent 2X magnified versions of the annular regions of staining on the cells.
t-CNRP1 reverts the increased excitability in Nf1-edited sensory neurons
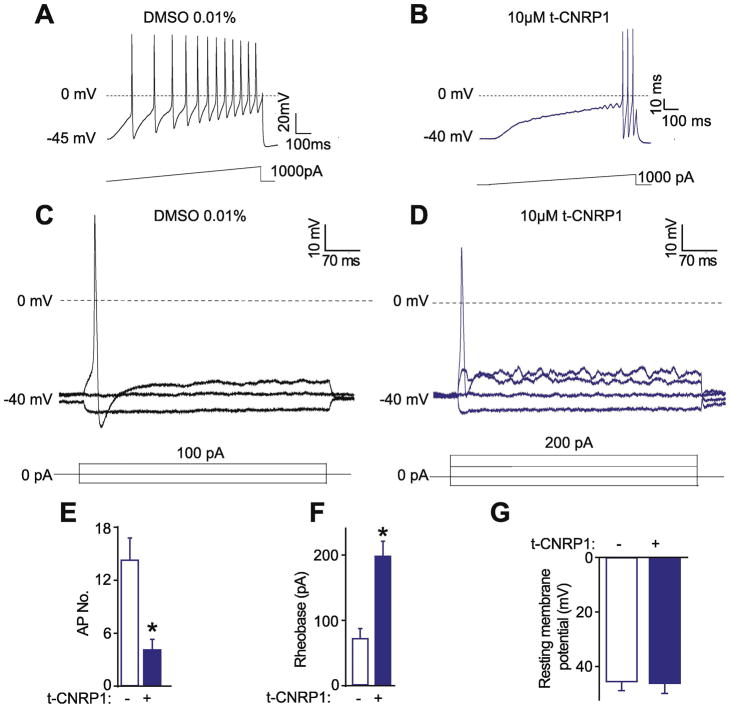
NaV1.7 is a voltage gated Na+ channel participating in the upstroke of action potential firing by sensory neurons (Dib-Hajj et al., 2013). In Nf1-edited DRG neurons, we recently reported that the increase of Na+ currents is related to a decreased rheobase and increased action potential firing (Moutal, et al., 2017f). Thus, we tested if t-CNRP1 could reduce Nf1-edited DRG neuron excitability. Nf1-edited neurons were treated overnight with 10μM of t-CNRP1 or vehicle (0.01% DMSO) and neuronal excitability was measured in ramp and step protocols (Figure 4A–D). t-CNRP1 treatment of Nf1-edited sensory neurons reversed the increased action potential (AP) firing (Figure 4A–D) to the level of non-edited DRG neurons (Figure 4E). The rheobase of t-CNRP1-treated Nf1 edited neurons was more than doubled versus that in vehicle treated cells (0.01% DMSO) (Figure 4F). The resting membrane potential, which is not altered by Nf1-gene editing (Moutal, et al., 2017f), was not changed by t-CNRP1. This data shows the CRMP2 neurofibromin interface is involved in increased excitability manifest as in increase AP and a decrease in rheobase in Nf1-edited DRG sensory neurons.
Figure 4. Hyperexcitability in Nf1 edited sensory neurons is controlled by t-CNRP1.
Representative recordings in response to a step of depolarizing current evoked action potentials (APs) in Nf1-edited sensory neurons treated with (A) DMSO 0.01% or (B) 10 μM t-CNRP1. Representative recordings in response to various steps of depolarizing current to measure rheobase in Nf1-edited sensory neurons treated with (C) DMSO 0.01% or (D) 10 μM t-CNRP1. Rheobase is the current required for eliciting the first AP. Summary of the number of action potentials (E), rheobase (F), and resting membrane potentials (G) recorded from Nf1-edited DRG neurons treated with 10 μM t-CNRP1 or DMSO 0.01%. Recordings are from small and medium DRG neurons.
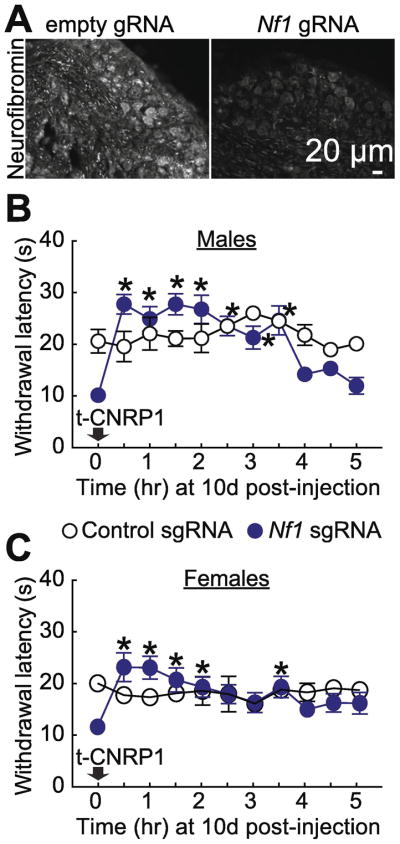
t-CNRP1 reverses Nf1-editing induced thermal hyperalgesia in both male and female rats
In vivo Nf1 gene-editing (Figure 5A) leads to the development of thermal hyperalgesia (Moutal, et al., 2017f), setting up a causal relationship between neurofibromin and pain. Our in vitro data shows that t-CNRP1 treatment of Nf1-edited cells normalizes the increased CaV2.2 and NaV1.7 currents as well as sensory neuronal excitability. Thus, we tested if t-CNRP1 could be beneficial in this new model of NF1 pain. Intraperitoneal injection (30mg/kg) of t-CNRP1 reversed the decreased paw withdrawal latency after 30 min, an effect that lasted for 4 hours in males (Figure 5B) and for 3.5 hours in females (Figure 5C). Although, some early time points demonstrated paw withdrawal latency above the baseline of unedited rats, this effect was not significant. Injecting t-CNRP1 in non-edited rats (control gRNA) did not elicit any analgesia. t-CNRP1 did not change the thermal withdrawal threshold in control animals where Nf1 is intact.
Figure 5. NF1-related thermal hyperalgesia can be reverted by t-CNRP1.
(A) Micrographs of a 10 μm section of adult DRG from animals having received intrathecal injection of either Nf1 gRNA containing or control gRNA plasmid (day 10) immunostained with neurofibromin antibody as indicated. Scale bar: 20 μm. Thermal hyperalgesia (paw withdrawal latency) of rats, 10 days after in vivo transfection with control or Nf1 gRNA containing plasmids. Paw withdrawal latency was followed for 5 hours after intraperitoneal injection of t-CNRP1 (30mg/kg). Paw withdrawal latency threshold was increased by t-CNRP1 injection in Nf1-edited males (B) and females (C) rats (n=6 each). *P < 0.05 versus control gRNA, 2-way ANOVA with a Student–Newman–Keuls post hoc test. Error bars represent mean ± s.e.m.
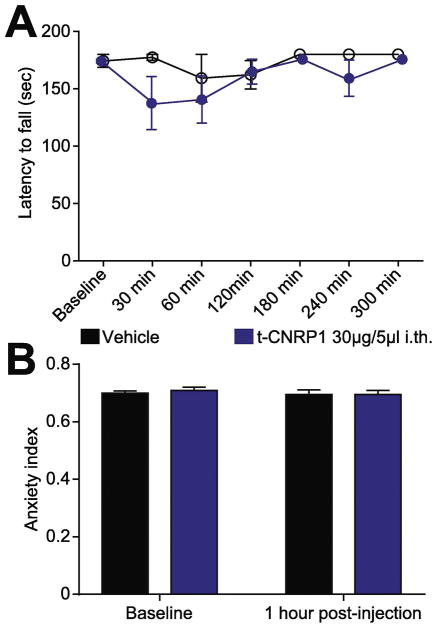
To rule out possible off-target effects of the CNRP1 peptide, we tested whether t-CNRP1 could be involved in motor coordination (rotarod) or anxiety (EPM). To ensure availability of the peptide in the spinal cord, we administered the drug intrathecally in these experiments. Intrathecal injection of t-CNRP1 (30μg/5μl) had no effect on the latency to fall (Figure 6A) or on the anxiety index (Figure 6B). Thus, targeting the CRMP2 neurofibromin interface specifically reduces NF1-related pain. Another inference from the behavioral results is that t-CNRP1 is likely acting on DRG neurons and not on motor neurons.
Figure 6. Comparison of anxiety index following targeted intrathecal editing of Nf1 in vivo.
Spinal administration of t-CNRP1 (30μg/5μl) or vehicle (0.9% saline) was evaluated for motor deficits using the rotarod performance test. No significant difference was measured between t-CNRP1 treated and vehicle treated animals, 2-way ANOVA with a Student–Newman–Keuls post hoc test. (B) The anxiety index, an integrated measure of times and entries into the arms of the elevated plus maze, was not different between vehicle (0.01% DMSO) and t-CNRP1 (30μg/5μl), before and after injection. Values are showed as mean ± s.e.m (n=6 each).
Discussion
The work presented here reveals additional insights into NF1-related pain. We had previously identified CRMP2 expression to be required for NF1-related pain (Moutal, et al., 2017a) and had reported that the CRMP2-neurofibromin interaction was an important mediator of post-surgical, inflammatory and neuropathic nociceptive behaviors [33]. The CRMP2-neurofibromin interaction likely regulates CRMP2 functions by (i) modifying CRMP2’s post-translational state (Moutal, et al., 2017f; Patrakitkomjorn, et al., 2008) and (ii) controlling CRMP2’s interactome [33]. These observations suggested that the loss of protein-protein interactions (PPIs) between CRMP2 and neurofibromin might explain NF1-related pain. We thus tested whether targeting the CRMP2-neurofibromin interface could normalize ion channel dysregulation and nociceptive behaviors in a CRISPR/Cas9–induced in vitro and in vivo model of NF1-related pain.
A CRMP2-derived peptide (CNRP1) was designed based on CRMP2’s interaction domain with neurofibromin. We previously found that this peptide inhibited CRMP2’s interaction with CaV2.2, neurofibromin, and syntaxin 1A. A cell-penetrant version of this peptide (t-CNRP1) inhibited CaV2.2 and NaV1.7 currents in Nf1-edited neurons. CRMP2 regulation of the NaV1.7 TTX-S Na+ channel was dependent on the CRMP2’s phosphorylation and SUMOylation state (Dustrude, et al., 2016; Dustrude et al., 2017b; Dustrude, et al., 2013). The results reported here appear independent of these modifications and identify a novel pathway of NaV1.7 regulation by the CRMP2 – neurofibromin interface.
How does the CRMP2 neurofibromin interaction bestow regulation on NaV1.7?
One possibility is via CRMP2’s interaction with syntaxin 1A. We previously reported that t-CNRP1 inhibits the CRMP2/syntaxin 1A interaction [11]. Syntaxin 1A is part of the SNARE (Soluble N-ethylmaleimide-sensitive-factor Attachment REceptor) protein complex mediating vesicular fusion at the synapse (Fergestad et al., 2001). Syntaxin 1A can interact with synaptotagmin, a protein known to interact with voltage-gated Na+ channels (Sampo et al., 2000) and possibly with NaV1.7 (Kanellopoulos et al., 2017). This CRMP2-Syntaxin 1A-synaptotagmin-NaV1.7 PPI complex might coordinate NaV1.7 membrane localization. By disrupting these interactions, t-CNRP1 could trigger the internalization of the Na+ channel. CRMP2 can also, in its unphosphorylated state, bind to tubulin and promote axonal growth (Fukata, et al., 2002; Gu et al., 2000). CRMP2 phosphorylation by Cdk5 (Uchida et al., 2005)or Fyn (Uchida et al., 2009) results in a decrease of CRMP2 binding to tubulin and inhibits axonal elongation. Since we found NaV1.7 function to be protected by CRMP2 phosphorylation by Cdk5 and inhibited after CRMP2 phosphorylation by Fyn (Dustrude, et al., 2016), the fraction of CRMP2 regulating NaV1.7 is unlikely to interact with tubulin. This rules out a potential function of t-CNRP1 in altering CRMP2 function on axonal growth.
In addition, this PPI complex, identified elsewhere (Dustrude, et al., 2016; Moutal, et al., 2017e), sets up a possible coupling of NaV1.7 activity to neurotransmitter release. Support for this comes from previous observations that t-CNRP1 inhibits the release of the nociceptive calcitonin gene related peptide (CGRP) (Moutal, et al., 2017e), a transmitter that is increased in Nf1-edited rats and Nf1+/− mice (Hingtgen, et al., 2006; Moutal, et al., 2017a). A function for NaV1.7 in CGRP release was suggested by the use of voltage-gated Na+ channel activating ciguatoxins (Touska et al., 2017). These toxins elicited a strong CGRP release, ~40% of which was dependent on NaV1.7 activation (Touska, et al., 2017). Thus, CRMP2’s interface with neurofibromin may recruit CaV2.2, NaV1.7 and the SNARE complex to link the trafficking function of these channels to neuropeptide release in NF1-related pain.
CRMP2 forms a tetramer in solution (Dustrude, et al., 2017a; Francois-Moutal, et al., 2015; Moutal, et al., 2016c; Stenmark et al., 2007; Wang et al., 1997) and thus presents multiple copies of PPI sites available to build potential PPI modules. A closer look at the sequence of CRMP2 reveals clues as to the organization of key binding motifs that may participate in a concerted fashion to link trafficking to transmitter release. Key CRMP2 domains include: (i) residues 460-474 that interact with neurofibromin or syntaxin 1A (Moutal, et al., 2017e); (ii) residue 484-498 (Brittain, et al., 2011), later refined as 484-489 (Moutal et al., 2017d) that mediates its binding to CaV2.2; (iii) residues 466-490 that bind to tubulin (Niwa et al., 2017); and (iv) residues 422-572 that interact with the molecular motor kinesin1 that allows vesicular transport towards the synapse (Kimura et al., 2005). Because of its tetrameric structure, CRMP2 could be a scaffold between all of these proteins and contribute to the assembly of a neurotransmitter release platform. CRMP2 interactions with tubulin and kinesin1 may coordinate the peregrination of CaV2.2 and NaV1.7 to the synapse, possibly explaining their respective increased current densities. Also, CRMP2’s interaction with syntaxin 1A, which is augmented when CRMP2’s interaction with neurofibromin is lost (Moutal, et al., 2017e), could aid in docking of synaptic vesicles directly to these channels. Syntaxin 1A’s interaction with synaptotagmin could, in turn, link CRMP2, CaV2.2 and NaV1.7 together. Such a model would accommodate a potential interplay between these channels. NaV1.7 activation would cause local membrane depolarization, readily opening CaV2.2. The release of the neurotransmitter containing vesicles, directly docked between both channels through CRMP2 and syntaxin 1A, would then be triggered by the influx of Ca2+ ions thus eliciting a nociceptive signal. If true, it follows that an increased formation of such a signaling platform could underlie the hyperalgesia observed upon Nf1 gene editing.
In summary, our findings identify a novel signaling hub, designated here as a neurotransmitter release platform that positions ion channels (i.e., CaV2.2, NaV1.7) adjacent to the release machinery (i.e., syntaxin 1A, synaptotagmin) for rapid nociceptive signal transmission. In this platform, neurofibromin interacts with CRMP2 to prevent recruitment of syntaxin 1A and CaV2.2, thus leading to block of CGRP and a reduction in pain behaviors. Our findings support future pharmacological targeting of the CRMP2/neurofibromin interface for NF1-related pain relief.
Highlights.
CRMP2/neurofibromin interface controls increased voltage-gated Ca2+ channel CaV2.2 currents after Nf1 gene editing
CNRP1 peptide reversed voltage-gated Na+ channel NaV1.7 currents and DRG excitability after Nf1 gene editing
Membrane localization of CaV2.2 and NaV1.7 is controlled by CRMP2/neurofibromin interface
Targeting CRMP2/neurofibromin interface reversed NF1-related nociceptive behavior without motor impairment
Acknowledgments
This work was supported by National Institutes of Health awards (1R01NS098772 from the National Institute of Neurological Disorders and Stroke and 1R01DA042852 from the National Institute on Drug Abuse); a Neurofibromatosis New Investigator Award from the Department of Defense Congressionally Directed Military Medical Research and Development Program (NF1000099); and a Children’s Tumor Foundation NF1 Synodos award (2015-04-009A) to R.K and a General Program of the National Science Foundation of China grant (no.81571231) to S.L. A.M. was supported by a Young Investigator’s Award from the Children’s Tumor Foundation (2015-01-011).
Glossary
- ANOVA
analysis of variance
- AP
action potential
- Cas9
CRISPR-associated protein-9 nuclease
- CaV2.2
N-type voltage gated calcium channel
- CGRP
calcitonin gene related peptide
- CNRP1
CRMP2-neurofibromin regulating peptide 1
- CRISPR
clustered regularly interspaced short palindromic repeats
- CRMP2
Collapsin response mediator protein 2
- DMEM
Dulbecco’s Modified Eagle’s medium
- DRG
Dorsal Root Ganglia
- EPM
Elevated plus maze
- GRNA
guide RNA
- NaV1.7
Na+ voltage-gated sodium channel 1.7
- NF1
Neurofibromatosis type 1
- PPIs
protein-protein interactions
- TTX-S
tetrodotoxin-sensitive
- TTX-R
tetrodotoxin-resistant
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bicudo NP, de Menezes Neto BF, da Silva de Avo LR, Germano CM, Melo DG. Quality of Life in Adults with Neurofibromatosis 1 in Brazil. J Genet Couns. 2016;25:1063–1074. doi: 10.1007/s10897-016-9939-8. [DOI] [PubMed] [Google Scholar]
- Brittain JM, Duarte DB, Wilson SM, Zhu W, Ballard C, Johnson PL, Liu N, Xiong W, et al. Suppression of inflammatory and neuropathic pain by uncoupling CRMP-2 from the presynaptic Ca(2)(+) channel complex. Nature medicine. 2011;17:822–829. doi: 10.1038/nm.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittain JM, Piekarz AD, Wang Y, Kondo T, Cummins TR, Khanna R. An atypical role for collapsin response mediator protein 2 (CRMP-2) in neurotransmitter release via interaction with presynaptic voltage-gated calcium channels. The Journal of biological chemistry. 2009;284:31375–31390. doi: 10.1074/jbc.M109.009951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittain JM, Wang Y, Eruvwetere O, Khanna R. Cdk5-mediated phosphorylation of CRMP-2 enhances its interaction with CaV2.2. FEBS letters. 2012;586:3813–3818. doi: 10.1016/j.febslet.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Chi XX, Schmutzler BS, Brittain JM, Wang Y, Hingtgen CM, Nicol GD, Khanna R. Regulation of N-type voltage-gated calcium channels (Cav2.2) and transmitter release by collapsin response mediator protein-2 (CRMP-2) in sensory neurons. J Cell Sci. 2009;122:4351–4362. doi: 10.1242/jcs.053280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe W, Messinger RB, Leach E, Eckle VS, Obradovic A, Salajegheh R, Jevtovic-Todorovic V, Todorovic SM. TTA-P2 is a potent and selective blocker of T-type calcium channels in rat sensory neurons and a novel antinociceptive agent. MolPharmacol. 2011;80:900–910. doi: 10.1124/mol.111.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford HA, Barton B, Wilson MJ, Berman Y, McKelvey-Martin VJ, Morrison PJ, North KN. The Impact of Neurofibromatosis Type 1 on the Health and Wellbeing of Australian Adults. J Genet Couns. 2015;24:931–944. doi: 10.1007/s10897-015-9829-5. [DOI] [PubMed] [Google Scholar]
- Creange A, Zeller J, Rostaing-Rigattieri S, Brugieres P, Degos JD, Revuz J, Wolkenstein P. Neurological complications of neurofibromatosis type 1 in adulthood. Brain. 1999;122:473–481. doi: 10.1093/brain/122.3.473. [DOI] [PubMed] [Google Scholar]
- Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The Na(V)1.7 sodium channel: from molecule to man. Nature reviews Neuroscience. 2013;14:49–62. doi: 10.1038/nrn3404. [DOI] [PubMed] [Google Scholar]
- Drouet A, Wolkenstein P, Lefaucheur JP, Pinson S, Combemale P, Gherardi RK, Brugieres P, Salama J, et al. Neurofibromatosis 1-associated neuropathies: a reappraisal. Brain: a journal of neurology. 2004;127:1993–2009. doi: 10.1093/brain/awh234. [DOI] [PubMed] [Google Scholar]
- Duan JH, Hodgdon KE, Hingtgen CM, Nicol GD. N-type calcium current, Cav2.2, is enhanced in small-diameter sensory neurons isolated from Nf1+/− mice. Neuroscience. 2014;270:192–202. doi: 10.1016/j.neuroscience.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustrude ET, Moutal A, Yang X, Wang Y, Khanna M, Khanna R. Hierarchical CRMP2 posttranslational modifications control NaV1.7 function. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E8443–E8452. doi: 10.1073/pnas.1610531113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustrude ET, Perez-Miller S, Francois-Moutal L, Moutal A, Khanna M, Khanna R. A single structurally conserved SUMOylation site in CRMP2 controls NaV1.7 function. Channels (Austin) 2017a:1–13. doi: 10.1080/19336950.2017.1299838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustrude ET, Perez-Miller S, Francois-Moutal L, Moutal A, Khanna M, Khanna R. A single structurally conserved SUMOylation site in CRMP2 controls NaV1.7 function. Channels. 2017b;11:316–328. doi: 10.1080/19336950.2017.1299838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustrude ET, Wilson SM, Ju W, Xiao Y, Khanna R. CRMP2 protein SUMOylation modulates NaV1.7 channel trafficking. The Journal of biological chemistry. 2013;288:24316–24331. doi: 10.1074/jbc.M113.474924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito T, Piluso G, Saracino D, Uccello R, Schettino C, Dato C, Capaldo G, Giugliano T, et al. A novel diagnostic method to detect truncated neurofibromin in neurofibromatosis 1. Journal of neurochemistry. 2015;135:1123–1128. doi: 10.1111/jnc.13396. [DOI] [PubMed] [Google Scholar]
- Fergestad T, Wu MN, Schulze KL, Lloyd TE, Bellen HJ, Broadie K. Targeted mutations in the syntaxin H3 domain specifically disrupt SNARE complex function in synaptic transmission. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2001;21:9142–9150. doi: 10.1523/JNEUROSCI.21-23-09142.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferner RE. Neurofibromatosis 1 and neurofibromatosis 2: a twenty first century perspective. Lancet Neurol. 2007;6:340–351. doi: 10.1016/S1474-4422(07)70075-3. [DOI] [PubMed] [Google Scholar]
- Ferner RE, Thomas M, Mercer G, Williams V, Leschziner GD, Afridi SK, Golding JF. Evaluation of quality of life in adults with neurofibromatosis 1 (NF1) using the Impact of NF1 on Quality Of Life (INF1-QOL) questionnaire. Health and quality of life outcomes. 2017;15:34. doi: 10.1186/s12955-017-0607-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois-Moutal L, Wang Y, Moutal A, Cottier KE, Melemedjian OK, Yang X, Wang Y, Ju W, et al. A membrane-delimited N-myristoylated CRMP2 peptide aptamer inhibits CaV2.2 trafficking and reverses inflammatory and postoperative pain behaviors. Pain. 2015;156:1247–1264. doi: 10.1097/j.pain.0000000000000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, Menager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, et al. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nature cell biology. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- Gu Y, Ihara Y. Evidence that collapsin response mediator protein-2 is involved in the dynamics of microtubules. JBiolChem. 2000;275:17917–17920. doi: 10.1074/jbc.C000179200. [DOI] [PubMed] [Google Scholar]
- Hargreaves K, Dubner R, Brown F, Flores C, Joris J. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain. 1988;32:77–88. doi: 10.1016/0304-3959(88)90026-7. [DOI] [PubMed] [Google Scholar]
- Hingtgen CM, Roy SL, Clapp DW. Stimulus-evoked release of neuropeptides is enhanced in sensory neurons from mice with a heterozygous mutation of the Nf1 gene. Neuroscience. 2006;137:637–645. doi: 10.1016/j.neuroscience.2005.09.030. [DOI] [PubMed] [Google Scholar]
- Huynh TN, Krigbaum AM, Hanna JJ, Conrad CD. Sex differences and phase of light cycle modify chronic stress effects on anxiety and depressive-like behavior. Behavioural brain research. 2011;222:212–222. doi: 10.1016/j.bbr.2011.03.038. [DOI] [PubMed] [Google Scholar]
- Ju W, Li Q, Allette YM, Ripsch MS, White FA, Khanna R. Suppression of pain-related behavior in two distinct rodent models of peripheral neuropathy by a homopolyarginine-conjugated CRMP2 peptide. Journal of neurochemistry. 2012;124:869–879. doi: 10.1111/jnc.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata T, Subleski M, Hara Y, Yuhki N, Kung H, Copeland NG, Jenkins NA, Yoshimura T, et al. Isolation and characterization of a bovine neural specific protein (CRMP-2) cDNA homologous to unc-33, a C. elegans gene implicated in axonal outgrowth and guidance. Brain research Molecular brain research. 1998;54:219–236. doi: 10.1016/s0169-328x(97)00332-x. [DOI] [PubMed] [Google Scholar]
- Kanellopoulos AH, Koenig J, Huang H, Pyrski M, Millet Q, Lolignier S, Morohashi T, Gossage SJ, et al. Mapping protein interactions of sodium channel NaV1.7 using epitope-tagged gene targeted mice. BioRxiv. 2017 doi: 10.15252/embj.201796692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Watanabe H, Iwamatsu A, Kaibuchi K. Tubulin and CRMP-2 complex is transported via Kinesin-1. Journal of neurochemistry. 2005;93:1371–1382. doi: 10.1111/j.1471-4159.2005.03063.x. [DOI] [PubMed] [Google Scholar]
- Kodra Y, Giustini S, Divona L, Porciello R, Calvieri S, Wolkenstein P, Taruscio D. Health-related quality of life in patients with neurofibromatosis type 1. A survey of 129 Italian patients. Dermatology. 2009;218:215–220. doi: 10.1159/000187594. [DOI] [PubMed] [Google Scholar]
- Lin YL, Hsueh YP. Neurofibromin interacts with CRMP-2 and CRMP-4 in rat brain. Biochemical and biophysical research communications. 2008;369:747–752. doi: 10.1016/j.bbrc.2008.02.095. [DOI] [PubMed] [Google Scholar]
- Mintz IM, Venema VJ, Swiderek KM, Lee TD, Bean BP, Adams ME. P-type calcium channels blocked by the spider toxin omega-Aga-IVA. Nature. 1992;355:827–829. doi: 10.1038/355827a0. [DOI] [PubMed] [Google Scholar]
- Moutal A, Cai S, Luo S, Voisin R, Khanna R. CRMP2 is necessary for Neurofibromatosis type 1 related pain. Channels (Austin) 2017a doi: 10.1080/19336950.2017.1370524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutal A, Chew LA, Yang X, Wang Y, Yeon SK, Telemi E, Meroueh S, Park KD, et al. (S)-lacosamide inhibition of CRMP2 phosphorylation reduces postoperative and neuropathic pain behaviors through distinct classes of sensory neurons identified by constellation pharmacology. Pain. 2016a;157:1448–1463. doi: 10.1097/j.pain.0000000000000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutal A, Dustrude ET, Khanna R. Sensitization of Ion Channels Contributes to Central and Peripheral Dysfunction in Neurofibromatosis Type 1. Mol Neurobiol. 2017b;54:3342–3349. doi: 10.1007/s12035-016-9907-1. [DOI] [PubMed] [Google Scholar]
- Moutal A, Dustrude ET, Largent-Milnes TM, Vanderah TW, Khanna M, Khanna R. Blocking CRMP2 SUMOylation reverses neuropathic pain. Molecular psychiatry. 2017c doi: 10.1038/mp.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutal A, Eyde N, Telemi E, Park KD, Xie JY, Dodick DW, Porreca F, Khanna R. Efficacy of (S)-Lacosamide in preclinical models of cephalic pain. Pain Rep. 2016b:1. doi: 10.1097/PR9.0000000000000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutal A, Francois-Moutal L, Perez-Miller S, Cottier K, Chew LA, Yeon SK, Dai J, Park KD, et al. (S)-Lacosamide Binding to Collapsin Response Mediator Protein 2 (CRMP2) Regulates CaV2.2 Activity by Subverting Its Phosphorylation by Cdk5. Molecular neurobiology. 2016c;53:1959–1976. doi: 10.1007/s12035-015-9141-2. [DOI] [PubMed] [Google Scholar]
- Moutal A, Li W, Wang Y, Ju W, Luo S, Cai S, Francois-Moutal L, Perez-Miller S, et al. Homology-guided mutational analysis reveals the functional requirements for antinociceptive specificity of collapsin response mediator protein 2-derived peptides. British journal of pharmacology. 2017d doi: 10.1111/bph.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutal A, Wang Y, Yang X, Ji Y, Luo S, Dorame A, Bellampalli SS, Chew LA, et al. Dissecting the role of the CRMP2-neurofibromin complex on pain behaviors. Pain. 2017e doi: 10.1097/j.pain.0000000000001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutal A, Yang X, Li W, Gilbraith KB, Luo S, Cai S, Francois-Moutal L, Chew LA, et al. CRISPR/Cas9 editing of Nf1 gene identifies CRMP2 as a therapeutic target in neurofibromatosis type 1 (NF1)-related pain that is reversed by (S)-Lacosamide. Pain. 2017f doi: 10.1097/j.pain.0000000000001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb R, Szoke B, Palma A, Wang G, Chen X, Hopkins W, Cong R, Miller J, et al. Selective peptide antagonist of the class E calcium channel from the venom of the tarantula Hysterocrates gigas. Biochemistry. 1998;37:15353–15362. doi: 10.1021/bi981255g. [DOI] [PubMed] [Google Scholar]
- Niwa S, Nakamura F, Tomabechi Y, Aoki M, Shigematsu H, Matsumoto T, Yamagata A, Fukai S, et al. Structural basis for CRMP2-induced axonal microtubule formation. Sci Rep. 2017;7:10681. doi: 10.1038/s41598-017-11031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrakitkomjorn S, Kobayashi D, Morikawa T, Wilson MM, Tsubota N, Irie A, Ozawa T, Aoki M, et al. Neurofibromatosis type 1 (NF1) tumor suppressor, neurofibromin, regulates the neuronal differentiation of PC12 cells via its associating protein, CRMP-2. The Journal of biological chemistry. 2008;283:9399–9413. doi: 10.1074/jbc.M708206200. [DOI] [PubMed] [Google Scholar]
- Piekarz AD, Due MR, Khanna M, Wang B, Ripsch MS, Wang R, Meroueh SO, Vasko MR, et al. CRMP-2 peptide mediated decrease of high and low voltage-activated calcium channels, attenuation of nociceptor excitability, and anti-nociception in a model of AIDS therapy-induced painful peripheral neuropathy. Molecular pain. 2012;8:54. doi: 10.1186/1744-8069-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riklin E, Talaei-Kheoi M, Merker VL, Sheridan MR, Jordan JT, Plotkin SR, Vranceanu AM. First report of factors associated with satisfaction in patients with neurofibromatosis. American journal of medical genetics Part A. 2017;173:671–677. doi: 10.1002/ajmg.a.38079. [DOI] [PubMed] [Google Scholar]
- Roy ML, Narahashi T. Differential properties of tetrodotoxin-sensitive and tetrodotoxin-resistant sodium channels in rat dorsal root ganglion neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1992;12:2104–2111. doi: 10.1523/JNEUROSCI.12-06-02104.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampo B, Tricaud N, Leveque C, Seagar M, Couraud F, Dargent B. Direct interaction between synaptotagmin and the intracellular loop I–II of neuronal voltage-sensitive sodium channels. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3666–3671. doi: 10.1073/pnas.97.7.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- Scroggs RS, Fox AP. Calcium current variation between acutely isolated adult rat dorsal root ganglion neurons of different size. J Physiol. 1992;445:639–658. doi: 10.1113/jphysiol.1992.sp018944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark P, Ogg D, Flodin S, Flores A, Kotenyova T, Nyman T, Nordlund P, Kursula P. The structure of human collapsin response mediator protein 2, a regulator of axonal growth. JNeurochem. 2007;101:906–917. doi: 10.1111/j.1471-4159.2006.04401.x. [DOI] [PubMed] [Google Scholar]
- Touska F, Sattler S, Malsch P, Lewis RJ, Reeh PW, Zimmermann K. Ciguatoxins Evoke Potent CGRP Release by Activation of Voltage-Gated Sodium Channel Subtypes NaV1.9, NaV1.7 and NaV1.1. Mar Drugs. 2017:15. doi: 10.3390/md15090269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Ohshima T, Sasaki Y, Suzuki H, Yanai S, Yamashita N, Nakamura F, Takei K, et al. Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3beta phosphorylation of CRMP2: implication of common phosphorylating mechanism underlying axon guidance and Alzheimer’s disease. Genes to cells: devoted to molecular & cellular mechanisms. 2005;10:165–179. doi: 10.1111/j.1365-2443.2005.00827.x. [DOI] [PubMed] [Google Scholar]
- Uchida Y, Ohshima T, Yamashita N, Ogawara M, Sasaki Y, Nakamura F, Goshima Y. Semaphorin3A signaling mediated by Fyn-dependent tyrosine phosphorylation of collapsin response mediator protein 2 at tyrosine 32. The Journal of biological chemistry. 2009;284:27393–27401. doi: 10.1074/jbc.M109.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Strittmatter SM. Brain CRMP forms heterotetramers similar to liver dihydropyrimidinase. JNeurochem. 1997;69:2261–2269. doi: 10.1046/j.1471-4159.1997.69062261.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Brittain JM, Wilson SM, Hingtgen CM, Khanna R. Altered calcium currents and axonal growth in Nf1 haploinsufficient mice. Translational neuroscience. 2010a;1:106–114. doi: 10.2478/v10134-010-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Duan JH, Hingtgen CM, Nicol GD. Augmented sodium currents contribute to enhanced excitability of small diameter capsaicin-sensitive Nf1+/− mouse sensory neurons. J Neurophysiol. 2010b doi: 10.1152/jn.01010.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nicol GD, Clapp DW, Hingtgen CM. Sensory neurons from Nf1 haploinsufficient mice exhibit increased excitability. JNeurophysiol. 2005;94:3670–3676. doi: 10.1152/jn.00489.2005. [DOI] [PubMed] [Google Scholar]
- Wolkenstein P, Zeller J, Revuz J, Ecosse E, Leplege A. Quality-of-life impairment in neurofibromatosis type 1: a cross-sectional study of 128 cases. Arch Dermatol. 2001;137:1421–1425. doi: 10.1001/archderm.137.11.1421. [DOI] [PubMed] [Google Scholar]
- Wolters PL, Burns KM, Martin S, Baldwin A, Dombi E, Toledo-Tamula MA, Dudley WN, Gillespie A, et al. Pain interference in youth with neurofibromatosis type 1 and plexiform neurofibromas and relation to disease severity, social-emotional functioning, and quality of life. American journal of medical genetics Part A. 2015;167A:2103–2113. doi: 10.1002/ajmg.a.37123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaksh TL, Rudy TA. Chronic catheterization of the spinal subarachnoid space. Physiology & behavior. 1976;17:1031–1036. doi: 10.1016/0031-9384(76)90029-9. [DOI] [PubMed] [Google Scholar]