Abstract
The retinoid visual cycle is an ocular retinoid metabolism specifically dedicated to support vertebrate vision. The visual cycle serves not only to generate light-sensitive visual chromophore 11-cis-retinal, but also to clear toxic byproducts of normal visual cycle (i.e. all-trans-retinal and its condensation products) from the retina, ensuring both the visual function and the retinal health. Unfortunately, various conditions including genetic predisposition, environment and aging may attribute to a functional decline of the all-trans-retinal clearance. To combat all-trans-retinal mediated retinal degeneration, we sought to slow down the retinoid influx from the RPE by inhibiting the visual cycle with a small molecule. The present study describes identification of CU239, a novel non-retinoid inhibitor of RPE65, a key enzyme in the visual cycle. Our data demonstrated that CU239 selectively inhibited isomerase activity of RPE65, with IC50 of 6 μM. Further, our results indicated that CU239 inhibited RPE65 via competition with its substrate all-trans-retinyl ester. Mice with systemic injection of CU239 exhibited delayed chromophore regeneration after light bleach, and conferred a partial protection of the retina against injury from high intensity light. Taken together, CU239 is a potent visual cycle modulator and may have a therapeutic potential for retinal degeneration.
Keywords: Age-related macular degeneration, enzyme inhibitor, retinal degeneration, RPE65, Stargardt’s disease, visual cycle
1. Introduction
Vertebrate visual function relies on vitamin A metabolism referred to as the visual cycle (Fig. 1) (1, 2). Various enzymes and retinoid-binding proteins located in the retinal pigment epithelium (RPE), inter-photoreceptor matrix (IPM), and photoreceptors participate in a biochemical cascade of the visual cycle, to generate and transport light sensitive vitamin A derivative namely 11-cis-retinaldehyde (11-cis-RAL).
Fig. 1. A schematic representation of the visual cycle and the target of CU239.
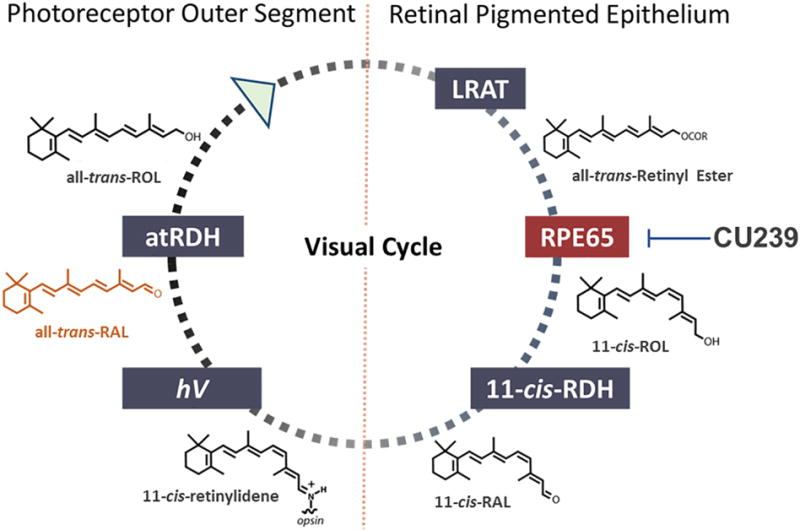
All-trans-ROL undergoes a cascade of enzymatic reactions in the retinal pigmented epithelium (RPE), producing a light sensitive chromophore 11-cis-RAL. 11-cis-RAL binds with opsin in photoreceptor outer segment that allows photon capture. All-trans-RAL, a photo-isomerized product of 11-cis-RAL, has disease implication when accumulated in excessive amounts. CU239 selectively inhibits RPE65 and ameliorates the retinal damage induced by accelerated accumulation of all-trans-RAL in LIRD model.
In order to trigger the phototransduction pathway, 11-cis-RAL, the chromophore of visual pigments, must first form a complex with opsin that resides in photoreceptor outer segment. Upon light activation, 11-cis-RAL rapidly photo-isomerizes to all-trans-retinal (all-trans-RAL), induces a conformational change of opsin, and subsequently activates G-protein transducin, triggering the phototransduction cascade. The generated all-trans-RAL ultimately dissociates from opsin. All-trans-RAL must be isomerized back to 11-cis-RAL through the visual cycle to serve as chromophore of visual pigments again.
Following its dissociation from opsin, all-trans-RAL is transported out of the lumen of photoreceptor disk membranes via ATP-cassette transporter 4 (ABCA4)(3). Subsequently, all-trans-retinol dehydrogenases (atRDH) reduce all-trans-RAL into all-trans-retinol (all-trans-ROL), and interphotoreceptor retinoid-binding protein (IRBP) facilitates its transportation back to the RPE (4, 5). In the RPE, all-trans-ROL is esterified by lecithin retinol acyltransferase (LRAT) to form all-trans-retinyl ester, the substrate of retinol isomerase RPE65. RPE65 converts all-trans-retinyl ester to 11-cis-ROL (6–8). Subsequently, 11-cis-ROL dehydrogenases (11-cis-RDH) oxidize it to 11-cis-RAL, which is then transported back to photoreceptors and recombines with opsin to form visual pigments.
Proper visual cycle function is the biochemical foundation for vertebrate vision and one of many pillars of retinal health. For example, the visual cycle efficiently clears all-trans-RAL from photoreceptor outer segment not only to restore visual sensitivity, but also to prevent all-trans-RAL from accumulating to a cytotoxic level (9, 10). An abundant amount of studies demonstrates that impaired clearance of all-trans-RAL can attribute to increased susceptibility to retinal degeneration and loss of retinal function in both humans and rodents (11–14), leading to various retinal dystrophies such as Stargardt’s disease (STGD) and AMD. In the case of STGD, excessive accumulation of lipofuscin in the retina seems to be the sole causative factor in the disease pathogenesis. Assuming that lipofuscin bisretinoids (exemplified by bisretinoid N-retinylidene-N-retinylethanolamine, A2E) mediate lipofuscin toxicity in the AMD and STGD retina, it has been hypothesized that pharmacological inhibition of bisretinoid formation in the retina may provide a means to delay or suppress retinal degenerative processes in dry AMD and STGD. Biosynthesis of lipofuscin bisretinoids occurs in the retina in a non-enzymatic manner from visual cycle retinoids such as all-trans-RAL (15) and, potentially, 11-cis-RAL (16). Since bisretinoids are formed from visual cycle retinoids as byproducts of the properly functioning visual cycle, partial pharmacological inhibition of the visual cycle was suggested as a treatment strategy for dry AMD and STGD. A critical step in the visual cycle is the conversion of all-trans-retinyl ester to 11-cis-ROL by RPE65. The RPE65 reaction is rate-limiting in visual cycle function (17), thus making it an important drug target for partial inhibition of the visual cycle. In light of this, visual cycle inhibitors have been an active area of research over the past decade (18–23). Most of the well-characterized RPE65 inhibitors are retinoids, which generally predict broad specificity and undesired off-target activities in vivo (24, 25). The most extensively studied non-retinoid RPE65 antagonist, emixustat, has significant pharmacokinetic liabilities, such as fast metabolism, high clearance and signs of retinal toxicity, which may complicate its clinical development (26, 27). Therefore, identification and evaluation of new classes of non-retinoid RPE65 inhibitors is of high importance. Here we demonstrate that our novel non-retinoid compound CU239 is able to specifically target and inhibit RPE65, both in vitro and in vivo. In doing so, CU239 confers a protective effect on the retina in light-induced retinal damage model, a commonly used model for retinal degeneration.
2. Materials and methods
2.1. Animal Care
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Oklahoma Health Sciences Center, and performed following the guidelines of the Association for Research in Vision and Ophthalmology (ARVO) statement for the “Use of Animals in Ophthalmic and Vision Research”. BALB/cJ mice which were homozygous for the Rpe65-Leu450 polymorphism were used for all experiments. The animals were maintained in standard housing with 12 h light/dark cycles and food and water ad libitum.
2.2. In silico screening of potential RPE65 inhibitors
Using the available crystallographic model of bovine RPE65 (PDB ID: 3KVC) (28), we generated a docking model of its ligand-binding pocket and conducted the in silico screen of 350,000 compounds from Life Chemicals Stock HTS Collection. The generation of the model included: building of the full protein structure by Blast and Modeller 9v7 (29, 30), and modeling of binding position of natural substrate (retinyl palmitate) in the active-site of RPE65 with genetic algorithm (GA) implemented in Gold software (31).
To simulate natural conditions, we preserved the metal ion bounded to histidine residues in the active site. Annealing parameters for H-bonds and vdW interactions were set to default values (3.0/6.0). Flexible search for ligand and protein side chains was used. Number of GA runs was 100. GoldScore scoring function was applied. Rescoring was performed within ASP function (astenic static potential derived from experimental data). Binding site was determined according to a list of residues mentioned in literature.
At the next step, a Unity query was created from the amino acids that were identified in binding site as critical for ligand interaction. Unity query included: 10 H-bond donor and accept features, 4 hydrophobic features and positive center feature (iron cofactor). Partial match constraints were as following: at least one H-bond donor, acceptor feature and 2 hydrophobic features must be matched.
Docking was performed with Unity Flex (SYBYL-X 1.1, Tripos, Inc). 5 search conformations for each ligand were set. Compounds with the highest query fit (QFIT) score were characterized experimentally. Docking experiments were conducted at Life Chemicals, Inc.
2.3. In vitro retinol isomerase assays
Compounds CU239, CU262, CU270 and CU223 were purchased from Life Chemicals (Niagara-on-the-Lake, Canada). To confirm the identity and purity of CU239, the compound was resynthesized following the procedure described in Supplementary Materials. All the experiments thereafter were performed using resynthesized CU239. All-trans-[11,12-3H]-retinol (1 mCi/mL, 50 Ci/mmol, Perkin Elmer, Waltman, MA, 2 μL), in N, N-dimethyl formamide (DMF) was used as the substrate. For each reaction, 20 μg of proteins from bovine RPE microsomes was added into 200 μL of reaction buffer (10 mM BTP, pH 8.0, 100 mM NaCl) containing 0.2 μM all-trans-ROL, 0.5% BSA, and 25 μM CRALBP. Alternatively, unlabeled all-trans-retinyl palmitate containing liposomes (250 μM lipids, 3.3 μM all-trans-retinyl palmitate) and 25 μg of purified recombinant chicken RPE65 were used as a substrate and enzyme in the presence or absence of the CU239 inhibitor. After 2 h of incubation at 37°C in the dark, the generated retinoids were extracted with 300 μL of methanol and 300 μL of hexane and subjected to HPLC as described previously (32). Each form of retinoids was identified based on comparison to retention time of known retinoid standards. The inhibition constant for CU239 was calculated from the following equation: Ki = [I]/(Kmi/Km −1) where [I] – concentration of inhibitor, Km – Michaelis constant in the absence of inhibitor, Kmi – Michaelis constant in the presence of inhibitor (33).
2.4. RPE65 fluorescence binding assay
We performed binding assay as previously described (34). In brief, we utilized the intrinsic tryptophan fluorescence of RPE65 and its quenching behavior as the binding occurs. Excitation was at 278 nm and emission at 340 nm. RPE65 (0.1 μM) in PBS (with 0.1 % CHAPS, pH7.4) was incubated with varying concentrations of CU239 for 20 minutes at 25°C. All measurements were performed using quartz cuvettes on PC-1 spectrofluorometer (path length = 1.0 cm; excitation and emission slit widths = 2 mm). The binding constant (Kd) and the number of binding sites were calculated by nonlinear regression fitting of the experimentally derived α values using the equation (35). Pα = Rα/n (1−α) − Kd/n where P = total protein concentration, α = (Fmax − F)/(Fmax − F0), n = number of independent binding sites, R = total CU239 concentration at each addition of ligand, Kd = dissociation constant, Fmax = fluorescence intensity at saturation, and F0 = initial fluorescence intensity.
2.5. Light-induced retinal damage (LIRD)
Mice were dark-adapted overnight with food and water ad libitum. On the following day, mice were intraperitoneally injected with freshly prepared CU239 or vehicle (Solutol HS 15 (BASF Corp, Florham Park, NJ, USA) and dimethyl sulfoxide (DMSO) dissolved in sterile saline (0.9% NaCl)). One hour after the systemic administration of CU239 or vehicle, mice were placed in a light box illuminated with white fluorescent tube lights (8,000 lux for 3 hours). The mice were returned to regular housing for 5 days. The retinal damage assessments were performed 5 days following the LIRD.
2.6. Histology
The superior side of the cornea was demarcated with green tattoo dye, and the eyes were carefully enucleated. The eyes were immersed in Davidson’s fixative (2% of 37-40% formaldehyde, 35% ethanol, 10% glacial acetic acid, 53% of water) for 24 hours, and kept in 70% ethanol until embedded in paraffin. Sagittal sections along the superior-inferior retinal axis were cut, and the slides were deparaffinized prior to hematoxylin and eosin staining. Light microscopy was performed with Olympus Provis Ax-70 microscope, and the acquired images were analyzed with Image J software (NIH, Bethesda).
2.7. HPLC retinoid profile assay
Dark-adapted mice were sacrificed, and their eyes enucleated under dim red light. The whole eyes were homogenized with a glass grinder in lysis buffer [10 mM NH2OH, 50% ethanol, 50% 2-(N-morpholino) ethanesulfonic acid, pH 6.5], and retinoids were extracted with hexane. Solvent was evaporated under argon gas, and dried retinoid samples were resuspended in 200 μl of HPLC mobile phase (11.2% ethyl acetate, 2.0% dioxane, 1.4% octanol, 85.4% hexane) and injected into HPLC (515 HPLC pump; Waters Corp., Milford, MA) with a normal phase Lichrosphere SI-60 (Alltech, Deerfield, IL) 5 μm column and isocratic mobile phase (1 ml/min).
2.8. ERG recording
Mice were dark-adapted overnight prior to ERG recording. Mice were anesthetized by an intraperitoneal injection of 2 μl/g body weight of 40 mg/ml ketamine and 3 mg/ml xylazine diluted with saline. Pupils were dilated with 1% cyclopentolate hydrochloride ophthalmic solution (Cyclogyl) (Sandoz Inc, Princeton, NJ) and 10% phenylephrine-HCl. Hypromellose ophthalmic demulcent solution (Goniovisc; 2.5%) (HUB pharmaceuticals LLC, Rancho Cucamonga, CA) was applied to each cornea, followed by the placement of gold wire electrodes. Stainless steel electrodes were placed into the right cheek and the tail to serve as a reference and the ground, respectively. Dark-adaptation recovery protocol was run using an Espion E3 system with a Ganzfeld ColorDome system (Diagnosys LLC, Lowell, MA). The rod photoreceptor function was measured by single flash stimuli (−2.4 to 1.6 log cd·s/m2), and the cone photoreceptor function was measured by 3 and 10 cd·s/m2 with a background light of 10 cd/m2. (Vehicle: n=8 and CU239: n=8).
2.9. Statistical Analyses
GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA) was used for statistical analyses. Paired Student’s t-test was performed to examine statistical significance (expressed as mean ± SEM or SD).
3. Results
3.1. Screening and identification of potential RPE65 inhibitors
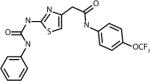
Docking of bovine RPE65 (PDB ID: 3KVC) with 350,000 structures comprising the Life Chemicals HTS collection was performed using the Unity Flex module in SYBYL×1.1 software (Tripos, Inc.) at multiple search conformations per compound (28). The QFIT score was used to rank the compounds, which reflects how close the ligand atoms match the query target coordinates within the range of a spatial constraint tolerance. A total of 65 compounds with the highest QFIT scores and significant diversity were selected for the experimental verification (i.e. in vitro isomerase assay). Significant diversity of 65 hits did not allow for identification of common structural elements among the primary hits. Although a select few of the identified hits do share structural motifs at their cores (i.e. purines, oxalamides and piperazines), these compounds have no commonality as the appendages they bear are significantly diverse. Structure of compounds selected for experimental verification is shown in Supplementary Material, along with compound docking scores. Out of 65 compounds representing primary in silico hits, only 4 analogs shown in the Table 1 were experimentally confirmed to act as isomerase inhibitor as described below for CU239. The four compounds with proven inhibitory activity had generally high QFIT scores (range: 58.7-70.6) indicating a good alignment between pharmacophore model and compound conformer. The docking score of four verified compounds was not significantly different from the scores of many other primary hits as all 65 compounds were selected for the follow-up characterization based on their high QFIT values. CU239, a compound shown to be most active, was re-synthesized as described in Supplementary Material to confirm its identity and purity.
Table 1.
| CUID | Structure | IC50(μM) |
|---|---|---|
| 239 |
|
5 |
| 262 |

|
10 |
| 270 |
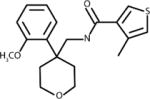
|
12 |
| 223 |
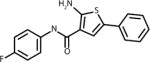
|
70 |
3.2. CU239 inhibits isomerase activity in vitro
To test whether CU239 is able to inhibit RPE65 isomerase activity, we used bovine RPE microsomes and performed in vitro isomerase assay with or without CU239. Bovine RPE microsomal proteins (20 μg) were incubated with 0.2 μM of all-trans-[3H] ROL at 37°C for two hours, and the experimental groups contained varying amounts of CU239 in their respective reaction mixtures. The generated retinoids were extracted and analyzed by HPLC retinoid profile analysis (Fig. 2A–C). As shown in Figure 2B, levels of 11-cis-ROL are markedly reduced when 25 μM CU239 was added to the reaction when compared to solvent control, suggesting an inhibitory effect of CU239 on isomerase activity in vitro. The production of retinyl ester did not decrease, suggesting that LRAT was not inhibited by CU239. In addition, CU239 exhibited a concentration-dependent inhibition of isomerase activity (Fig. 2C) with apparent IC50 of 6 μM. Even though the solubility of CU239 in aqueous solutions is limited, it allowed evaluation of the compound inhibitory activity in concentrations up to 50 μM (Fig. 2C). Analysis of resynthesized CU239 confirmed its activity as an inhibitor of isomerohydrolase activity mediated by RPE65 (data not shown).
Fig. 2. Inhibition of the isomerase activity by CU239.
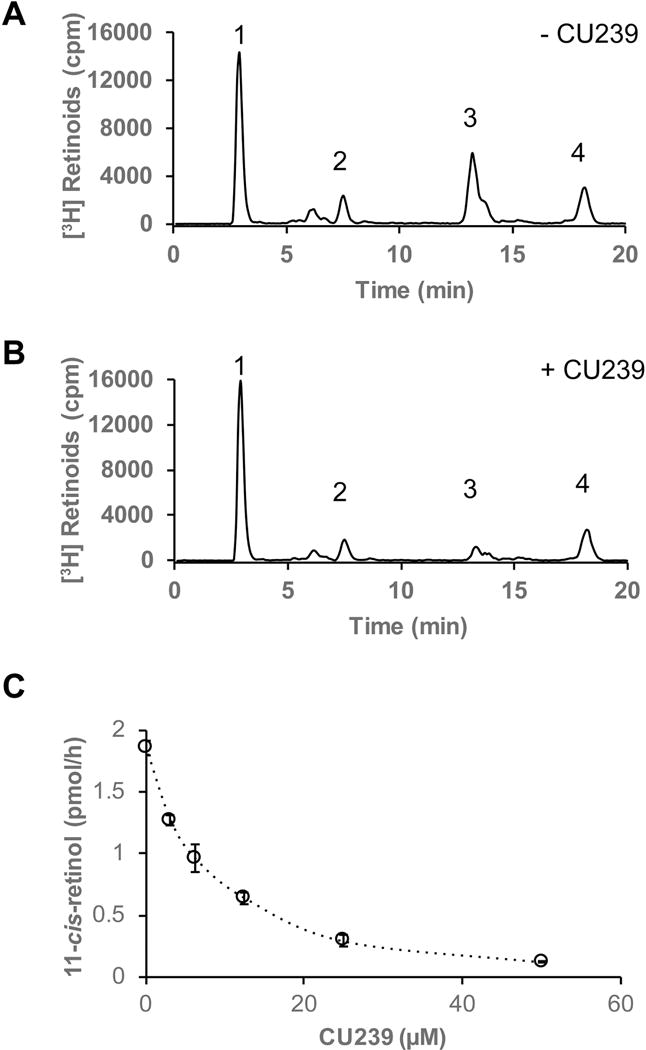
Bovine RPE microsomes (20 μg) were incubated with 0.2 μM of all-trans-[3H]-ROL in the presence or absence of CU239 for 2 h at 37°C. The retinoids generated were analyzed by HPLC and flow scintillation counter. (A) HPLC elution profile without CU239; (B) with 25 μM of CU239. Peak 1, retinyl esters; 2, all-trans-RAL; 3, 11-cis-ROL; 4, all-trans-ROL. (C) Concentration-dependent inhibition of 11-cis-[3H]-ROL generation by CU239 in the isomerohydrolase assay (mean ± SEM, n=3).
3.3. Measurement of CU239 binding to RPE65 by fluorescent titration
Having observed inhibitory effect of CU239 on isomerase activity, we then wanted to understand the binding mechanism of CU239 to the target. Using the available crystal structure of bovine RPE65 (PBD ID: 3KVC), we performed in silico binding prediction for CU239 with the Genetic Optimization for Ligand Docking (GOLD) software, to observe how CU239 may interact with RPE65. A unity query was created from the amino acids that were identified in the binding site as critical for ligand interaction. The unity query included 10 H-bond donors and acceptors features, 4 hydrophobic features and a positive center feature (i.e. iron cofactor). As shown in Figure 3A, our prediction model demonstrates that a docking of CU239 (shown in colored sticks) closely resembles that of the docking of retinyl palmitate, the native substrate of RPE65 (shown in black lines). This suggests that CU239 may inhibit isomerase activity of RPE65 by directly competing with its native substrate. To confirm that CU239 directly binds with RPE65, we performed fluorescent titration assay of purified chicken RPE65 with CU239. Our results demonstrated that CU239 quenched intrinsic tryptophan fluorescence of RPE65 in a concentration-dependent manner, indicating a ligand-protein binding activity. A representative titration curve is shown in Figure 3B. The binding constant can be derived using mass law equation. The inner filter effect due to absorbance of CU239 was very small and thus can be neglected. The apparent dissociation curve calculated from the titration data was 230±48 nM. The calculated number of binding sites suggests that 1:1 molecular complex formed between the inhibitor and RPE65. Taken together, these data suggest that CU239 may exert its inhibitory effect by directly binding to RPE65 and competing with its natural substrate. Specificity of RPE65 binding for CU239 was assessed in a binding assay measuring compound interaction with another retinoid-binding protein, Retinol-Binding Protein 4 (RBP4). Consistent with high RPE65 selectivity, CU239 (as well as CU262, CU270, and CU223) had no activity in the RBP4 binding assay (data not shown).
Fig. 3. CU239 is able to bind RPE65.
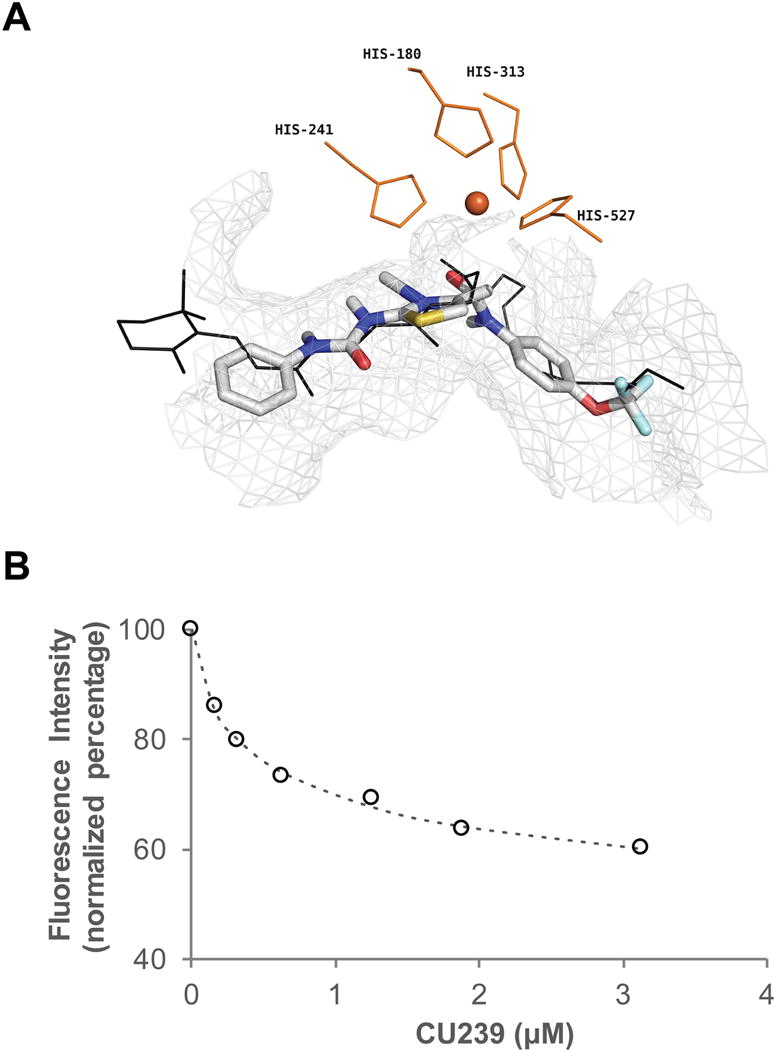
(A) Binding pose of CU239 obtained from a Unity Model in comparison with binding of retinyl palmitate to the bovine RPE65. CU239: colored stick; Retinyl palmitate: black line. Grey mesh outlines the residues within 4Å of CU239. Shown in orange are the iron co-factor and the four histidine residues that bind iron. (B) Fluorescence measurement of binding of CU239 to RPE65. Titration of chicken RPE65 with CU239 as measured by the quenching of protein fluorescence. Excitation was recorded at wavelength 278 nm, emission at 340 nm. The titration system consisted of 2 ml of 0.1μM of RPE65 in 0.1% CHAPS in PBS, pH 7.4.
3.4. CU239 is a competitive inhibitor of RPE65
Based on our computer modeling and the binding assay result, we wanted to investigate whether CU239 is indeed inhibiting RPE65 through substrate competition. To test this, we measured the inhibition of RPE65 in a one-step reaction with its direct substrate retinyl ester. In this assay we used recombinant chicken RPE65, which has higher isomerase activity as compared to that from rod dominant species. Since retinyl ester is insoluble in water, we employed a liposome-based isomerohydrolase assay; the substrate for RPE65 all-trans-retinyl palmitate was incorporated into liposomes (1,2-dioleoyl-sn-glycero-3-phosphocholin: 1,2-dilauroyl-sn-glycero-3-phosphocholine, 85:15) (36), and purified RPE65 was incubated with varying amounts of the substrate-laden liposomes with or without CU239. As shown in the graph, CU239 significantly reduced the reaction rate at lower substrate concentrations, but had less inhibitory activity when higher concentrations of substrate were used (Fig. 4A–C). As shown in the Lineweaver-Burk plot (Fig. 4C), apparent Km increased in the presence of CU239 without changing Vm, demonstrating a hallmark of competitive inhibition (33). Analysis of these results according to the formula for competitive inhibition yielded Ki = 9.9 ± 0.8 μM for CU239. Taken together, our data support that CU239 is a competitive inhibitor of RPE65.
Fig. 4. Competitive inhibition of the RPE65 isomerase by CU239 in a liposome-based isomerase assay.
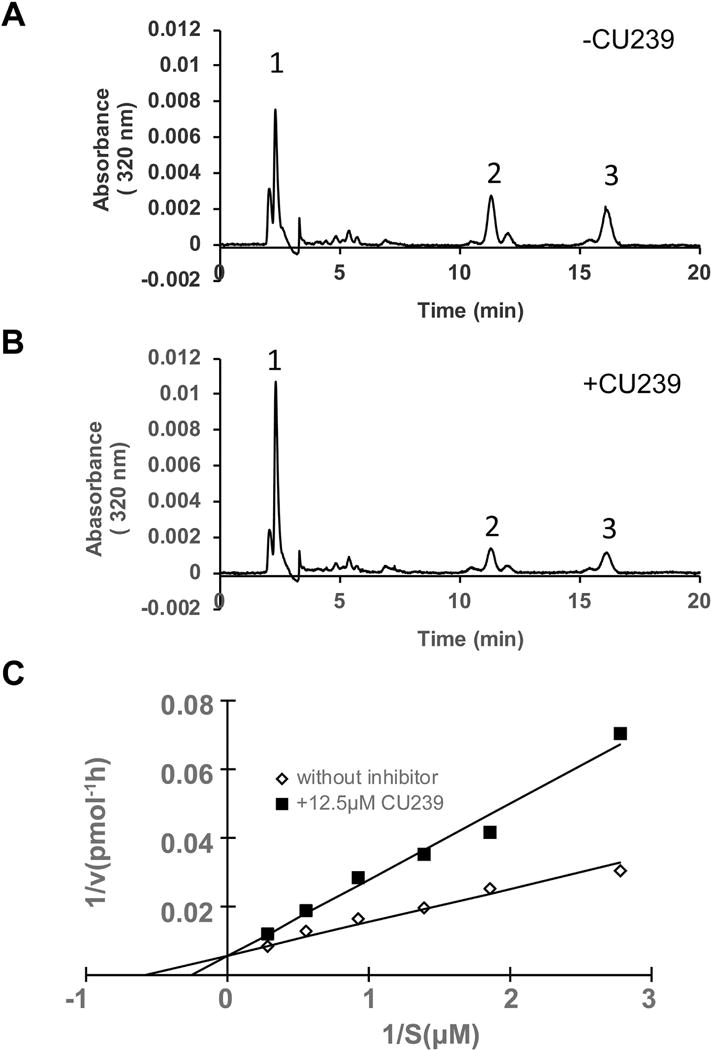
All-trans-retinyl ester incorporated in liposomes was used as a substrate for purified RPE65 in the isomerase assay. (A) HPLC elution profile without CU239; (B) with 12.5 μM CU239; Peak 1, retinyl esters; 2, 11-cis-ROL; 3, all-trans-ROL. (C) Lineweaver-Burk plot of 11-cis-ROL generation by RPE65. Liposomes with increasing concentrations (S) of all-trans-retinyl ester were incubated with equal amounts (125 μg) of chicken RPE65 in the absence (◊) or in the presence of CU239 (12.5 μM) (■).
3.5. Systemic administration of CU239 delays visual chromophore regeneration after bleaching in mice
To evaluate the efficacy of CU239 as an inhibitor of RPE65 in vivo, we systemically delivered CU239 into BALB/cJ mice by intraperitoneal injection, and examined visual chromophore regeneration after bleaching under 5,000 lux fluorescent light for 30 minutes. We have chosen BALB/c mice as a model because BALB/c mice have higher endogenous levels and activities of RPE65 as compared to other mouse strains (32). Hence, the effect of inhibitor of visual cycle can be measured more accurately (32). Following the light exposure for 30 minutes, mice were returned to the dark for 30 minutes, to allow visual chromophore regeneration prior to harvesting the eyeballs. The retinoids were extracted from the harvested eyeballs under dim red light, and the levels of 11-cis-RAL, all-trans-RAL and all-trans-retinyl ester were determined by HPLC analysis (Fig. 5A–C). The results showed that CU239 exhibited a dose-dependent inhibition of chromophore regeneration, and mice injected with 52 mg/kg of CU239 demonstrated the most substantial decrease in 11-cis-RAL regeneration (<40% of vehicle control) (Fig. 5A). Mice injected with 35 and 52 mg/kg of CU239 also exhibited increased levels of all-trans-retinyl ester, the substrate of RPE65, further indicating decreased RPE65 activities (Fig. 5C). Since there are no significant levels of unbound 11-cis-RAL in the mouse eye, the measured 11-cis-RAL equals rhodopsin amount (37, 38).Taken together, these data suggest that CU239 administration inhibits 11-cis-RAL and rhodopsin regeneration in the mouse eye.
Fig. 5. Systemic injection of CU239 delays chromophore regeneration in BALB/cJ mice.
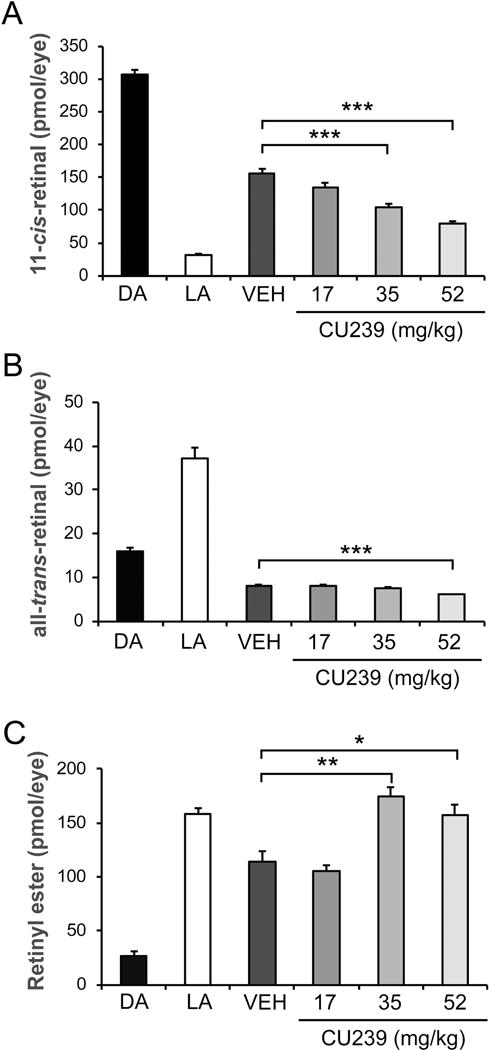
Dark-adapted mice were injected with varying amounts of CU239, and subjected to light-adaptation under 5,000 lux of fluorescent light for 30 minutes. The eyes were harvested for HPLC retinoid profiling after 30 minutes of dark-adaptation, to measure levels of (A) 11-cis-RAL, (B) all-trans-RAL, and (C) all-trans-retinyl ester. Student’s t-test. *P<0.05 and ***<0.001. (Mean ± SEM).
3.6. CU239 partially prevents light-induced retinal damage (LIRD) by suppressing the visual cycle
LIRD is a commonly used model to study various types of retinal degeneration (39, 40). A growing body of evidence supports that LIRD can be prevented when the visual cycle is suppressed—most notably by inhibiting the isomerase RPE65 (20, 21). In fact, Grimm et. al have demonstrated that Rpe65−/− mice are resistant to LIRD, validating a proof of concept for visual cycle modulation as a therapeutic strategy (41). Moreover, mice with lower abundance of RPE65 with Leu450Met mutation have slower regeneration kinetics of rhodopsin and increased resistance to LIRD (42). These findings suggest that generation of visual chromophore is essential for LIRD. Based on its selective inhibition of RPE65, we tested whether CU239 can confer a neuroprotective effect on the retina in the LIRD model. To test this, fully dark-adapted mice were injected with either CU239 (35 mg/kg, IP) or vehicle 1 hour prior to LIRD (8,000 lux for 3 hours). Post-LIRD assessments were performed 5 days after LIRD, which included outer nuclear layer (ONL) thickness measurement on H&E stained retinal sections, and retina functional assessment by electroretinography (ERG) analysis. The retinas of vehicle injected mice exhibited greatly reduced ONL thickness compared to no LIRD control (Fig. 6A and B), while the retinas with CU239 treatment showed a milder reduction of ONL thickness (Fig. 6C). In addition to our histological observation, ERG analysis on the mice injected with CU239 prior to LIRD revealed significantly higher visual sensitivities compared to the LIRD mice treated with vehicle, suggesting that inhibition of RPE65 with CU239 partially prevented rod photoreceptors from LIRD (Fig. 7A–C). In addition, significantly higher photopic responses from the CU239 injected mice with LIRD suggest that CU239 also confers its protective effect on cone photoreceptors as well (Fig. 7D and E). Taken together, the data presented herein clearly demonstrate that suppression of the visual cycle with CU239 can confer a neuroprotective effect on the retina challenged with LIRD.
Fig. 6. Histological analysis of the retinas from CU239 injected mice 5 days post-LIRD.
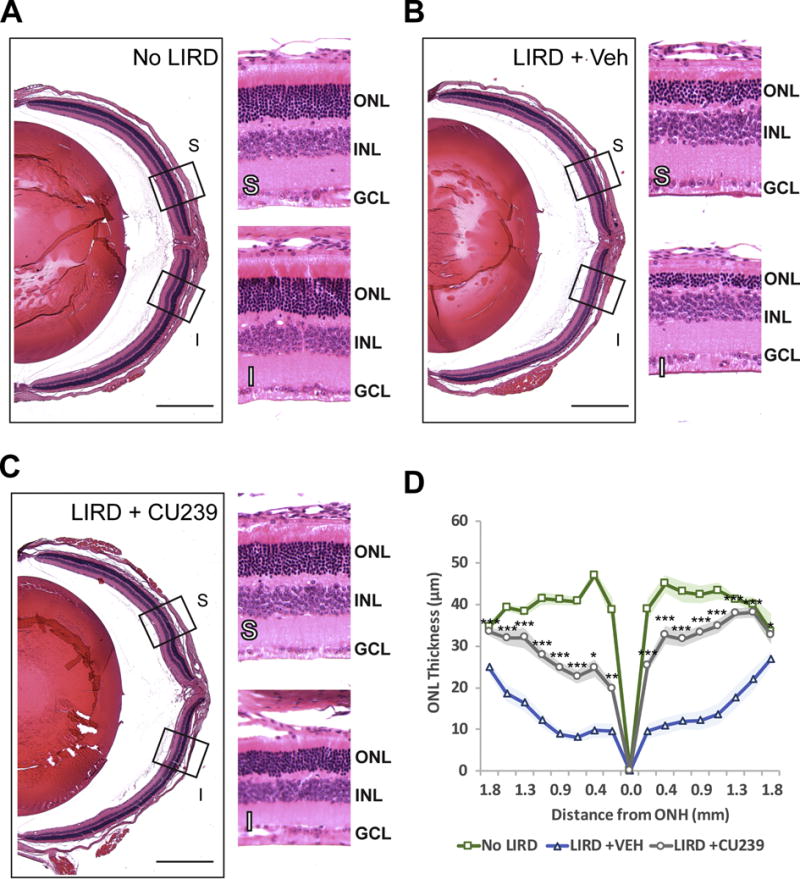
Retinal cross-sections of (A) No LIRD Control, (B) LIRD + Vehicle, and (C) LIRD + CU239 were stained with H&E for morphological comparison. (D) A graph showing ONL thickness measured in cross sections of the retina. Student’s t-test. *P<0.05 and ***<0.001. (Mean ± SEM). S: Superior retina; I: inferior retina. Scale bar: 500 μm (4× images). Boxed areas on 4× images were zoomed under 20× magnification.
Fig. 7. ERG responses of CU239 injected mice 5 days post-LIRD.
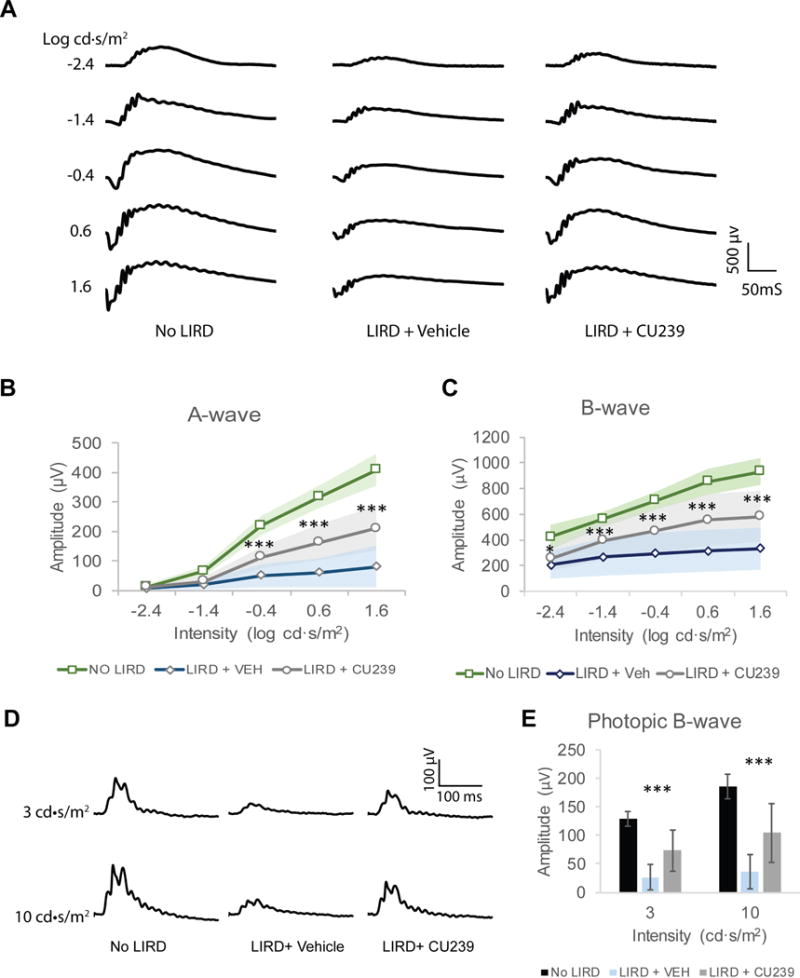
(A) Representative scotopic ERG waveforms from mouse groups No LIRD control, LIRD + Vehicle and LIRD + CU239. Five different stimulus intensities were used, ranging from −2.4 to 1.6 log cd·s/m2. (B) Comparison of the averaged A-wave amplitude from the three groups. Student’s t-test was used (n=8, ***P<0.001, mean ± SEM). (C) Comparison of the averaged B-wave amplitude from the three groups. Student’s t-test (n=8, *P<0.05 and ***<0.001; mean ± SEM). (D) Representative photopic ERG waveforms from groups No LIRD control, LIRD + Vehicle or LIRD + CU239. (E) Comparison of the averaged photopic B-wave amplitude from the three groups (n=8, mean ± SEM).
4. Discussion
Ocular retinoid metabolism involves many retinoid processing enzymes and retinoid-binding proteins, and their concerted activities not only enable photoreception but also ensure retinal health. As evident from both clinical studies and experimental animal models of human blindness, a key aspect of the retinoid homeostasis in the retina is to efficiently clear all-trans-RAL, a toxic byproduct of the visual cycle. For example, loss of clearance mechanism due to mutations in ABCA4, a gene that encodes ATP-binding cassette transmembrane protein, which removes all-trans-RAL out from rod photoreceptors—leads to a juvenile macular degeneration known as Stargardt’s disease. Studies using Abca4−/− and/or Rdh8−/− mice demonstrated functional declines of retinas with corresponding retinal degeneration, which is attributed to an abnormal accumulation of bisretinoids such as A2E (43, 44). Clearance impediments, and consequentially abnormal accumulation of bisretinoids, may also arise from aging (45). Various studies have demonstrated that abnormal accumulation of bisretinoid condensation products have multiple disease implications for both retina and the RPE, including but not limited to DNA damage, oxidative stress, complement activation, and mitochondrial dysfunction (46–50).
Gaining further insights into retinoid dynamics in visual function, especially in diseased state, is imperative for identifying targetable molecules, proteins and pathway(s), and ultimately for developing rational therapeutic strategies to prevent visual impairment. For instance, AMD is the third most prevalent cause of blindness across the world after glaucoma and cataract, and 90% of AMD patients develop the dry form of AMD, for which there is no treatments available at present. As of year 2000, nearly 2 million people in the U.S. alone were affected by AMD, and the number is projected to double by 2020 due to increasing geriatric population (51). Since distorted visual cycle has disease implication for various retinal dystrophies such as AMD, many attempts using gene therapy and visual cycle modulators have been explored for the past decade as a means to restore the retinoid dynamics in visual function (18, 21, 52–54).
RPE65 is an ideal target for visual cycle modulation because it a key enzyme of the visual cycle, catalyzes the rate-limiting step of the visual cycle, and is almost exclusively expressed in the RPE (6, 55–57). A non-retinoid RPE65 inhibitor emixustat has demonstrated a dose-dependent reduction of A2E levels in Abca4−/− mice after 3 months of oral administration (18). Furthermore, Rpe65−/− mice are resistant to LIRD, as are the mice and rats treated with small-molecule inhibitors of RPE65 (18, 21, 41, 54). Hence, visual cycle modulation provides valuable insights into developing potential therapeutic application for retinal degeneration. Of interesting note, a recent article by Zhang et al. (2015) has reported that MB-002, a diol derivative of emixustat, is unable to confer protective effect against LIRD despite its inhibitory effect on RPE65 (23). The authors account that such finding may be due to the inability of MB-002 to sequester all-trans-RAL via Schiff base formation, a feature common to RPE65 inhibitors with a primary amine group. However, the inability of MB-002 to protect against light damage also might be explained by its lower bioavailability in the RPE. No pharmacokinetics data was shown for this compound. On the contrary, we have demonstrated a complete protection against LIRD in rats by administration of a-phenyl-N-tert-butylnitrone (PBN), which inhibits RPE65 without the ability of forming Schiff base with all-trans-RAL due to its lack of primary amine group (21). Another well-known visual cycle inhibitor, 13-cis-retinoic acid, also does not contain a primary amine and protects the retina from light damage (58). Hence, the neuroprotective effect of visual cycle modulation warrants more mechanistic insights.
The data presented herein demonstrate that CU239, a novel non-retinoid compound, can also effectively inhibit a key enzyme of the visual cycle enzyme RPE65, and confer a partial protective effect on the retinas from LIRD.
From our in vitro isomerase assay, using all-trans-ROL and endogenous LRAT to create retinyl ester as a substrate for RPE65, we have shown that CU239 have IC50 of 6 μM. Moreover, we found that CU239 did not inhibit retinyl ester synthesis catalyzed by LRAT, suggesting that CU239 selectively inhibits the isomerization step of the visual cycle.
To determine the mode of CU239 inhibition, we used a one-step reaction of conversion of retinyl ester incorporated in liposomes. This assay provided a direct measurement of the conversion of all-trans-retinyl palmitate to 11-cis-ROL in the presence of CU239, from which the inhibition constant was calculated. The inhibition of this reaction was found to be competitive with the inhibition constant of 9.9 μM.
To confirm that CU239 inhibition occurs through direct binding to RPE65, we used fluorescence quenching of purified RPE65 by CU239. The fluorescent method is well suited for measurements of binding of ligands to RPE65 because RPE65 contains multiple tryptophan residues which are the major fluorescent amino acids. Our fluorescence quenching assay revealed that CU239 was able to directly bind RPE65 with Kd = 230 nM, likely at the substrate binding site of RPE65, as our ligand docking model have predicted. Our results from the competition assay supports that CU239 binds RPE65 at the substrate-binding site.
We have also investigated whether CU239 can inhibit RPE65 in vivo. As shown by the HPLC retinoid profile analysis, mice injected with CU239 showed delayed chromophore regeneration after photobleach in a dose-dependent manner, suggesting a slowed visual cycle. In addition, we have administered 35 mg/kg of CU239 in mice to examine whether CU239-mediated visual cycle inhibition could protect the retina from light-induced retinal damage. As expected, our post-LIRD assessments— histological analyses and functional analysis with ERG—demonstrated a protective effect of CU239 against LIRD. As our understanding of the catalytic mechanism of RPE65 remains incomplete, it is difficult to determine the precise mechanism underlying the inhibitory effect of CU239. However, our ligand docking prediction coupled with the isomerase assay strongly suggest that CU239 is a competitive inhibitor of RPE65.
Future studies will require co-crystallization of RPE65-CU239 to determine the structural basis of CU239-RPE65 interaction, and to better understand the catalytic mechanism of RPE65.
5. Conclusions
In conclusion, we demonstrate that CU239, a novel non-retinoid compound, is a specific and potent inhibitor of RPE65 that directly competes with the substrate all-trans-retinyl ester. Our compound may be useful for not only future studies on the catalytic mechanisms of RPE65, but also for developing a potential approach to offset the retinoids imbalance caused by a defective visual cycle.
Supplementary Material
Highlights.
Visual cycle can produce vitamin A-based toxins harmful to the retinal health
AMD and Stargardt’s diseases exhibit accumulation of vitamin A-based toxins
Suppression of visual cycle can cripple the production of vitamin A-based toxins
CU239 suppresses visual cycle and prevents retinal damage in LIRD model
Acknowledgments
We would like to thank the Diabetes Animal Core of the Diabetes COBRE (The University of Oklahoma Health Sciences Center) and the histology core at Dean McGee Eye Institute.
Funding:
This study was supported by NIH grants EY018659, EY012231, EY019309, GM104934, GM122744, a Juvenile Diabetes Research Foundation (JDRF) grant (2-SRA-2014-147-Q-R), an Oklahoma Center for the Advancement of Science and Technology (OCAST) grant (HR16-041) and Presbyterian Health Foundation bridge grant.
Abbreviations
- RPE
retinal pigment epithelium
- IPM
inter-photoreceptor matrix
- 11-cis-RAL
11-cis-retinaldehyde
- all-trans-RAL
all-trans-retinaldehyde
- ABCA4
ATP-cassette transporter 4
- atRDH
all-trans-retinol dehydrogenases
- all-trans-ROL
all-trans-retinol
- IRBP
interphotoreceptor retinoid-binding protein
- LRAT
lecithin retinol acyltransferase
- 11-cis-RDH
11-cis-ROL
- STGD
Stargardt’s disease
- AMD
age-related macular degeneration
- A2E
N-retinylidene-N-retinylethanolamine, LIRD, light-induced retinal damage
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest:
The authors have no conflict of interest.
References
- 1.Wald G. The chemistry of rod vision. Science. 1951;113:287–91. doi: 10.1126/science.113.2933.287. [DOI] [PubMed] [Google Scholar]
- 2.Wald G. The molecular basis of visual excitation. Nature. 1968;219:800–807. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- 3.Sun H, Molday RS, Nathans J. Retinal stimulates ATP hydrolysis by purified and reconstituted ABCR, the photoreceptor-specific ATP-binding cassette transporter responsible for Stargardt disease. J Biol Chem. 1999;274:8269–81. doi: 10.1074/jbc.274.12.8269. [DOI] [PubMed] [Google Scholar]
- 4.Fong SL, Liou GI, Landers RA, Alvarez RA, Gonzalez-Fernandez F, Glazebrook PA, Lam DM, Bridges CD. Characterization, localization, and biosynthesis of an interstitial retinol-binding glycoprotein in the human eye. J Neurochem. 1984;42:1667–76. doi: 10.1111/j.1471-4159.1984.tb12758.x. [DOI] [PubMed] [Google Scholar]
- 5.Rattner A, Smallwood PM, Nathans J. Identification and characterization of all-trans-retinol dehydrogenase from photoreceptor outer segments, the visual cycle enzyme that reduces all-trans-retinal to all-trans-retinol. J Biol Chem. 2000;275:11034–43. doi: 10.1074/jbc.275.15.11034. [DOI] [PubMed] [Google Scholar]
- 6.Moiseyev G, Chen Y, Takahashi Y, Wu BX, Ma J-X. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc Natl Acad Sci. 2005;102:12413–12418. doi: 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin M, Li S, Moghrabi WN, Sun H, Travis GH. Rpe65 Is the Retinoid Isomerase in Bovine Retinal Pigment Epithelium. Cell. 2005;122:449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redmond TM, Poliakov E, Yu S, Tsai JY, Lu Z, Gentleman S. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc Natl Acad Sci U S A. 2005;102:13658–13663. doi: 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Okano K, Maeda T, Chauhan V, Golczak M, Maeda A, Palczewski K. Mechanism of All-trans-retinal Toxicity with Implications for Stargardt Disease and Age-related Macular Degeneration. J Biol Chem. 2012;287:5059–5069. doi: 10.1074/jbc.M111.315432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda A, Golczak M, Maeda T, Palczewski K. Limited roles of Rdh8, Rdh12, and Abca4 in all-trans-retinal clearance in mouse retina. Investig Ophthalmol Vis Sci. 2009;50:5435–5443. doi: 10.1167/iovs.09-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrault I, Hanein S, Gerber S, Barbet F, Ducroq D, Dollfus H, Hamel C, Dufier JL, Munnich A, Kaplan J, Rozet JM. Retinal Dehydrogenase 12 (RDH12) Mutations in Leber Congenital Amaurosis. Am J Hum Genet. 2004;75:639–646. doi: 10.1086/424889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suter M, Remé C, Grimm C, Wenzel A, Jäättela M, Esser P, Kociok N, Leist M, Richter C. Age-related macular degeneration: The lipofuscin component N-retinyl-N-retinylidene ethanolamine detaches proapoptotic proteins from mitochondria and induces apoptosis in mammalian retinal pigment epithelial cells. J Biol Chem. 2000;275:39625–39630. doi: 10.1074/jbc.M007049200. [DOI] [PubMed] [Google Scholar]
- 13.Lamb LE, Simon JD. A2E: a component of ocular lipofuscin. Photochem Photobiol. 2004;79:127–136. doi: 10.1562/0031-8655(2004)079<0127:aacool>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Holz FG, Strauss EC, Schmitz-Valckenberg S, Van Lookeren Campagne M. Geographic atrophy: Clinical features and potential therapeutic approaches. Ophthalmology. 2014;121:1079–1091. doi: 10.1016/j.ophtha.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 15.Sparrow JR, Wu Y, Kim CY, Zhou J. Phospholipid meets all-trans-retinal: the making of RPE bisretinoids. J Lipid Res. 2010;51:247–61. doi: 10.1194/jlr.R000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyer NP, Higbee D, Currin MB, Blakeley LR, Chen C, Ablonczy Z, Crouch RK, Koutalos Y. Lipofuscin and N-retinylidene-N-retinylethanolamine (A2E) accumulate in retinal pigment epithelium in absence of light exposure: their origin is 11-cis-retinal. J Biol Chem. 2012;287:22276–86. doi: 10.1074/jbc.M111.329235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gollapalli DR, Rando RR. The specific binding of retinoic acid to RPE65 and approaches to the treatment of macular degeneration. Proc Natl Acad Sci U S A. 2004;101:10030–5. doi: 10.1073/pnas.0401936101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bavik C, Henry SH, Zhang Y, Mitts K, McGinn T, Budzynski E, Pashko A, Lieu KL, Zhong S, Blumberg B, Kuksa V, Orme M, Scott I, Fawzi A, Kubota R. Visual Cycle Modulation as an Approach toward Preservation of Retinal Integrity. PLoS One. 2015;10:e0124940. doi: 10.1371/journal.pone.0124940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeda A, Maeda T, Golczak M, Imanishi Y, Leahy P, Kubota R, Palczewski K. Effects of potent inhibitors of the retinoid cycle on visual function and photoreceptor protection from light damage in mice. Mol Pharmacol. 2006;70:1220–1229. doi: 10.1124/mol.106.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golczak M, Maeda A, Bereta G, Maeda T, Kiser PD, Hunzelmann S, von Lintig J, Blaner WS, Palczewski K. Metabolic basis of visual cycle inhibition by retinoid and nonretinoid compounds in the vertebrate retina. J Biol Chem. 2008;283:9543–54. doi: 10.1074/jbc.M708982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandal MNA, Moiseyev GP, Elliott MH, Kasus-Jacobi A, Li X, Chen H, Zheng L, Nikolaeva O, Floyd RA, Ma J-X, Anderson RE. -Phenyl-N-tert-butylnitrone (PBN) Prevents Light-induced Degeneration of the Retina by Inhibiting RPE65 Protein Isomerohydrolase Activity. J Biol Chem. 2011;286:32491–32501. doi: 10.1074/jbc.M111.255877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eroglu A, Gentleman S, Poliakov E, Redmond TM. Inhibition of RPE65 Retinol Isomerase Activity by Inhibitors of Lipid Metabolism. J Biol Chem. 2016;291:4966–4973. doi: 10.1074/jbc.M115.685651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Kiser PD, Badiee M, Palczewska G, Dong Z, Golczak M, Tochtrop GP, Palczewski K. Molecular pharmacodynamics of emixustat in protection against retinal degeneration. J Clin Invest. 2015;125:2781–2794. doi: 10.1172/JCI80950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radu RA, Mata NL, Nusinowitz S, Liu X, Sieving PA, Travis GH. Treatment with isotretinoin inhibits lipofuscin accumulation in a mouse model of recessive Stargardt’s macular degeneration. Proc Natl Acad Sci U S A. 2003;100:4742–7. doi: 10.1073/pnas.0737855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maiti P, Kong J, Kim R, Sparrow JR, Allikmets R, Rando RR. Small Molecule RPE65 Antagonists Limit the Visual Cycle and Prevent Lipofuscin Formation. doi: 10.1021/bi0518545. [DOI] [PubMed] [Google Scholar]
- 26.Kubota R, Boman NL, David R. Safety and effect on rod function of acu-4429, a novel small-molecule visual cycle modulator. Retina. 2012;32:183–188. doi: 10.1097/IAE.0b013e318217369e. [DOI] [PubMed] [Google Scholar]
- 27.Kubota R, Al-Fayoumi S, Mallikaarjun S, Patil S, Bavik C, Chandler JW. Phase 1, dose-ranging study of emixustat hydrochloride (acu-4429), a novel visual cycle modulator, in healthy volunteers. Retina. 2014;34:603–609. doi: 10.1097/01.iae.0000434565.80060.f8. [DOI] [PubMed] [Google Scholar]
- 28.Golczak M, Kiser PD, Lodowski DT, Maeda A, Palczewski K. Importance of membrane structural integrity for RPE65 retinoid isomerization activity. J Biol Chem. 2010;285:9667–82. doi: 10.1074/jbc.M109.063941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–5. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 31.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking 1 1Edited by F. E. Cohen. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 32.Moiseyev G, Crouch RK, Goletz P, Oatis J, Redmond TM, Ma J xing. Retinyl esters are the substrate for isomerohydrolase. Biochemistry. 2003;42:2229–2238. doi: 10.1021/bi026911y. [DOI] [PubMed] [Google Scholar]
- 33.Fersht A. Enzyme Structure and Mechanism. W.H. Freeman and Co; New York: 1985. pp. 108–109. [Google Scholar]
- 34.Moiseyev G, Nikolaeva O, Chen Y, Farjo K, Takahashi Y, Ma J. Inhibition of the visual cycle by A2E through direct interaction with RPE65 and implications in Stargardt disease. Proc Natl Acad Sci U S A. 2010;107:17551–6. doi: 10.1073/pnas.1008769107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cogan U, Kopelman M, Mokady S, Shinitzky M. Binding Affinities of Retinol and Related Compounds to Retinol Binding Proteins. Eur J Biochem. 1976;65:71–78. doi: 10.1111/j.1432-1033.1976.tb10390.x. [DOI] [PubMed] [Google Scholar]
- 36.Nikolaeva O, Takahashi Y, Moiseyev G, Ma JX. Purified RPE65 shows isomerohydrolase activity after reassociation with a phospholipid membrane. FEBS J. 2009;276:3020–3030. doi: 10.1111/j.1742-4658.2009.07021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmerman WF. The distribution and proportions of vitamin A compounds during the visual cycle in the rat. Vision Res. 1974;14:795–802. doi: 10.1016/0042-6989(74)90143-6. [DOI] [PubMed] [Google Scholar]
- 38.Lee KA, Nawrot M, Garwin GG, Saari JC, Hurley JB. Relationships among visual cycle retinoids, rhodopsin phosphorylation, and phototransduction in mouse eyes during light and dark adaptation. Biochemistry. 2010;49:2454–63. doi: 10.1021/bi1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rózanowska M, Sarna T. Light-induced damage to the retina: role of rhodopsin chromophore revisited. Photochem Photobiol. 2005;81:1305–1330. doi: 10.1562/2004-11-13-IR-371. [DOI] [PubMed] [Google Scholar]
- 40.Grimm C, Remé CE. Light Damage as a Model of Retinal Degeneration. Methods in molecular biology (Clifton, NJ) 2012;935:87–97. doi: 10.1007/978-1-62703-080-9_6. pp 87–97. [DOI] [PubMed] [Google Scholar]
- 41.Grimm C, Wenzel A, Hafezi F, Yu S, Redmond TM, Remé CE. Protection of Rpe65-deficient mice identifies rhodopsin as a mediator of light-induced retinal degeneration. Nat Genet. 2000;25:63–66. doi: 10.1038/75614. [DOI] [PubMed] [Google Scholar]
- 42.Wenzel A, Reme CE, Williams TP, Hafezi F, Grimm C. The Rpe65 Leu450Met variation increases retinal resistance against light-induced degeneration by slowing rhodopsin regeneration. J Neurosci. 2001;21:53–8. doi: 10.1523/JNEUROSCI.21-01-00053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeda A, Golczak M, Maeda T, Palczewski K. Limited roles of Rdh8, Rdh12, and Abca4 in all-trans-retinal clearance in mouse retina. Invest Ophthalmol Vis Sci. 2009;50:5435–43. doi: 10.1167/iovs.09-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radu RA, Yuan Q, Hu J, Peng JH, Lloyd M, Nusinowitz S, Bok D, Travis GH. Accelerated accumulation of lipofuscin pigments in the RPE of a mouse model for ABCA4-mediated retinal dystrophies following vitamin A supplementation. Investig Ophthalmol Vis Sci. 2008;49:3821–3829. doi: 10.1167/iovs.07-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murdaugh LS, Wang Z, Del Priore LV, Dillon J, Gaillard ER. Age-related accumulation of 3-nitrotyrosine and nitro-A2E in human Bruch’s membrane. Exp Eye Res. 2010;90:564–571. doi: 10.1016/j.exer.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Sparrow JR, Zhou J, Ben-Shabat S, Vollmer H, Itagaki Y, Nakanishi K. Involvement of oxidative mechanisms in blue-light-induced damage to A2E-laden RPE. Invest Ophthalmol Vis Sci. 2002;43:1222–7. [PubMed] [Google Scholar]
- 47.Sparrow JR, Zhou J, Cai B. DNA is a target of the photodynamic effects elicited in A2E-laden RPE by blue-light illumination. Invest Ophthalmol Vis Sci. 2003;44:2245–51. doi: 10.1167/iovs.02-0746. [DOI] [PubMed] [Google Scholar]
- 48.Zhou J, Jang YP, Kim SR, Sparrow JR. Complement activation by photooxidation products of A2E, a lipofuscin constituent of the retinal pigment epithelium. Proc Natl Acad Sci. 2006;103:16182–16187. doi: 10.1073/pnas.0604255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vives-Bauza C, Anand M, Shirazi AK, Magrane J, Gao J, Vollmer-Snarr HR, Manfredi G, Finnemann SC, Finnemann SC. The Age Lipid A2E and Mitochondrial Dysfunction Synergistically Impair Phagocytosis by Retinal Pigment Epithelial Cells. J Biol Chem. 2008;283:24770–24780. doi: 10.1074/jbc.M800706200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng J, Chen X, Sun X, Wang F, Sun X. Expression of Endoplasmic Reticulum Stress Markers GRP78 and CHOP Induced by Oxidative Stress in Blue Light-Mediated Damage of A2E-Containing Retinal Pigment Epithelium Cells. Ophthalmic Res. 2014;52:224–233. doi: 10.1159/000363387. [DOI] [PubMed] [Google Scholar]
- 51.Friedman DS, O’Colmain BJ, Muñoz B, Tomany SC, McCarty C, de Jong PTVM, Nemesure B, Mitchell P, Kempen J, Eye Diseases Prevalence Research Group Prevalence of Age-Related Macular Degeneration in the United States. Arch Ophthalmol. 2004;122:564. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 52.Han Z, Conley SM, Makkia RS, Cooper MJ, Naash MI, Naash M. DNA nanoparticle-mediated ABCA4 delivery rescues Stargardt dystrophy in mice. J Clin Invest. 2012;122:3221–6. doi: 10.1172/JCI64833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong J, Kim SR, Binley K, Pata I, Doi K, Mannik J, Zernant-Rajang J, Kan O, Iqball S, Naylor S, Sparrow JR, Gouras P, Allikmets R. Correction of the disease phenotype in the mouse model of Stargardt disease by lentiviral gene therapy. Gene Ther. 2008;15:1311–1320. doi: 10.1038/gt.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Golczak M, Kuksa V, Maeda T, Moise AR, Palczewski K. Positively charged retinoids are potent and selective inhibitors of the trans-cis isomerization in the retinoid (visual) cycle. Proc Natl Acad Sci U S A. 2005;102:8162–7. doi: 10.1073/pnas.0503318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicoletti A, Wong DJ, Kawase K, Gibson LH, Yang-Feng TL, Richards JE, Thompson DA. Molecular characterization of the human gene encoding an abundant 61 kDa protein specific to the retinal pigment epithelium. Hum Mol Genet. 1995;4:641–9. doi: 10.1093/hmg/4.4.641. [DOI] [PubMed] [Google Scholar]
- 56.Hamel CP, Tsilou E, Pfeffer BA, Hooks JJ, Detrick B, Redmond TM. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J Biol Chem. 1993;268:15751–7. [PubMed] [Google Scholar]
- 57.Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma JX, Crouch RK, Pfeifer K. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat Genet. 1998;20:344–351. doi: 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- 58.Sieving PA, Chaudhry P, Kondo M, Provenzano M, Wu D, Carlson TJ, Bush RA, Thompson DA. Inhibition of the visual cycle in vivo by 13-cis retinoic acid protects from light damage and provides a mechanism for night blindness in isotretinoin therapy. Proc Natl Acad Sci U S A. 2001;98:1835–40. doi: 10.1073/pnas.041606498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


