Abstract
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides found throughout the cell that lack protein-coding function. Their functions are closely linked to their interaction with RNA-binding proteins (RBPs) and nucleic acids. Nuclear lncRNAs have been studied extensively, revealing complexes with structural and regulatory roles that enable gene organization and control transcription. Cytoplasmic lncRNAs are less well understood, but accumulating evidence indicates that they also form complexes with diverse structural and regulatory functions. Here, we review our current knowledge of cytoplasmic lncRNAs and the different levels of gene regulation controlled by cytoplasmic lncRNA complexes, including mRNA turnover, translation, protein stability, sponging of cytosolic factors, and modulation of signaling pathways. We conclude by discussing areas of future study needed to investigate comprehensively the biology of lncRNAs, to further understand the impact of lncRNAs on physiology and design lncRNA-centered therapeutic strategies.
Keywords: noncoding RNA, post-transcriptional gene regulation, ribonucleoprotein complexes, lncRNA, circRNA, miRNA
Introduction
Long noncoding RNAs (lncRNAs)
The identification of complete transcriptomes in a wide range of species, including human, revealed that a vast portion of transcripts do not encode protein (Carninci et al., 2005; Derrien et al., 2012; Hangauer, Vaughn, & McManus, 2013). Among those noncoding (nc)RNAs, long noncoding (lnc) RNAs are defined as being >200 nt in length and reside throughout the cell. Recently, de novo transcriptome assembly of next-generation RNA sequencing (RNA-seq) data collected from various human tissues identified >50,000 lncRNAs present at one copy or more per cell, and transcribed from intergenic regions (lincRNAs) or from introns and/or exons of protein-coding genes in the sense or antisense direction (Derrien et al., 2012; Montes & Lund 2016). Through their primary sequence and secondary structure, lncRNAs can bind to other nucleic acids and/or proteins to regulate gene expression programs, in turn controlling a growing number of cellular processes, such as cell division, the stress response, differentiation, survival, and senescence (Grammatikakis, Panda, Abdelmohsen, & Gorospe, 2014; Li, Tian, Yang & Gong, 2016; Audas & Lee S, 2016; Chen, Satpathy, & Chang, 2017). By affecting these processes, there is increasing appreciation that lncRNAs have a direct impact on the physiology of tissues and organs, and on a growing number of disease processes (e.g. muscle disease, cancer, and cardiovascular pathologies) (Greco, Gorospe, & Martelli, 2015; Schmitt, & Chang, 2016; Ballarino, Morlando, Fatica, & Bozzoni, 2016; Alvarez-Dominguez, & Lodish. 2017; Wang, Xiao, and Wang; 2017). Despite intense efforts, however, only a small number of lncRNAs have been characterized functionally, while the vast majority of lncRNAs have no known functions at present.
Nuclear lncRNAs and their nuclear functions have been studied quite extensively, revealing a number of nucleus-specific lncRNA functions (i.e. chromosome scaffolding, chromatin remodeling, alternative splicing, epigenetic control of transcription, etc), most often serving key regulatory roles for transcriptional programs and subcellular structures (Derrien et al., 2012; Kugel, & Goodrich, 2012; Tripathi et al., 2013; Hung et al., 2011; Clemson et al., 2009; Zhao, Sun, Erwin, Song, & Lee, 2008; Lee, 2012; Mercer et al., 2011). These structural and functional roles were found to require, almost universally, the interaction of lncRNAs with RNA-binding proteins (RBPs), forming nuclear lncRNA-associated ribonucleoprotein complexes (lncRNPs).
Cytoplasmic lncRNAs, on the other hand, also form complexes with RBPs but are substantially less well understood. Recent studies showed that cytoplasmic lncRNPs can comprise lncRNAs transcribed from nuclear DNA or expressed locally in the cytoplasm (e.g. mitochondrial DNA-encoded lncRNAs) (Mercer et al., 2011). These cytoplasmic lncRNPs can govern cytoplasmic events essential for maintaining cellular structure and functions (Yoon, Abdelmohsen, & Gorospe, 2013; Rashid, Shah, & Shan, 2016), including protein localization and turnover, mRNA translation and stability, availability of cytoplasmic factors, and scaffolding of proteins operating in a shared pathway. In this review, we focus on the major cytoplasmic lncRNPs studied to-date (Table 1), discuss their functions in different cellular contexts (Figure 1), and suggest directions of future research that will advance our knowledge of lncRNP function.
Table 1. Cytoplasmic lncRNAs, interacting factors, and function of lncRNA complexes.
Table includes the functions of the cytoplasmic lncRNAs reviewed (column 1), the lncRNAs (column 2) and interacting RBPs and RNAs (column 3), and the function of the ensuing complexes (column 4).
| Functional Category | Long noncoding RNAs (lncRNAs) | Interacting Factors | Function | Refs. |
|---|---|---|---|---|
| mRNA turnoverand translation | 1/2-sbsRNAs | STAU1 | Degradation of 1/2-sbsRNA target mRNAs | Gong & Maquat, 2011 |
| TINCR | STAU1 | Stabilization of epidermal differentiation mRNAs | Kretz et al., 2013 | |
| CUGexp (CUG repeats) | STAU1 | Induction of translation of mRNAs bearing CUGexp | Ravel-Chapuis et al., 2012 | |
| LincRNA-p21 | HuR | Degradation of lincRNA-p21, suppression of TNNB1 and JUNB translation | Chu & Rana, 2006; Yoon et al., 2012 | |
| BACE1AS | HuD | Stabilization of BACE1 and APP mRNAs | Kang et al., 2014 | |
| GAS5 | eIF4E | Suppression of MYC translation | Hu, Lou, & Gupta, 2014 | |
| Uchl1-AS1 | Uchl1 5′UTR | Induction of UCHL1 translation | Carrieri et al., 2012 | |
|
| ||||
| Protein turnover | LincRNA-p21 | HIF1A | Prevention of VHL-mediated HIF1A ubiquitination | Yang, Zhang, Mei, & Wu, 2014 |
| HOTAIR | DZIP3, MEX3B | Degradation of Snurportin-1 and Ataxin-1 | Yoon et al., 2013 | |
| NRON | CUL4B, PSMD11 | Degradation of HIV protein Tat | Li et al., 2016 | |
|
| ||||
| RBPdecoy | Gadd7 | TDP-43 | Degradation of Cdk6 mRNA | Buratti & Baralle, 2008; Liu, Li, Zhang, Guo, & Zhan, 2012 |
| OIP5-AS1 | HuR | Prevent HuR binding to targets (CCNA2, CCND1, SIRT1 mRNAs) | Kim et al., 2016 | |
| OIP5-AS1 | GAK mRNA | Degradation of GAK mRNA | Kim et al., 2017 | |
| 7SL | TP53 mRNA | Repression of TP53 translation | Abdelmohsen et al., 2014 | |
| LncMyoD | IMP2 | Reduction of NRAS and MYC protein | Gong et al., 2015 | |
| GAS5 | GR | Prevention of GR mobilization to the nucleus | Mourtada-Maarabouni et al., 2009 | |
| H19 | KSRP (KHSRP) | Destabilization of Myog mRNA | Giovarelli et al., 2014 | |
| UCA1 | HNRNPI | Prevention of HNRNPI binding to p27 mRNA, repression of p27 translation | Huang et al., 2014 | |
|
| ||||
| MicroRNA decoy | HULC | miR-372 | Induction of PRKACB translation | Wang et al., 2010 |
| lincRNA-RoR | miR-145 | Derepression of NANOG, OCT, SOX2 production | Loewer et al., 2010; Wang et al., 2013 | |
| H19 | let-7 | Interference with let-7 activity | Kallen et al., 2013 | |
| Linc-MD1 | HuR, miR-133b | Inhibition of miR-133b generation | Cesana et al., 2011; Legnini et al., 2014 | |
| PTENP1 | miR-21,-20a,-214,-19b,-26a,-499-5p | Derepression of PTEN production | Tang, Ning, Zeng, & Li, 2016 | |
|
| ||||
| LncRNP mobilization | NRON | NFAT | NFAT nuclear translocation | Willingham et al., 2005 |
| MALAT1 | HNRNPC | Cytoplasmic translocation of MALAT1 during G2/M transition | Yang, Yi, Han, Du, & Liang, 2013 | |
| RMRP | HuR, GRSF1 | Transport of RMRP to cytoplasm, maintenance of mitochondrial function | Noh et al., 2016 | |
| RMRP, RPPH1 | PNPASE | Transport of RMRP and RPPH1 through mitochondrial inner membrane | Wang et al., 2010 | |
|
| ||||
| Cellular signaling | LINK-A | BRK | Activation of kinase BRK | Lin et al., 2016 |
| LINK-A | LRRK2 | Modification of BRK, normoxic HIF1A stabilization | Lin et al., 2016 | |
| Lnc-DC | STAT3 | Promotion of STAT3 phosphorylation | Wang et al., 2014 | |
| NKILA | NF-κB/IκB complex | Inhibition of NF-κB signaling | Liu et al., 2015 | |
| Lethe | RELA | Reduction in RELA transcriptional activity | Rapicavoli et al., 2013 | |
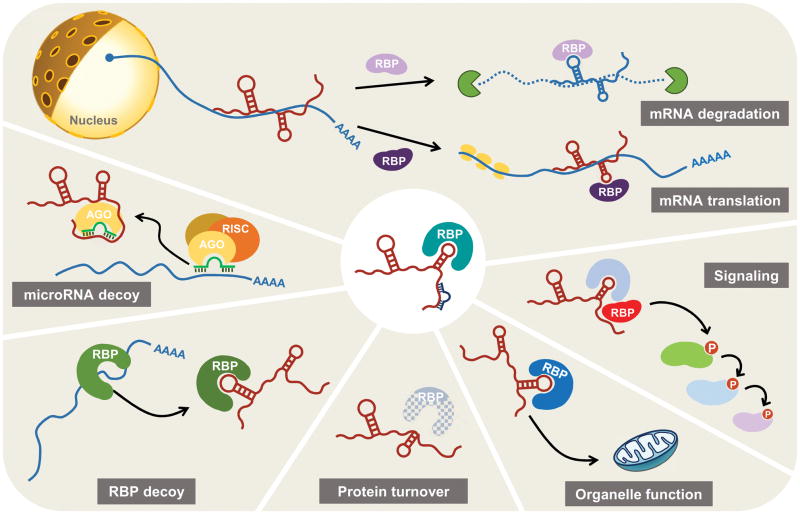
Figure 1. Different levels of regulation of gene expression by cytoplasmic lncRNAs.
(Clockwise). Top, following export to the cytoplasm, lncRNAs can associate with RNA-binding proteins (RBPs) or with partially complementary mRNAs to regulate the stability and/or translation of specific mRNAs. Signaling, association of RBPs with lncRNAs can lead to conformational changes that activate signaling molecules (e.g., kinases). Organelle function, RBPs can chaperone the mobilization of lncRNAs to cellular organelles where they carry out specific functions. Protein stability, lncRNAs can serve as platforms that facilitate the presentation of specific RBPs to the protein degradation machinery. RBP decoy and microRNA decoy, lncRNAs binding to RBPs and microRNAs can reduce the availability of these factors to mRNAs, in turn modulating mRNA fate. See text and Table 1 for details.
LncRNAs controlling mRNA stability and translation
Cytoplasmic lncRNAs can modulate two cytoplasmic processes closely interconnected that impact profoundly on protein production: the turnover and translation of specific mRNAs. As discussed below, this influence may be elicited through the binding of lncRNAs to mRNAs leading to the recruitment of RBPs that promote decay, RBPs that suppress translation, or factors that initiate translation. It may also be mediated via lncRNA-mRNA complexes that prevent microRNAs from binding the intended target mRNAs. These regulatory paradigms were found to influence specific physiologic processes, including muscle homeostasis, Alzheimer’s pathogenesis, and T lymphocyte function. We anticipate that additional mechanisms of lncRNA-mediated translation and turnover of mRNAs will be elucidated as studies in these areas continue to advance.
1/2-sbsRNAs-STAU1, TINCR-STAU1
Intermolecular base-pairing was reported between an Alu element present in a 3′-untranslated region (UTR) and a partially complementary Alu sequence present within a class of lncRNAs named 1/2-sbsRNAs (half STAU1-binding site RNAs). The ensuing double-stranded RNA can recruit the RBP STAU1 (Staufen1), resulting in the degradation of the target mRNAs via a process called STAU1-mediated mRNA decay (SMD, reviewed by Park & Maquat, 2013), which additionally implicates the RNA helicase UPF1. Depletion of 1/2-sbsRNAs led to the upregulation of the respective target mRNAs (Gong & Maquat, 2011).
Terminal differentiation-induced ncRNA (TINCR) is another cytoplasmic lncRNA associating directly with STAU1. Recently, the TINCR-STAU1 lncRNP complex was found to be necessary for epidermal differentiation (Kretz et al., 2013). TINCR interacts with various mRNAs encoding differentiation proteins through a 25-nucleotide ‘TINCR box’ motif that is highly enriched in such mRNAs, and renders the mRNAs stable. Notably, the lack of change in TINCR levels by STAU1 depletion and the direct binding of TINCR to STAU1 without other RNAs such as 1/2-sbsRNA indicates that TINCR is not a direct degradation target of STAU1 (Kretz et al., 2013). This finding also suggests alternative, UPF1/2-independent ways in which STAU1 regulates mRNAs, possibly involving TINCR-STAU1 lncRNP complexes in the cytoplasm.
Recently, Ravel-Chapuis et al. suggested novel rescue functions for STAU1 in myotonic dystrophy type 1 (DM1). Overexpressed STAU1 was found to induce the nuclear export and translation of mRNAs bearing expanded CUG repeats (CUGexp) in noncoding segments, and it rescued the alternative splicing of insulin receptor (IR) pre-mRNA, both of which provide beneficial effects to DM1 (Ravel-Chapuis et al., 2012). Additional transcriptome-wide studies are needed to understand the mechanisms whereby STAU1 affects the fate of coding and noncoding RNAs.
LincRNA-p21-HuR
Although nuclear lincRNA-p21 had been shown to regulate transcription (Huarte et al., 2010; Dimitrova et al. 2014), Yoon et al. reported a model of translational suppression mediated by lincRNA-p21. Cytoplasmic lincRNA-p21 was found to bind the RBP HuR [human antigen R, also known as ELAV-like protein (ELAVL)1] and this interaction led to the recruitment of microRNA let-7 along with the RNA-induced silencing complex (RISC) onto lincRNA-p21, triggering its degradation. In the absence of HuR, lincRNA-p21 was stable, accumulated in the cytoplasm, and was proposed to form partial hybrids through multiple regions of complementarity with target mRNAs including CTNNB1 mRNA [encoding CTNNB1 (β-catenin)] and JUNB mRNA (encoding the transcription factor JUNB). These regions of partial base-paring at coding and noncoding mRNA segments led to the recruitment of translational suppressors RCK/p54 and FMRP and lowered the translation of CTNNB1 and JUNB (Chu & Rana, 2006; Yoon et al., 2012).
BACE1-AS-HuD
BACE1 [β-site amyloid precursor protein (APP)-cleaving enzyme 1] is a crucial enzyme in the pathophysiology of Alzheimer’s disease (AD), as it cleaves APP to generate the toxic peptide Aβ, which can then lead to the formation of amyloid plaques that are the hallmark of AD (Faghihi, Mottagui-Tabar, &Wahlestedt, 2004). As reported earlier, the cytoplasmic function of lncRNA BACE1-AS (BACE1 antisense) is to enhance the stability of BACE1 mRNA through a region of partial complementarity, following exposure to various cell stressors including Aβ42 (Faghihi et al., 2008). Kang and colleagues subsequently found that the cytoplasmic RBP HuD (ELAVL4) (Kasashima, Terashima, Yamamoto, Sakashita, & Sakamoto, 1999), increased the stability of BACE1-AS, as well as the stabilities of BACE1 and APP mRNAs, thereby coordinating several steps that converge on the generation of amyloidogenic Aβ in the AD brain (Kang et al., 2014). Faghihi and coworkers discovered that the base-pairing between BACE1 mRNA and BACE1-AS resulted in the masking of a binding site for miR-485-5p, which would otherwise suppress BACE1 biosynthesis (Faghihi et al., 2010). Together, these findings indicate that the mRNP and lncRNP complexes involving HuD (with APP mRNA, BACE1 mRNA, and BACE1-AS) jointly regulate the processing of APP to Aβ.
GAS5-eIF4E
The cytoplasmic lncRNA GAS5 was previously known to be upregulated in human T cells in response to the mTOR antagonist rapamycin (Mourtada-Maarabouni, Hasan, Farzaneh, & Williams, 2010). Considering the crucial role of mTOR in protein translation, this finding suggested that GAS5 might affect gene expression at the translational level.
GAS5 was subsequently found to regulate negatively the translation of the protein MYC (c-Myc) without affecting the levels of MYC mRNA or the stability of MYC protein. The authors reported that GAS5 was recruited to the translation initiation complex via direct binding to eIF4E. The direct interaction between MYC mRNA and GAS5 in the non-polysomal fraction suggested that GAS5 might decrease the efficiency of the initiation of MYC mRNA translation (Hu, Lou, & Gupta, 2014).
Uchl1-AS1-Uchl1 mRNA
The antisense lncRNA Uchl1-AS1 is predominantly nuclear, but shuttles to the cytoplasm under stress conditions to bind the first 73 nt in the 5′UTR of Uchl1 mRNA. This interaction triggers increased translation of the protein UCHL1 (Carrieri et al., 2012). However, it is not clear if specific RBPs are involved in either the export of Uchl1-AS1 to the cytoplasm or the enhanced Uchl1 mRNA translation.
LncRNAs regulating protein stability
Cytoplasmic lncRNAs may also influence protein expression programs by controlling protein degradation pathways, notably by hindering or enhancing access to the ubiquitin/proteasome machinery. As discussed in this section, examples of processes modulated by lncRNA-regulated protein stability include cellular senescence, the hypoxia response, and viral replication.
LincRNA-p21-HIF-1α
In addition to its function in repressing translation of partially complementary mRNAs (as mentioned above; Yoon et al., 2012), lncRNA-p21 elevation by hypoxic conditions contributed to hypoxia-enhanced glycolysis. Yang and colleagues found that the hypoxia-induced protein HIF1A (HIF-1α) transcriptionally activated lncRNA-p21, which in turn bound to both HIF1A and the von Hippel-Lindau (VHL) protein, thereby preventing VHL-mediated HIF1A ubiquitination and allowing HIF1A to accumulate. They suggested that a positive feedback loop between HIF1A and lincRNA-p21 promoted glycolysis under hypoxia (Yang, Zhang, Mei, & Wu, 2014).
HOTAIR-Snurportin-1, HOTAIR-Ataxin-1
Similar to the above-mentioned mechanisms whereby HuR binding to lncRNA-p21 led to the recruitment of let-7/RISC, HuR binding to the lncRNA HOTAIR, a lncRNA that resides both in the nucleus and the cytoplasm, also led to let-7/RISC-mediated degradation of HOTAIR.38 In conditions of low HuR abundance in cells (e.g., during senescence), HOTAIR was stable and accumulated in the cytosol, serving as a physical substrate for other proteins, including two ubiquitin ligases (DZIP3 and MEX3B) and their respective substrates [Snurportin 1 (SNUPN) and Ataxin-1 (ATXN1)]. Accordingly, when HOTAIR levels were high, SNUPN and ATXN1 were ubiquitinated and degraded, while reductions in HOTAIR allowed SNUPN and ATXN1 to accumulate in human fibroblasts and cancer cells (Yoon et al., 2013).
NRON-CUL4B, NRON-PSMD11
Recently, Li et al. reported that the cytoplasmic lncRNA NRON controlled HIV latency by suppressing the activation of the HIV-1 promoter. However, the authors also found that NRON interacted with the HIV protein Tat and promoted its degradation by recruiting the ubiquitin/proteasome components CUL4B (Cullin 4B) and PSMD11 (Proteasome 26S Subunit, Non-ATPase 11), which also associate with NRON. By forming the lncRNP complex NRON-CUL4B-PSMD11-Tat, NRON promotes the degradation of the essential HIV-1 regulatory protein Tat via the ubiquitin/proteasome system (Li et al., 2016).
LncRNAs regulating RBP availability
Accumulating evidence suggests that cytoplasmic lncRNAs can also serve as ‘decoys’ for RBPs, dissociating RBPs from target mRNAs, and thereby influencing the abundance and translation of such mRNAs. In some cases, the sequestration of RBPs by lncRNAs in the cytoplasm affected the actions of such RBPs in other locales (e.g., transcription in the nucleus). LncRNAs that modulate the molecular and spatial availability of RBPs were found to affect the cell division cycle, the cellular response to damaging agents, the oncogenic response, and the process of muscle differentiation.
gadd7-TDP-43
The lncRNA gadd7 (growth arrest- and DNA damage-inducible gene 7) was first identified in mammalian cells responding to growth inhibition and different types of damage (Fornace, Alamo, & Hollander, 1988; Hollander, Alamo, & Fornace, 1996; Brookheart, Michel, Listenberger, Ory, & Schaffer, 2009). Gadd7 binds TAR DNA-binding protein 43 (TDP-43), an RBP involved in mRNA transcription, processing, and stabilization (Buratti, & Baralle, 2008). Induction of gadd7 in Chinese hamster ovary (CHO)-K1 cells following ultraviolet light irradiation increased gadd7-TDP-43 complexes, in turn preventing the binding of TDP-43 to cyclin-dependent kinase 6 (Cdk6) mRNA and triggering Cdk6 mRNA decay (Liu, Li, Zhang, Guo, & Zhan, 2012).
OIP5-AS1-HuR
OIP5-AS1 (OIP5 antisense RNA 1) is a mammalian lncRNA spanning ~1.9 kb that is abundant in the cytoplasm (van Heesch et al., 2014). First identified as Cyrano in zebrafish, this lncRNA was found to be involved in brain and eye development (Ulitsky, Shkumatava, Jan, Sive, & Bartel, 2011). Recently, Kim et al. found that OIP5-AS1 interacted with cytoplasmic HuR in human cervical carcinoma cells. The authors uncovered a possible sponging function for OIP5-AS1, as it prevented HuR binding to target mRNAs (including those that encode proteins CCNA2, CCND1, and SIRT1), and thus suppressed the HuR-elicited proliferative phenotypes (Kim et al., 2016). In addition, OIP5-AS1 was found to interact with GAK mRNA, promoting GAK mRNA decay and hence reducing GAK protein levels and lowering cell proliferation (Kim et al., 2017). These findings indicate that OIP5-AS1, in HuR-dependent and -independent ways, is capable of regulating the levels of target mRNAs, and thus suppresses cell division.
7SL-HuR
The noncoding RNA 7SL is an essential component of the signal recognition particle (SRP) which transports proteins for secretion or membrane insertion (Walter, Blobel, 1982). 7SL interacts with the 3′UTR of the mRNA encoding the transcription factor and tumor suppressor TP53 (p53), thereby preventing HuR binding to TP53 mRNA and repressing TP53 translation. Downregulation of 7SL enhanced HuR binding to TP53 mRNA, leading to increased TP53 translation and hence triggering senescence, autophagy, and cell cycle arrest (Abdelmohsen et al., 2014). This example illustrates a regulatory paradigm whereby noncoding RNAs associate with mRNAs to modulate RBP binding and influence mRNA fate.
LncMyoD-IMP2
As reported recently by Gong and colleagues, murine lncRNA lncMyoD was directly activated by its neighboring gene Myod and controlled cell-cycle exit during myoblast differentiation. The formation of a novel lncRNP comprised of lncMyoD and IMP2 (IGF2-mRNA-binding protein 2) caused a reduction in IMP2-mediated translation of proliferative proteins such as NRAS and MYC (Gong et al., 2015).
GAS5-GR
Besides the aforementioned function as an inhibitor of translation initiation (Hu G, Lou Z, Gupta M, 2014), lncRNA GAS5 was found to be involved in regulating cell growth by acting as a decoy for glucocorticoid receptor (GR). The transcription factor GR was inhibited through binding to GAS5 in the cytoplasm, which prevented the mobilization of GR to the nucleus and suppressed the transcriptional program mediated by GR (Mourtada-Maarabouni, Pickard, Hedge, Farzaneh, & Williams, 2009).
H19-KSRP
The lncRNA H19 controls imprinting of a conserved cluster of genes that includes IGF2 (insulin-like growth factor 2), which encodes a protein important for cell proliferation, survival, fat metabolism, and fat deposition (Jones, Levorse, &Tilghman, 2001). A recent study revealed that H19 interacts with K homology-type splicing regulatory protein (KHSRP or KSRP) in the cytoplasm of mouse C2C12 myoblasts, in turn causing the destabilization of the Myog mRNA [encoding the muscle protein myogenin (MYOG)], a labile KSRP-target transcript. This lncRNP complex was dissociated by AKT activation, thereby permitting KSRP to bind and stabilize Myog mRNA, and inducing muscle differentiation (Giovarelli et al., 2014).
UCA1-HNRNPI
UCA1 (Urothelial cancer-associated 1) was first identified as an oncogenic lncRNA that was highly expressed in human bladder cancer (Wang et al., 2006). Recently, UCA1 was found to promote breast cancer cell growth by lowering the expression of the tumor suppressor CDKN1B (p27), a protein that is translationally upregulated by HNRNPI (hnRNP I). The reduction in p27 levels was proposed to be mediated, at least in part, via UCA1 binding to HNRNPI, as this interaction prevented the binding of HNRNPI to the 5′UTR of p27 mRNA and suppressed p27 translation. Silencing UCA1 promoted growth arrest by enabling the restoration of p27 translation by HNRNPI (Huang et al., 2014).
In addition, many other lncRNPs modulate the activity of the RBPs in the complex, and thus indirectly affect the outcome of the mRNAs that are targets of such RBPs. For example, Gumireddy and colleagues identified a translational repressor RNA (‘treRNA’) that reduced E-cadherin (CDH1) translation via the actions of three proteins (hnRNPK, FXR1, and FXR2) (Gumireddy et al, 2013). The authors postulated that treRNA was instrumental in assembling these three proteins on the 3′UTR of E-cadherin mRNA, thereby reducing E-cadherin translation, even though treRNA and E-cadherin mRNA did not interact. In another example, Tichon and colleagues linked the actions of lncRNA NORAD (Lee et al., 2016) to the half-lives of mRNAs which are targets of PUM1 and PUM2; the observed effects appeared mediated via the classic mRNA decay-promoting function of Pumilio proteins, and only indirectly by NORAD (Tichon et al., 2016).
LncRNAs as microRNA decoys
Cytoplasmic lncRNAs may also function broadly as competing endogenous (ce)RNAs. By sequestering miRNAs, they reduce their availability to AGO2/RISC and hence to target mRNAs. In this manner, cytoplasmic lncRNAs can relieve numerous instances of miRNA-mediated translational repression. LncRNAs that modulate the availability of microRNAs have been implicated in carcinogenesis and in differentiation programs including myogenesis.
HULC
The lncRNA HULC (‘highly upregulated in liver cancer’) is processed like mRNA as it undergoes splicing and polyadenylation (Panzitt et al., 2007). The microRNA miR-372 is known to bind to PRKACB (protein kinase cAMP-activated catalytic subunit beta) mRNA and repress its translation. HULC acts as a microRNA decoy or sponge for miR-372 and thereby enhances PRKACB mRNA translation. Interestingly, PRKACB phosphorylates and activates the transcription factor CREB (cAMP-responsive element-binding protein), which promotes the transcription of HULC (Wang et al., 2010).
LincRNA-RoR
LincRNA-RoR also functions as a microRNA sponge in embryonic stem cells, controlling the levels of miR-145 available to repress production of the transcription factors and regulators of differentiation NANOG, OCT4, and SOX2 (Loewer et al., 2010; Wang et al., 2013).
H19
Besides binding RBPs, as mentioned in the previous section (Giovarelli et al., 2014), H19 acts as a sponge for microRNA let-7 in HEK293 cells, and was thus proposed to be pro-oncogenic. Accordingly, depleting H19 increased let-7 activity and muscle differentiation in C2C12 cells (Kallen et al., 2013). Interestingly, H19 is also the host of microRNA miR-675, which suppresses the production of the tumor suppressor PTEN, further enhancing the oncogenic function of H19 (Cai & Cullen, 2007).
Linc-MD1
The cytoplasmic, muscle-specific lncRNA linc-MD1 was previously known to act as a competitor endogenous (ce)RNA for miR-133 and miR-135; miR-133 reduces the levels of the protein mastermind-like-1 (MAML1), miR-135 lowers the abundance of myocyte-specific enhancer factor 2C (MEF2C). In this paradigm, expression of linc-MD1 increased the levels MAML1 and MEF2C through the inhibition of microRNA activity (Cesana et al., 2011). Recently, Legnini et al. proposed a novel feedforward regulatory mechanism involving HuR and linc-MD1 in early phases of muscle differentiation. Intriguingly, HuR was found to interact with both linc-MD1 and miR-133b and was proposed to recruit miR-133b onto linc-MD1 in the cytoplasm. This paradigm was further complicated by the repression of HuR production by miR-133. Cytoplasmic HuR increased the sponging activity of linc-MD1 and inhibited the generation of miR-133b from a precursor sequence embedded in linc-MD1 by interfering with the access of DROSHA, an RNase involved in processing microRNAs. In later differentiation stages, HuR levels declined as miR-133 levels rose, triggering its exit from the regulatory circuit (Legnini, Morlando, Mangiavacchi, Fatica, & Bozzoni, 2014).
PTENP1
LncRNA PTENP1, expressed from a PTEN pseudogene, is a well-recognized sponge of several microRNAs, including miR-21, miR-20a, miR-214, miR-19b, miR-26a, and miR-499-5p. As reviewed by Tang, Ning, Zeng, & Li (2016), these microRNAs also target the PTEN 3′UTR and repress production of the tumor suppressor PTEN. By sponging this group of microRNAs, lncRNA PTENP1 derepresses PTEN production and enables its tumor suppressor function.
Subcellular mobilization of lncRNAs
LncRNAs can shuttle to various subcellular locations, sometimes leading to their accumulation in specific cellular compartment (van Heesch et al. 2014; Cabili et al. 2015). The studies discussed in this section reveal that the local interaction of lncRNAs with RBPs is critical for retaining structural and functional integrity of subcellular organelles such as mitochondria, as well as for T cell function.
NRON-NFAT
A decade ago, Willingham et al. suggested a novel function for a cytoplasmic lncRNA (now named NRON) that regulated the nuclear translocation of nuclear factor of activated T cells (NFAT) and involved importin-β (Willingham et al. 2005). This example was among the first to illustrate the shuttling of lncRNAs among subcellular compartments. As mentioned above, NRON was subsequently implicated in HIV replication (Li et al., 2016).
MALAT1-HNRNPC
Recent efforts to create a ‘subcellular atlas’ have made it possible to classify a number of lncRNAs according to their subcellular localization. MALAT1 was known to be predominantly nuclear, but MALAT1 transcripts were recently found to enter the cytoplasm in the G2/M phase. In this regard, the RBP HNRNPC was found to form a lncRNP with MALAT1 that mediated its cytoplasmic translocation during the G2/M phase transition (Yang, Yi, Ha, Du, & Liang, 2013), providing an example of lncRNPs that promote the timely mobilization of lncRNAs and RBPs to elicit appropriate cellular functions.
RMRP-HuR, RMRP-GRSF1
In another recent study, Noh et al. reported two RBPs that contributed to the subcellular mobilization of the organelle-specific localization of lncRNP complexes. A lncRNA abundant in the nucleus, RMRP (an RNA component of the mitochondrial RNA-processing endoribonuclease) associated with the RBP HuR in the nucleus, and the resulting lncRNP was mobilized into the cytoplasm through CRM1 (chromosome region maintenance 1)-dependent nuclear export. This exported RMRP was selectively targeted to the innermost compartment of mitochondria (the matrix), where the mitochondria resident protein GRSF1 (G-rich RNA sequence-binding factor 1) associated with RMRP and formed a mitochondrial lncRNP complex, RMRP-GRSF1, which was proposed to contribute to the maintenance of mitochondrial structure as well as functions such as oxidative phosphorylation and mitochondrial DNA replication (Noh et al., 2016).
RMRP-PNPASE, RPPH1-PNPASE
The protein PNPASE, which resides in the mitochondrial intermembrane space (IMS), was found to mobilize noncoding RNAs such as RPPH1 and RMRP, encoded by nuclear DNA but found in mitochondria (Mercer et al., 2011). PNPASE recognized a particular stem-loop structure embedded in RPPH1 and RMRP and promoted RNA import into the mitochondrial matrix (Wang et al., 2010). It is still unclear what factors mediate the physical import of RNA through the inner membrane (IM), but PNPASE was proposed to capture RNAs in the IMS and transferred them to complexes that delivered RNAs through the IM into the matrix.
LncRNAs modulating signaling pathways
Cytoplasmic lncRNAs capable of modulating signal transduction pathways by binding specific signaling molecules and/or altering their phosphorylation status are also beginning to be recognized. Although the proteins implicated in these regulatory paradigms notably lack canonical RNA-binding domains (RBDs), they can play prominent functions in physiological and pathological conditions. Their functions have been studied primarily in the immune and inflammatory responses, as well as in cancer.
LINK-A-BRK and LINK-A-LRRK2
A novel function was recently proposed for the cytoplasmic lncRNA LINK-A (long intergenic non-coding RNA for kinase activation) in the signal transduction triggered by heparin-binding epithelial growth factor (HB-EGF). LINK-A recruited the breast tumor kinase BRK to the dimer EGFR-GPNMB (EGF receptor-transmembrane glycoprotein NMB), helping to activate BRK. LINK-A also interacted with the kinase LRRK2, in turn triggering the conformational change, phosphorylation, and activation of BRK; these changes subsequently led to normoxic HIF1A stabilization in triple-negative breast cancer cells (Lin et al., 2016).
Lnc-DC-STAT3
Wang et al. reported a novel lncRNP complex that affected cytoplasmic signaling. The lncRNA lnc-DC, which is essential for the differentiation of dendritic cells, directly bound to cytoplasmic STAT3 (signal transducer and activator of transcription 3) and promoted phosphorylation of STAT3 by preventing the inhibitory function of SHP1 [tyrosine phosphatase Src-homology 2 domain (SH2)-containing PTP-1] (Wang et al., 2014).
NKILA-NF-κB, Lethe-NF-κB
The NF-κB pathway is essential for the transcriptional induction of several mRNAs encoding proteins involved in inflammation and cell survival. Liu et al. reported a novel cytoplasmic lncRNA, NKILA (NF-κB-interacting long noncoding RNA), which is upregulated by inflammatory cytokines and by NF-κB activity in tumor cells. They also found that NKILA negatively regulated NF-κB signaling by associating with the NF-κB/IκB complex, masking the phosphorylation sites of IκB and preventing NF-κB signaling (Liu et al., 2015). Another lncRNA called Lethe was found to bind RELA (a member of the NF-κB/Rel family), which inhibited RELA binding to DNA, thus reducing the transcriptional activity of RELA (Rapicavoli et al., 2013). These findings indicate that several lncRNAs can interfere with the NF-κB pathway and thus influence gene expression in inflammation and carcinogenesis.
Remaining challenges in cytoplasmic lncRNA biology
Despite much progress in elucidating the constituents and functions of cytoplasmic lncRNPs, major questions remain. In this section, we point to key emerging areas of lncRNP biology awaiting in-depth study.
LncRNAs associated with ribosomes
A number of lncRNAs (e.g. H19 and TUG1) have been found enriched in the polysomal fraction.45 For example, the C. elegans lncRNA tts-1 (transcribed telomeric sequence 1) was found to extend the life span of the long-lived daf-2 mutant (reviewed by Kenion, 2010) in a manner dependent on its ribosome-binding activity (Essers et al., 2015), and Ingolia and co-workers found many lncRNAs directly associated with the translation machinery in mammalian cells (Ingolia, Lareau, Weissman, 2011). Subsequent studies similarly identified lncRNAs associated with ribosomes and likely to be translated into short peptides (van Heesch et al., 2014;. Bazzini et al., 2014; Ruiz-Orera, Messeguer, Subirana, & Alba, 2014; Ingolia et al., 2014). Carlevaro-Fita and colleagues recently identified ~70% of cytoplasmic lncRNAs in polysomes, ~30% in the free cytoplasmic fraction (Carlevaro-Fita et al., 2016). They also found that the presence of exonic transposable elements (TEs) in cytoplasmic lncRNAs correlates with their ribosome-free localization, supporting previous observations that TEs are functional lncRNA sequences (Carlevaro-Fita et al., 2016; Kelley & Rinn, 2012; Johnson & Guigo R; 2014). It will be important to identify systematically the lncRNAs associated with polysomes, elucidate the lncRNA regions that control these associations, and assess systematically whether they are translated and/or have other functions.
Circular RNAs
In eukaryotes, numerous circular (circ)RNAs have been identified, many of them derived from back-spliced exons (Ebbesen, Kjems, & Hansen, 2016), capable of binding cytoplasmic molecules including RBPs and microRNAs. For example, CircPABPN1 was a strong target of the RBP HuR; accordingly, high levels of CircPABPN1 reduced HuR’s ability to bind PABPN1 mRNA, in turn lowering PABPN1 expression in cancer cells (Abdelmohsen et al., 2017). Although the exact mechanisms governing these interactions are not known, CircPABPN1 originates from exon 6 of PABPN1 mRNA and the sequence similarity might be linked to their competitive binding (Abdelmohsen et al., 2017). In another example, circRNA ciRS-7, which contains ~70 sites of interaction with miR-7, sponged and suppressed miR-7 function in brain and islet cells (Memczak et al., 2013; Xu, Guo, Li, & Yu, 2015). Similarly, circRNA Sry (sex-determining region Y) was found to sponge miR-138, suppressing miR-138 effect on target mRNAs (Hansen et al., 2013).
Examples are also emerging of circRNAs that originate from linear lncRNAs. For instance, CircPVT1, generated from the oncogenic lncRNA PVT1, was recently identified as a senescence-associated circular RNA (SAC-RNA). CircPVT1 was low in senescent WI-38 human fibroblasts, and its high abundance in proliferating WI-38 cells was linked to the binding and hence the functional suppression of let-7, the inhibition of cell senescence, and the enhancement of cell division (Panda et al., 2017). CircPVT1 also regulates chondrocyte apoptosis in osteoarthritis by acting as a sponge for miR-488-3p (Li, Li, Luo, Liu, & Yu, 2017).87 Given the important functions elicited by circRNAs, building a complete catalog of circRNPs will be critical towards a fuller understanding of lncRNP biology.
Dynamic, tissue-specific maps of lncRNPs
Similar to the proposal by Batista and Chang that lncRNAs are key components of nuclear architecture and function (Batista and Chang, 2013), spatio-temporal changes of cytoplasmic lncRNAs are essential for regulating various cellular events. As summarized above, many cytoplasmic lncRNAs function by interacting with RBPs, indicating that their various cytoplasmic roles are mostly attributed to lncRNP activity. However, the complete catalogs of RBPs interacting with each lncRNA, and their competitive, cooperative, or sequential binding to form diverse lncRNPs, are unknown. Considering that the functions of most lncRNAs are not yet known, creating a comprehensive inventory of lncRNAs and RBPs that constitute cytoplasmic lncRNPs will critically advance our knowledge of the cytoplasmic lncRNA functions.
Resources to study lncRNP identity and function
Even though mRNA-RBP interactions (mRNPs) have been identified for decades (Gerstberger, Hafner, & Tuschl, 2014), comprehensive maps of lncRNPs will require advances in both wet-lab and computational methods. Crosslinking and immunoprecipitation (CLIP, including its variants HITS-CLIP, PAR-CLIP, iCLIP, eCLIP, etc), followed by RNA-seq analysis, allows state-of-the-art mapping transcriptome-wide RNA-binding sites of an RBP of interest (Konig, Zarnack, Luscombe, & Ule, 2011). Large CLIP-derived datasets were generated recently from over 100 RBPs (http://clipdb.ncrnalab.org) (Licatalosi et al., 2008; Hafner et al., 2010; Konig et al., 2010; Yang et al., 2015). For example, TARDBP (TAR DNA-binding protein 43, TDP-43), a DNA/RNA-binding protein associated with the neurodegenerative diseases amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Onesto et al., 2016), showed increased disease-associated localization in mitochondria and strong binding to mitochondrial ND3 and ND6 mRNAs, suppressing their expression and causing disassembly of the mitochondrial complex I. The authors proposed that interfering with the mitochondrial localization of TDP-43 could have therapeutic benefit in neurodegeneration (Wang et al., 2016). Together with other CLIP datasets it is clear that under certain conditions, TDP-43 can interact with cytoplasmic lncRNAs, with nucleus-retained lncRNAs (e.g., MALAT1 and NEAT1), and with mRNAs encoding neurodegeneration-associated proteins (e.g., FUS, progranulin) (Tollervey et al., 2011; Polymenidou et al., 2011).
Individual lncRNPs can be identified experimentally, but this is still a costly proposition and depends on the availability of good antibodies. To identify lncRNPs computationally, many databases have been developed to provide information on RBP target RNAs and RNA-binding motifs using validated protein-RNA interactions (Cook, Kazan, Zuberi, Morris, & Hughes, 2011; Giulietti et al., 2013; Ray et al., 2013; Park, Yu, Choi, Kim, & Lee, 2014; Giudice, Sanchez-Cabo, Torroja, & Lara-Pezzi E, 2016). A recent attempt to construct a global lncRNA-protein network (LPN) based on experimentally verified functional interactions among lncRNAs and interacting RBPs revealed that lncRNA-binding proteins have distinct topological properties (Shang et al., 2015). We propose that developing accurate computational algorithms to predict lncRNP interactions will greatly enhance our understanding of cytoplasmic lncRNA functions systematically.
Subcellular detection of endogenous lncRNAs
Eukaryotic cells have evolved many cellular domains and compartments, including membrane-enclosed cytoplasmic organelles. Given that RNA functions are closely linked to their subcellular localization, it is critical to investigate their subcellular distribution. Recent advances in fractionation and RNA-seq technologies have uncovered vast numbers of lncRNAs in specific cellular compartments, suggesting that lncRNAs may be mobilized into different cytoplasmic regions, including structured subcellular organelles (e.g. ribosomes, endoplasmic reticulum, exosomes, mitochondria, etc.) and more dynamic domains such as processing bodies or stress granules (Mercer et al., 2011; Guttman, & Rinn, 2012; Guttman, Russell, Ingolia, Weissman, & Lander, 2013; Halic et al., 2004; Rackham et al., 2011; Turner, Galloway, & Vigorito, 2014).
However, the accurate localization of cytoplasmic lncRNAs requires in situ detection methods. Nelles et al. detected endogenous RNAs by using a nuclease-inactive CRISPR/Cas9 system that comprised an RNA-directed Cas9 (RCas9) and an RNA-specific guide RNA (sgRNA) that tracked endogenous mRNAs (Nelles et al., 2016). Compared with earlier methods that employed genetically manipulated aptamers such as MS2-MCP (MS2-coat protein; reviewed by Weil, Parton, & Davis, 2010), RCas9 has the advantage of tracking ‘tag-less’ endogenous RNAs. It remains to be seen whether this method can detect low-abundance lncRNAs and whether it is superior for live-cell imaging compared with conventional RNA FISH or tagged RNA detection. An alternative form of RNA tagging was recently suggested by using another partner of CRISPR, Cas13a (Abudayyeh et al., 2016; East-Seletsky, O’Connell, Burstein, Knott, & Doudna, 2017; Gootenberg et al., 2017). Considering that its major advantage over RCas9 is that the Cas13a system does not require the additional oligonucleotide ‘PAMmer’, Cas13a appears likely to become a powerful tool for targeting and detecting endogenous lncRNAs.
Subcellular and suborganellar distribution of lncRNPs
An earlier study revealed that all organisms can import tRNAs from the cytoplasm to mitochondria (Rubio et al., 2008). tRNAs appeared to be actively delivered to the mitochondrial surface instead of passively reaching mitochondria after being released into the cytoplasm (Yoshihisa, Yunoki-Esaki, Ohshima, Tanaka, & Endo, 2003). A recent proteomic analysis of tRNA export to the cytosol identified sets of proteins necessary for mediating the nuclear export and mitochondrial import of tRNAs, including nuclear export machineries, actin cytoskeleton components, and mitochondrial outer membrane proteins (Wu, Bao, Chatterjee, Wan, & Hopper, 2015). For lncRNAs, it is still unknown what RBPs might mobilize them out of the nucleus and onto the surface of organelles such as mitochondria, or into organelles. The specific lncRNA sequences required for subcellular transport are also unknown.
Exosomes are small vesicles, with diameters of 30–120 nm, secreted by most cell types and present in all body fluids. They have been found to carry lncRNAs, proteins, DNA, microRNAs, and other molecules (Nolte-‘t Hoen et al., 2012; Gezer, Ozgur, Cetinkaya, Isin, & Dalay, 2014; Ahadi, Brennan, Kennedy, Hutvagner, & Tran, 2016). Although it is not yet known how the contents of this membrane-enclosed structure are loaded, specific RBPs are believed to form lncRNPs in order to deliver cytoplasmic lncRNAs to exosomes.
As we gain more information about the specific cellular distribution of lncRNAs, we will be able to integrate and predict the functions of specific lncRNPs. Guo et al. assembled all the known public databases for protein subcellular location (Guo, Liu, Ju, Wang, & Wang, 2016), and Itzhak et al. proposed a ‘global dynamic map’, which included physical distribution and movement of proteins in cells. This proteomic view of the cell, which includes large protein complexes (ribosomes and proteasome) and cellular organelles (e.g. membrane, ER, Golgi apparatus, endosome, lysosome, peroxisome and mitochondria) (Itzhak, Tyanova, Cox, & Borner, 2016), can guide our efforts to map cellular lncRNPs in dynamic spaces.
Structural and chemical aspects of lncRNAs
The secondary and tertiary structures of lncRNAs are more highly conserved than their primary sequence, and thus they are considered to be more relevant to lncRNA biological function (Johnsson, Lipovich, Grander, & Morris, 2014; Mercer, & Mattick, 2013). Once mobilized to the cytoplasm, structural changes in lncRNAs may affect their function and/or binding to target proteins. Therefore, analyzing their structures in each compartment appears to be essential for understanding lncRNA function. Some computational methods have been developed to predict RNA secondary (or tertiary) structure (Yan et al., 2016), and experimental techniques have been designed to probe RNA structures in the whole transcriptome (i.e., the ‘RNA structurome’) (Wan, Kertesz, Spitale, Segal, & Chang, 2011; Ding et al., 2014; Novikova, Dharap, Hennelly, & Sanbonmatsu, 2013. As we use this approach to study lncRNPs, we should be mindful of post-transcriptional modifications of RNAs. For example, a recent report uncovered that N6-methyladenosine (m6A) altered the local structure of mRNA and lncRNA globally, thereby modifying their interaction with an RBP, HNRNPC (Liu et al., 2015).
Closing remarks
Several thousand human lncRNAs have been identified, but their potential roles and subcellular localizations are largely unknown. Many studies in recent years have solidified the idea that nuclear lncRNPs function as ‘localization elements’ to define nuclear domains with key roles in maintaining structural and functional integrity of the nucleus. As discussed above, these concepts can be extended to cytoplasmic lncRNA functions, such as maintaining cytoplasmic structure, defining cytoplasmic domains, and promoting organelle activity. In other words, the distribution of cytoplasmic lncRNPs may be controlled dynamically in time and space, with lncRNPs delivered to specific cytoplasmic locations requiring certain enzymatic activities and/or structural platforms.
A comprehensive understanding of the composition and localization of cytoplasmic lncRNPs will set the stage for exploring the biological functions of cytoplasmic lncRNAs. As we understand the cellular processes controlled by lncRNAs, we can begin to devise strategies to intervene therapeutically in pathological situations in which such processes become impaired.
Acknowledgments
This work was supported entirely by the National Institute on Aging Intramural Research Program, National Institutes of Health. We thank J. L. Martindale for critical reading of the manuscript.
Footnotes
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- Abdelmohsen K, Panda AC, Kang MJ, Guo R, Kim J, Grammatikakis I, … Gorospe M. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Research. 2014;42:10099–10111. doi: 10.1093/nar/gku686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, … Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biology. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, … Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahadi A, Brennan S, Kennedy P, Hutvagner G, Tran N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Scientific Reports. 2016;6:24922. doi: 10.1038/srep24922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Dominguez JR, Lodish HF. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130:1965–1975. doi: 10.1182/blood-2017-06-788695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audas TE, Lee S. Stressing out over long noncoding RNA. Biochim Biophys Acta. 2016;1859:184–191. doi: 10.1016/j.bbagrm.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballarino M, Morlando M, Fatica A, Bozzoni I. Non-coding RNAs in muscle differentiation and musculoskeletal disease. Journal of Clinical Investigation. 2016;126:2021–2030. doi: 10.1172/JCI84419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, … Giraldez AJ. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO Journal. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookheart RT, Michel CI, Listenberger LL, Ory DS, Schaffer JE. The non-coding RNA gadd7 is a regulator of lipid-induced oxidative and endoplasmic reticulum stress. Journal of Biological Chemistry. 2009;284:7446–7454. doi: 10.1074/jbc.M806209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, … Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biology. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlevaro-Fita J, Rahim A, Guigo R, Vardy LA, Johnson R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA. 2016;22:867–882. doi: 10.1261/rna.053561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, … Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, … Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, … Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nature Immunology. 2017;18:962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biology. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Molecular Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic Acids Research. 2011;39:D301–308. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, … Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, … Jacks T. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Molecular Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochimica Biophysica Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, Doudna JA. RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Molecular Cell. 2017;66:373–383. e373. doi: 10.1016/j.molcel.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers PB, Nonnekens J, Goos YJ, Betist MC, Viester MD, Mossink B, … MacInnes AW. A Long Noncoding RNA on the Ribosome Is Required for Lifespan Extension. Cell Reports. 2015;S2211:1247. doi: 10.1016/j.celrep.2014.12.029. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, … Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nature Medicine. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Mottagui-Tabar S, Wahlestedt C. Genetics of neurological disorders. Expert Reviews in Molecular Diagnostics. 2004;4:317–332. doi: 10.1586/14737159.4.3.317. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, … Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biology. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace AJ, Jr, Alamo I, Jr, Hollander MC. DNA damage-inducible transcripts in mammalian cells. Proceedings of the National Academy of Sciences, USA. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nature Review Genetics. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gezer U, Ozgur E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biology International. 2014;38:1076–1079. doi: 10.1002/cbin.10301. [DOI] [PubMed] [Google Scholar]
- Giovarelli M, Bucci G, Ramos A, Bordo D, Wilusz CJ, Chen CY, … Gherzi R. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proceedings of the National Academy of Sciences USA. 2014;111:E5023–5028. doi: 10.1073/pnas.1415098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice G, Sanchez-Cabo F, Torroja C, Lara-Pezzi E. Database. Oxford: 2016. ATtRACT-a database of RNA-binding proteins and associated motifs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulietti M, Piva F, D’Antonio M, D’Onorio De Meo P, Paoletti D, Castrignano T, … Pesole G. SpliceAid-F: a database of human splicing factors and their RNA-binding sites. Nucleic Acids Research. 2013;41:D125–131. doi: 10.1093/nar/gks997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S, Glass DJ. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Developmental Cell. 2015;34:181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, … Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikakis I, Panda AC, Abdelmohsen K, Gorospe M. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging. 2014;6:992–1009. doi: 10.18632/aging.100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco S, Gorospe M, Martelli F. Noncoding RNA in age-related cardiovascular diseases. Journal of Molecular and Cellular Cardiology. 2015;83:142–155. doi: 10.1016/j.yjmcc.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Yan J, Setoyama T, Johannes GJ, Orom UA, … Huang Q. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO Journal. 2013;32:2672–84. doi: 10.1038/emboj.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Liu F, Ju Y, Wang Z, Wang C. Human protein subcellular localization with integrated source and multi-label ensemble classifier. Scientific Reports. 2016;6:28087. doi: 10.1038/srep28087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, … Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genetics. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hollander MC, Alamo I, Fornace AJ., Jr A novel DNA damage-inducible transcript, gadd7, inhibits cell growth, but lacks a protein product. Nucleic Acids Research. 2996;24:1589–1593. doi: 10.1093/nar/24.9.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Lou Z, Gupta M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS One. 2014;9:e107016. doi: 10.1371/journal.pone.0107016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, Mo YY. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death and Disease. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, … Rinn J. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, … Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature Genetics. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, … Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Reports. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhak DN, Tyanova S, Cox J, Borner GH. Global, quantitative and dynamic mapping of protein subcellular localization. Elife. 2016;5 doi: 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochimica Biophysica Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Guigo R. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. RNA. 2014;20:959–976. doi: 10.1261/rna.044560.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BK, Levorse J, Tilghman SM. Deletion of a nuclease-sensitive region between the Igf2 and H19 genes leads to Igf2 misregulation and increased adiposity. Human Molecular Genetics. 2001;10:807–814. doi: 10.1093/hmg/10.8.807. [DOI] [PubMed] [Google Scholar]
- Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, … Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Molecular Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Abdelmohsen K, Hutchison ER, Mitchell SJ, Grammatikakis I, Guo R, … Gorospe M. HuD regulates coding and noncoding RNA to induce APP-->Abeta processing. Cell Reports. 2014;7:1401–1409. doi: 10.1016/j.celrep.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasashima K, Terashima K, Yamamoto K, Sakashita E, Sakamoto H. Cytoplasmic localization is required for the mammalian ELAV-like protein HuD to induce neuronal differentiation. Genes Cells. 1999;4:667–683. doi: 10.1046/j.1365-2443.1999.00292.x. [DOI] [PubMed] [Google Scholar]
- Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biology. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kim J, Noh JH, Lee SK, Munk R, Sharov A, Lehrmann E, … Gorospe M. LncRNA OIP5-AS1/cyrano suppresses GAK expression to control mitosis. Oncotarget. 2017;8:49409–49420. doi: 10.18632/oncotarget.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Abdelmohsen K, Yang X, De S, Grammatikakis I, Noh JH, Gorospe M. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Research. 2016;44:2378–2392. doi: 10.1093/nar/gkw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nature Reviews Genetics. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, … Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature Structural & Molecular Biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, … Khavari PA. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel JF, Goodrich JA. Non-coding RNAs: key regulators of mammalian transcription. Trends in Biochemical Sciences. 2012;37:144–151. doi: 10.1016/j.tibs.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, … Mendell JT. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Molecular Cell. 2014;53:506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen C, Ma X, Geng G, Liu B, Zhang Y, … Zhang H. Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation. Nature Communications. 2016;7:11730. doi: 10.1038/ncomms11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO Journal. 2012;31:4415–4427. doi: 10.1038/emboj.2012.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tian H, Yang J, Gong Z. Long Noncoding RNAs Regulate Cell Growth, Proliferation, and Apoptosis. DNA Cell Biology. 2016;35:459–470. doi: 10.1089/dna.2015.3187. [DOI] [PubMed] [Google Scholar]
- Li Y, Li S, Luo Y, Liu Y, Yu N. LncRNA PVT1 Regulates Chondrocyte Apoptosis in Osteoarthritis by Acting as a Sponge for miR-488-3p. DNA Cell Biology. 2017;36:571–580. doi: 10.1089/dna.2017.3678. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, … Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, … Yang L. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nature Cell Biology. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, … Song E. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, … Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genetics. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, … Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, … Mattick JS. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature Structure & Molecular Biology. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Montes M, Lund AH. Emerging roles of lncRNAs in senescence. FEBS Journal. 2016;283:2414–2426. doi: 10.1111/febs.13679. [DOI] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Hasan AM, Farzaneh F, Williams GT. Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5) Molecular Pharmacology. 2010;78:19–28. doi: 10.1124/mol.110.064055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 2016;165:488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, … Gorospe M. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes & Development. 2016;30:1224–1239. doi: 10.1101/gad.276022.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte-‘t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, ‘t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Research. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova IV, Dharap A, Hennelly SP, Sanbonmatsu KY. 3S: shotgun secondary structure determination of long non-coding RNAs. Methods. 2013;63:170–177. doi: 10.1016/j.ymeth.2013.07.030. [DOI] [PubMed] [Google Scholar]
- Onesto E, Colombrita C, Gumina V, Borghi MO, Dusi S, Doretti A, … Ratti A. Gene-specific mitochondria dysfunctions in human TARDBP and C9ORF72 fibroblasts. Acta Neuropathologica Communications. 2016;4:47. doi: 10.1186/s40478-016-0316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda AC, Grammatikakis I, Kim KM, De S, Martindale JL, Munk R, … Gorospe M. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Research. 2017;45:4021–4035. doi: 10.1093/nar/gkw1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, … Zatloukal K. lncRNAtor: a comprehensive resource for functional investigation of long non-coding RNAs. Bioinformatics. 2014;30:2480–2485. doi: 10.1093/bioinformatics/btu325. [DOI] [PubMed] [Google Scholar]
- Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscipinary Reviews RNA. 2013;4:423–435. doi: 10.1002/wrna.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, … Cleveland DW. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nature Neuroscience. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O, Shearwood AM, Mercer TR, Davies SM, Mattick JS, Filipovska A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA. 2011;17:2085–2093. doi: 10.1261/rna.029405.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid F, Shah A, Shan G. Long Non-coding RNAs in the Cytoplasm. Genomics Proteomics Bioinformatics. 2016;14:73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel-Chapuis A, Belanger G, Yadava RS, Mahadevan MS, DesGroseillers L, Cote J, Jasmin BJ. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. Journal of Cell Biology. 2012;196:699–712. doi: 10.1083/jcb.201108113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, … Hughes TR. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MA, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Soll D, Alfonzo JD. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proceedings of the National Academy of Sciences USA. 2008;105:9186–9191. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. Long non-coding RNAs as a source of new peptides. Elife. 2014;3:e03523. doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang D, Yang H, Xu Y, Yao Q, Zhou W, Shi X, … Li X. A global view of network of lncRNAs and their binding proteins. Molecular Biosystems. 2015;11:656–663. doi: 10.1039/c4mb00409d. [DOI] [PubMed] [Google Scholar]
- Tang J, Ning R, Zeng B, Li Y. Molecular Evolution of PTEN Pseudogenes in Mammals. PLoS ONE. 2016;11:e0167851. doi: 10.1371/journal.pone.0167851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichon A, Gil N, Lubelsky Y, Havkin Solomon T, Lemze D, Itzkovitz S, Stern-Ginossar N, Ulitsky I. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nature Communications. 2016;7:12209. doi: 10.1038/ncomms12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, … Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genetics. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, … Ule J. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nature Neuroscience. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nature Immunology. 2014;15:484–491. doi: 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, … Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biology. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nature Reviews Genetics. 2011;12:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, … Teitell MA. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–67. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, … Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Research. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Xiao L, Wang JY. Posttranscriptional regulation of intestinal epithelial integrity by noncoding RNAs. Wiley Interdisciplinary Reviews RNA. 2017:8. doi: 10.1002/wrna.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, … Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- Wang W, Wang L, Lu J, Siedlak SL, Fujioka H, Liang J, … Wang X. The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nature Medicine. 2016;22:869–878. doi: 10.1038/nm.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, … Chen WF. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clinical Cancer Research. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, … Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Develomental Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Weil TT, Parton RM, Davis I. Making the message clear: visualizing mRNA localization. Trends in Cell Biology. 2010;20:380–390. doi: 10.1016/j.tcb.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Wu J, Bao A, Chatterjee K, Wan Y, Hopper AK. Genome-wide screen uncovers novel pathways for tRNA processing and nuclear-cytoplasmic dynamics. Genes & Development. 2015;29:2633–2644. doi: 10.1101/gad.269803.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Scientific Reports. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Arfat Y, Li D, Zhao F, Chen Z, Yin C, … Qian A. Structure Prediction: New Insights into Decrypting Long Noncoding RNAs. International Journal Molecular Science. 2016:17. doi: 10.3390/ijms17010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Yi F, Han X, Du Q, Liang Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Letters. 2013;587:3175–3181. doi: 10.1016/j.febslet.2013.07.048. [DOI] [PubMed] [Google Scholar]
- Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Molecular Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Yang YC, Di C, Hu B, Zhou M, Liu Y, Song N, … Lu ZJ. CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC Genomics. 2015;16:51. doi: 10.1186/s12864-015-1273-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Molecular Biology of the Cell. 2003;14:3266–3279. doi: 10.1091/mbc.E02-11-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. Journal of Molecular Biology. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, … Gorospe M. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nature Communications. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, … Gorospe M. LincRNA-p21 suppresses target mRNA translation. Molecular Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]