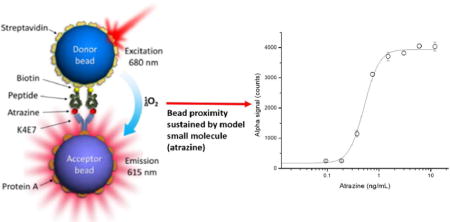
Abstract
Our group has previously developed immunoassays for noncompetitive detection of small molecules based on the use of phage borne anti-immunocomplex peptides. Recently, we substituted the phage particles by biotinylated synthetic anti-immunocomplex peptides complexed with streptavidin, and named these constructs nanopeptamers. In this work, we report the results of combining AlphaLisa, a commercial luminescent oxygen channeling bead system, with nanopeptamers for the development of a noncompetitive homogeneous assay for the detection of small molecules. The signal generation of AlphaLisa assays relies on acceptor-donor bead proximity induced by the presence of the analyte (a macromolecule) simultaneously bound by antibodies immobilised on the surface of these beads. In the developed assay, termed as nanoAlphaLisa, bead proximity is sustained by the presence of a small model molecule (Atrazine, MW = 215) using an anti-atrazine antibody captured on the acceptor bead, and an atrazine nanopeptamer on the donor bead. Atrazine, is one of the most used pesticides worldwide and its monitoring in water has relevant human health implications. NanoAlphaLisa allowed the homogeneous detection of atrazine down to to 0.3 ng/mL in undiluted water samples in one hour, which is ten-fold below the accepted limit in drinking water. NanoAlphaLisa has the intrinsic advantages for automation, high-throughput, simple, and fast homogeneous detection of target analytes that AlphaLisa assay provides.
Graphical abstract
INTRODUCTION
Immunoassays are simple, robust, inexpensive analytical techniques based on the use of antibodies for detecting molecules of interest. One of the most known immunoassay formats is the Enzyme Linked Immunosorbent Assay (ELISA), which is considered a heterogeneous method because it requires several washing steps in between addition of reagents. On the other hand, mix and measure assays, which do not require washing steps because the reagents and sample are mixed and the readout measured after a short incubation period are classified as ’homogeneous’. Homogeneous assays are simpler to perform, take less time and are easier to adapt to high throughput and automation than their heterogeneous counterparts.
In 2008, Perkin Elmer Inc, first commercialized a luminescent oxygen channeling chemistry assay1,2 termed Amplified Luminescent Proximity Homogeneous Assay (AlphaLISA). In this assay, acceptor and donor beads bound to antibodies that recognize different epitopes of the antigen (typically a macromolecule) are brought together when the antigen is present, in a “sandwich” like format. Laser irradiation of donor beads at 680 nm generates a flow of singlet oxygen, triggering a cascade of chemical events in nearby acceptor beads, which results in a chemiluminescent emission at 615 nm. This assay has been widely used for the detection of macromolecules,3-11 and particles (spores)12 but there are few references about its use for the detection of small molecules.13
We have previously developed immunoassays for small molecule detection (haptens) using analyte peptidomimetics in a competitive format14-16 and analyte-antibody anti-immunocomplexes peptides17,18 displayed on M13 viral particles in a noncompetitive format. These peptides were isolated by phage display technology and have been used in conventional ELISA,15,18 real-time immuno-PCR assays,16 and electrochemical biosensors.14,19 Recently we developed assays devoid of viral particles in which biotinylated anti-immunocomplex synthetic peptides complexed with a commercial streptavidin-peroxidase conjugate (referred to as nanopeptamers) were used as immunoassay reagents.20 Also, nanopeptamers of recombinant nature where the anti-immunocomplex peptide is produced as a fusion with a multimeric protein have been used in conventional ELISA and lateral flow immunochromatography.20-22 Nanopeptamers are more suitable reagents for the immunoassay industry than viral-based reagents, since E.coli infecting phages could be a matter of concern in some laboratories.
In this work, we report the results of combining AlphaLisa technology with an anti-immunocomplex nanopeptamer with the aim of developing a novel noncompetitive homogeneous immunoassay for the detection of small molecules. As a model target analyte we chose to use atrazine, since it is one of the most heavily used pesticides worldwide and its detection in water has relevant human health implications: it is reported to be an endocrine disruptive chemical and a potential carcinogen.23,24. The atrazine nanopeptamer based AlphaLisa, which we called nanoAlphaLisa, was successfully developed and exhibited excellent recoveries in river water samples, robustness, and sensitivities below the accepted limits in drinking water.23,24 Atrazine determination using this strategy was performed in one hour. Given the toxicological, environmental and medical analytical relevance of small molecules, this novel immunoassay prone to automation, high-throughput screening, and short incubation time can be adapted for the detection of these analytes.
Materials and Methods
Materials
Atrazine and related triazines were a gift from Shirley Gee.
The biotinylated synthetic peptides were supplied by a commercial manufacturer, Peptron, Inc (Daejeon, South Korea). These peptides were synthesized to 80% of purity by high-pressure liquid chromatography (HPLC), with intramolecular disulfides bonds between cysteines, an N-terminal biotin and amidated C-terminus. AlphaLisa acceptor beads and streptavidin-coated donor beads were obtained from PerkinElmer (San Jose, CA, USA). High-sensitivity streptavidin peroxidase (SPO) and sodium cyanoborohydride were purchased from Thermo Scientific, Pierce (Rockford, IL). Bovine serum albumin (BSA), gelatin, protein A, Tween 20, 3,3, 5,5′-tetramethylbenzidine (TMB), and Corning White 96-well microplates (half area) were purchased from Sigma (St. Louis, MO). Enzyme-linked immunosorbent assay (ELISA) and dilution microtiter polystyrene plates were purchased from Greiner (Solingen, Germany).
Atrazine NanoAlphaLisa protocol
The noncompetitive nanoAlphaLisa assay was performed using protein A-coated acceptor beads, streptavidin-coated donor beads, the purified monoclonal anti-atrazine antibody (MoAb) K4E7 and a biotinylated synthetic anti-immunocomplex peptide “13A” specific for the atrazine-K4E7 immunocomplex (Biotin-ASGSACTPVRWFDMC-NH2).18 Protein A was conjugated to acceptor beads as described in supporting information. Atrazine standards and samples spiked with atrazine were prepared by adding 100 μL of 10x AlphaLisa Buffer (250 mM HEPES, pH 7.4, 1% casein, 10 mg/mL Dextran-500, 5% Triton X-100) to 900 μL of milliQ water or water sample.
Based on the manufacturer’s instructions and as a starting point, assay performance was explored by keeping constant the amount of donor (7.5 μg/mL) and protein A conjugated acceptor beads (10 μg/mL), and testing different amounts of K4E7 MoAb captured on acceptor beads, in the final assay. Protein A coated acceptor beads at 100 μg/mL were prepared for assays by pre-incubation with K4E7 MoAb at 24 μg/mL, 12 μg/mL, 0.6 μg/mL, 0.3 μg/mL and 0.1 μg/mL for 30 minutes at 25 °C, (mixes 1) in 1X Alphalisa buffer. At the same time, the streptavidin donor beads were pre-incubated with an excess of biotinylated peptide in 1x AlphaLisa buffer. For each 10 μg of streptavidin (MW=58 kDa) donor bead, 15 μg of biotinylated peptide (MW~ 1.8 kDa) (3 μL) were added in a final volume of 30 μL (mix 2). The pre-incubation was performed on ice and protected from light exposure. Assuming that 10 μg of streptavidin coated bead was all streptavidin (0.172 nmoles), there was at least a 50-fold molar excess of biotinylated peptide. However, since only the surface of the bead is occupied by streptavidin the molar excess of peptide to streptavidin was much higher than 50-fold. The reaction was scaled-up or down depending on the required amount for each assay.
Five microliters of protein A acceptor beads pre-incubated with different amounts of K4E7 MoAb (mixes 1), and 5 microliters of streptavidin coated donor beads preincubated with synthetic biotinylated “13A” anti-immunocomplex peptide (mix 2) were dispensed in a white microtiter plate (Greiner) containing 5 μL of 10X AlphaLisa buffer and 35 μL of milliQ or water samples spiked with atrazine. After 15 - 60 minutes of incubation at 25 °C the reaction was read on the AlphaLisa signal reader (Fusion ™ α-FP. Packard, PerkinElmer).
NanoAlphaLisa assay Cross-reactivity with other triazines
The selectivity of the nanoAlphaLisa was explored by determining the cross-reactivity with related triazines. Cross-reactant concentrations in the 0–10,000 ng/mL range were used in the noncompetitive nanoAlphaLisa. The molar compound concentration corresponding to the inflection point of the curve, which corresponds to the concentration of analyte producing 50% saturation of the signal (SC50), was used to calculate the cross-reactivity of the assay according to the equation:
NanoAlphaLisa assay with undiluted river water samples, intra- and inter-assay reproducibility
For the analysis of matrix effect, different river and mineral water samples from California, USA, with no registered use of atrazine, were spiked with known amounts of analyte and assayed by nanoAlphaLisa in the 0.3 – 1 ng/mL range, using 35 μL of undiluted water samples plus 5 μL of 10x AlphaLisa Buffer. Five microliters of each bead pre-incubated with K4E7 MoAb and biotinylated peptide, respectively, were then added to the samples and incubated for one hour. The intra-assay and inter-assay parameters were tested by performing three consecutive measurements of the same sample and repeated for three consecutive days, respectively.
Results and Discussion
Development of nanoAlphaLisa assay and optimization
NanoAlphaLisa assay is represented in Figure 1a; in the presence of analyte, donor and acceptor beads are brought together. Anti-immunocomplex synthetic biotinylated peptides bound to streptavidin-coated donor bead recognize the atrazine-K4E7 MoAb immunocomplex (which has been previously captured by protein A-coated acceptor bead). Upon laser excitation (680 nm), the donor bead converts ambient oxygen to an excited singlet state. Singlet oxygen diffuses up to 200 nm to produce a chemiluminescent reaction in the acceptor bead, leading to light emission (at 615 nm). Acceptor beads were conjugated to protein A with the objective of assaying different amounts of anti-atrazine MoAb antibody K4E7 captured on the surface of the beads Nanopeptamer was formed on the surface of the donor beads by incubating biotinylated synthetic anti-immunocomplex peptide for atrazine (13A) with streptavidin coated donor beads using an excess of the biotinylated peptide (see materials and methods). In the first experimental step for the set up of nanoAlphalisa assay, we chose to use different concentrations of K4E7 MoAb captured on the surface of the acceptor beads, from 2.4 μg/mL to 0.1 μg/mL. Standard non-competitive atrazine curves were performed in these conditions. Taking into account the SC50 (50 % of saturation signal) as an indicator of the assay sensitivity, as antibody concentrations were lowered, luminescence signals decreased, but assay sensitivities increased regarding their SC50 (Figure 1b). The most sensitive assays were achieved when 0.3 μg/mL (SC50 = 1.4 ± 0.1 ng/mL) and 0.1 μg/mL (SC50 = 0.84 ± 0.4 ng/mL) of K4E7 MoAb were captured on the surface of the acceptor bead. In light of these results we chose to continue our work with 0.3 μg/mL of K4E7 MoAb captured on the surface of the acceptor bead, since this condition resulted in higher signals and more reproducible assays (data not shown).
Figure 1.
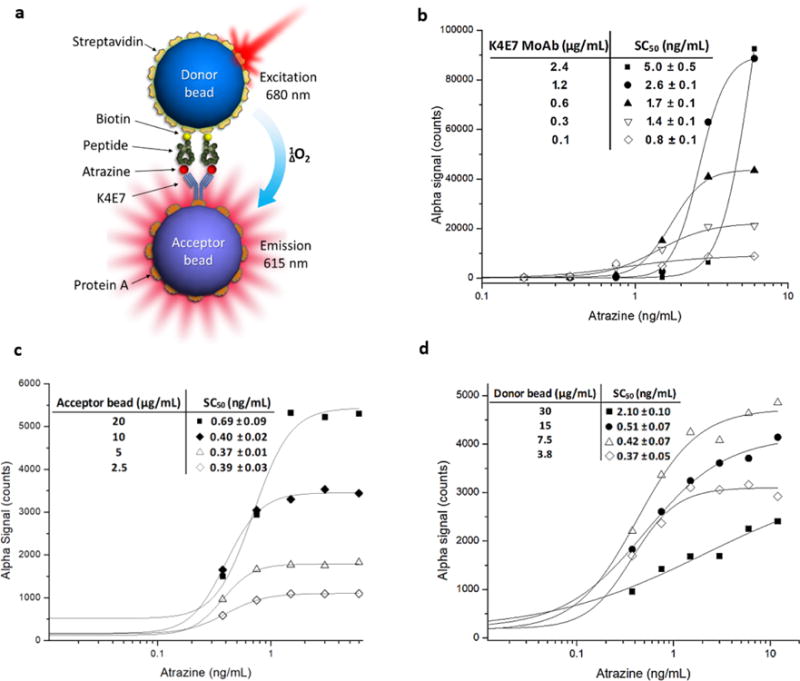
Optimization of conditions for developing sensitive nanoAlphaLisa assays for atrazine detection. (a) Scheme of noncompetitive nanoAlphaLisa assay. (b) Curves obtained using fixed amounts of streptavidin donor (7.5 μg/mL), protein A conjugated acceptor beads (10 μg/mL), and different amounts of K4E7MoAb: 2.4 μg/mL (black squares), 1.2 μg/mL (black circles), 0.6 μg/mL (black triangles), 0.3 μg/mL (white triangles), and 0.1 μg/mL (white diamonds). The obtained SC50 values are shown in the inset. (c) Curves obtained using fixed amounts of K4E7MoAb (0.3 μg/mL) and donor bead (7.5 μg/mL) and different amounts of protein A conjugated acceptor beads: 20 μg/mL (black squares), 10 μg/mL (black diamonds), 5 μg/mL (white triangles), and 2.5 μg/mL (white diamonds). The obtained SC50 values are shown in the inset. (d) Curves obtained using fixed amounts of K4E7MoAb (0.3 μg/mL) and protein A conjugated acceptor bead (10 μg/mL) and different amounts of streptavidin donor beads: 30 μg/mL (black squares), 15 μg/mL (black circles), 7.5 μg/mL (white triangles), and 3.8 μg/mL (white diamonds). The obtained SC50’s are shown in the inset.
Next, we studied the effect of the amount of acceptor beads in the assay performance by keeping constant both, the amount of captured K4E7 MoAb on the surface of the acceptor bead (0.3 μg/mL) and streptavidin donor beads (7.5 μg/mL). Protein A-coated acceptor beads were assayed at 20, 10, 5 and 2.5 μg/mL (final concentration). As shown in Figure 1c, signal intensities increased in a direct proportional relation to the concentration of acceptor beads, but no significant changes in assay sensitivities were found. We decided to continue our assays with 10 μg/mL of the acceptor beads as it presented a better signal to noise ratio than the 20 μg/mL assay. The last parameter that was explored was the influence of streptavidin coated donor bead concentration in assay performance by keeping constant the protein A acceptor bead and K4E7 MoAb concentrations at 10 μg/mL and 0.3 μg/mL, respectively. Streptavidin beads were assayed at 30, 15, 7.5, and 3.8 μg/mL. The results of this last evaluation are shown in Figure 1d; in most cases assay performances were satisfactory and similar among assays except when 30 μg/mL of donor beads were used which yielded a poor assay. Subsequent assays revealed that the concentration of 7.5 μg/mL of streptavidin facilitated development of the most robust and reproducible assays.
It is worth to note that in all the cases in which different conditions were explored for nanoAlphaLisa assay optimization, bead proximity only occurred in the presence of atrazine. This differs with previous results obtained in standard ELISA in which background noise due to residual interaction between the anti-immunocomplex peptide and the unliganded antibody was observed during assay setup, as shown in Figure S1.18 Taking into account the data obtained from the optimization experiments, we chose to perform an atrazine standard curve following the same procedure as described above. In a final volume of 50 μL, atrazine standards at 2-fold dilution concentration from 12 to 0.09 ng/mL were detected using the defined bead concentrations (donor bead at 7.5 μg/mL and acceptor bead at 10 μg/mL) and 0.3 μg/mL of K4E7 MoAb. The final optimized assay for atrazine is shown in Figure 2, and performed with an SC50 = 0.5 ± 0.02 ng/mL and limit of detection, defined as the amount of atrazine that results in a signal of 10% of the saturation signal, (LOD) = 0.3 ng/mL. This sensitivity is similar to that obtained with the original hapten-based competitive ELISA performed with K4E7 MoAb, which exhibited an IC50=0.64 ± 0.06 ng/mL and a LOD= 0.18 ng/mL.25 While the original ELISA took four hours with washing steps, nanoAlphaLisa was completed within one hour in a mix and measure mode with no washing steps.
Figure 2.

NanoAlphaLisa assay for atrazine. The assay was performed in 50 μL of AlphaLisa buffer containing K4E7 MoAb at 0.3 μg/mL, protein A acceptor beads at 10 μg/mL and atrazine at 2-fold dilution concentrations. Donor beads pre-incubated with an excess of biotinylated synthetic peptide 13A were added and the mixture was incubated for one hour at 25 °C. After that, beads were irradiated at 680 nm and signal was registered at 615 nm. The atrazine standard curve shows a SC50 = 0.5 ± 0.02 ng/mL and a LOD = 0.3 ng/mL.
It is well known that AlphaLisa assays display a loss of signal when an excess of analyte is present in the sample and surpass a saturation concentration (hook point), a phenomenon known as “hook effect” and described at the manufacturer website26. Beyond the hook point the excess of analyte disrupts associations between donor and acceptor beads causing a decrease in signal. However, in the nanoAlphaLisa assay, no hook effect was observed from 1 to 200 ng/mL of atrazine, as shown in supporting Figure S2. This showed that free atrazine is not bound by nanopeptamers and only atrazine complexed with K4E7 MoAb on the surface of the acceptor bead is recognized by nanopeptamers. The absence of hook effect has practical implications, since samples containing high amounts of analyte will not result in a false negative, which would be the case in a standard AlphaLisa.
In order to reduce the assay time, we explored lowering the incubation time with the objective of assessing the shortest incubation time of beads and atrazine that could still yield accurate assays. For this, we first performed standard atrazine curves in buffer prepared with MilliQ water. As shown in Figure S3, appropriate calibration curves were obtained when irradiating beads at 15, 30, and 45 minutes of incubation. However, when analyzing real water samples, the recoveries were affected with short incubation times and it was necessary to extend the incubation to 1 h.
Atrazine determination in river water samples using the nanoAlphaLisa assay
The performance of the assay with real samples was tested using undiluted samples obtained from river and mineral water. Atrazine was spiked with 1, 0.75, 0.5 and 0.3 ng/mL and each sample was analyzed in triplicates. The recoveries are shown in Table 1a. Very good recoveries ranging from 79 ± 2 to 124 ± 7 % were obtained for all the samples, showing that the nanoAlphaLisa can be used for detection of atrazine in undiluted water samples.
Table 1a.
Recovery (%, n=3) of atrazine in undiluted spiked water samples measured with nanoAlphaLisa assay
| Spiked atrazine (ng/mL) |
River 1 | River 2 | River 3 | Mineral Water |
|---|---|---|---|---|
| 1 | 79 ± 2 | |||
| 0.75 | 95 ± 2 | |||
| 0.5 | 102 ± 4 | 124 ± 7 | 117 ± 4 | 113 ± 6 |
| 0.3 | 105 ± 6 |
The intra-assay and inter-assay parameters were tested by performing three consecutive measurements of the same sample and repeated for three consecutive days, respectively. As shown in Table 1b. NanoAlphaLisa for atrazine showed a very good accuracy and reproducibility. Cross-reactivities with other compounds of the triazine family were very similar to those obtained in conventional ELISA when using M13 phage particles bearing anti-immunocomplex peptides as immunoassay reagents are shown in Table 2.
Table 1b.
Accuracy and reproducibility of the nanoAlphaLisa assay (% of recovery)
| Atrazine concentration (ng/mL) |
Intra-assay Mean |
Inter-assay Mean |
|---|---|---|
| 1 | 90 ± 6 | 110 ± 6 |
| 0.35 | 95 ± 10 | 107 ± 7 |
Table 2.
Cross reactivity (%) of atrazine in nanoAlphaLisa assay with related compounds from the triazine family
| Compound | Structure | Competitive ELISA15 | PHAIA18 | nanoAlphaLisa | ||
|---|---|---|---|---|---|---|
|
| ||||||

|
R1 | R2 | R3 | |||
| atrazine | Cl | C2H5 | CH(CH3)2 | 100 | 100 | 100 |
| simazine | Cl | C2H5 | C2H5 | 48 | 0 | 0 |
| propazine | Cl | CH(CH3)2 | CH(CH3)2 | 116 | 144 | 94 |
| cyanazine | Cl | C2H5 | C2N(CH3)2 | 91 | 0 | 0 |
| terbutryn | SCH3 | C2H5 | C(CH3)3 | 0 | 0 | 0 |
| ametryn | SCH3 | C2H5 | CH(CH3)2 | 0 | 0 | 0 |
| simetryn | SCH3 | C2H5 | C2H5 | 0 | 0 | 0 |
| prometryn | SCH3 | CH(CH3)2 | CH(CH3)2 | 0 | 0 | 0 |
All data are the mean of two independent experiments. A value of 0 means that there was no observable cross-reactivity with the highest concentration tested, 104 ng/mL.
CONCLUSIONS
AlphaLisa assay signal generation relies on acceptor-donor bead proximity induced by the presence of the analyte, which is typically bound by antibody pairs immobilized on the surface of the beads. Small analytes cannot be bound simultaneously by two antibodies and hence, they cannot be used to bridge the acceptor and donor beads. In this study we demonstrated that anti-immunocomplex peptides, which react selectively with small analyte-antibody complexes, provide sufficient stabilization of bead proximity to enable the generation of strong luminescent oxygen channeling signals. Using atrazine as a model small molecule analyte, we developed a novel immunoassay (nanoAlphaLisa) for this herbicide that allowed the sensitive mix-and-read detection of atrazine in undiluted water samples within one hour. Given the toxicological, environmental and medical analytical relevance of small molecules, and considering that anti-immunocomplex peptides can be isolated in a straightforward manner from phage display peptide libraries, the nanoAlphaLisa presented here constitutes a valuable addition to the toolbox of rapid methods for the detection of these compounds. This methodology also has the intrinsic advantages of automation and high-throughput processing that homogeneous luminescent oxygen channeling assays provide.
Supplementary Material
Acknowledgments
This work was supported by project CSIC 149 (UDELAR). We are thankful to Dr. Judy Callis and Dr. John Riggs of Molecular and Cell Biology Department in UC Davis for kindly providing access to perform experiments in the AlphaLisa reader Fusion ™ α-FP. Packard, PerkinElmer instrument. Gabriel Lassabe is a recipient of a doctorate fellowship from CAP (Comisión Académica de Posgrado) and performed this research in The Entomology Department in UC Davis with a fellowship from ANII, MOV_CA_2015_1_107451. Partial support from the NIEHS Superfund Program Research Program P42 ES004699
Footnotes
Supporting information
Three figures and supplementary methods. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Ullman EF, Kirakossian H, Switchenko AC, Ishkanian J, Ericson M, Wartchow CA, Pirio M, Pease J, Irvin BR, Singh S, Singh R, Patel R, Dafforn A, Davalian D, Skold C, Kurn N, Wagner DB. Clin Chem. 1996;42:1518–1526. [PubMed] [Google Scholar]
- 2.Beaudet LR-SR, Venne MH, Caron M, Bédard J, Brechler V, Parent S, Bielefeld-Sévigny M. Nature Methods. 2008;5 [Google Scholar]
- 3.Zhao H, Lin G, Liu T, Liang J, Ren Z, Liang R, Chen B, Huang W, Wu Y. J Immunol Methods. 2016;437:64–69. doi: 10.1016/j.jim.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Bracht T, Molleken C, Ahrens M, Poschmann G, Schlosser A, Eisenacher M, Stuhler K, Meyer HE, Schmiegel WH, Holmskov U, Sorensen GL, Sitek B. J Transl Med. 2016;14:201. doi: 10.1186/s12967-016-0952-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Nefzi A, Fields GB, Lakshmana MK, Minond D. Anal Biochem. 2014;459:24–30. doi: 10.1016/j.ab.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Steinsbo O, Dorum S, Lundin KE, Sollid LM. Gastroenterology. 2015;149:1530–1540 e1533. doi: 10.1053/j.gastro.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Burlein C, Bahnck C, Bhatt T, Murphy D, Lemaire P, Carroll S, Miller MD, Lai MT. Anal Biochem. 2014;465:164–171. doi: 10.1016/j.ab.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Liu TC, Huang H, Dong ZN, He A, Li M, Wu YS, Xu WW. Clin Chim Acta. 2013;426:139–144. doi: 10.1016/j.cca.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 9.He A, Liu TC, Dong ZN, Ren ZQ, Hou JY, Li M, Wu YS. J Clin Lab Anal. 2013;27:277–283. doi: 10.1002/jcla.21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou LP, Liu TC, Lin GF, Dong ZN, Hou JY, Li M, Wu YS. J Immunoassay Immunochem. 2013;34:134–148. doi: 10.1080/15321819.2012.690358. [DOI] [PubMed] [Google Scholar]
- 11.Chau DM, Shum D, Radu C, Bhinder B, Gin D, Gilchrist ML, Djaballah H, Li YM. Comb Chem High Throughput Screen. 2013;16:415–424. doi: 10.2174/1386207311316060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mechaly A, Cohen N, Weiss S, Zahavy E. Anal Bioanal Chem. 2013;405:3965–3972. doi: 10.1007/s00216-013-6752-1. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Huang B, Zhang J, Wang K, Jin J. J Sci Food Agric. 2012;92:1944–1947. doi: 10.1002/jsfa.5566. [DOI] [PubMed] [Google Scholar]
- 14.Arevalo FJ, Gonzalez-Techera A, Zon MA, Gonzalez-Sapienza G, Fernandez H. Biosens Bioelectron. 2012;32:231–237. doi: 10.1016/j.bios.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Cardozo S, Gonzalez-Techera A, Last JA, Hammock BD, Kramer K, Gonzalez-Sapienza GG. Environ Sci Technol. 2005;39:4234–4241. doi: 10.1021/es047931l. [DOI] [PubMed] [Google Scholar]
- 16.Kim HJ, Gonzalez-Techera A, Gonzalez-Sapienza GG, Ahn KC, Gee SJ, Hammock BD. Environ Sci Technol. 2008;42:2047–2053. doi: 10.1021/es702219a. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Techera A, Kim HJ, Gee SJ, Last JA, Hammock BD, Gonzalez-Sapienza G. Anal Chem. 2007;79:9191–9196. doi: 10.1021/ac7016713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Techera A, Vanrell L, Last JA, Hammock BD, Gonzalez-Sapienza G. Anal Chem. 2007;79:7799–7806. doi: 10.1021/ac071323h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Techera A, Zon MA, Molina PG, Fernandez H, Gonzalez-Sapienza G, Arevalo FJ. Biosens Bioelectron. 2015;64:650–656. doi: 10.1016/j.bios.2014.09.046. [DOI] [PubMed] [Google Scholar]
- 20.Vanrell L, Gonzalez-Techera A, Hammock BD, Gonzalez-Sapienza G. Anal Chem. 2013;85:1177–1182. doi: 10.1021/ac3031114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlomagno M, Lassabe G, Rossotti M, Gonzalez-Techera A, Vanrell L, Gonzalez-Sapienza G. Anal Chem. 2014;86:10467–10473. doi: 10.1021/ac503130v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lassabe G, Rossotti M, Gonzalez-Techera A, Gonzalez-Sapienza G. Anal Chem. 2014;86:5541–5546. doi: 10.1021/ac500926f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organisation, W. H. O. 2011 http://apps.who.int/iris/bitstream/10665/44584/1/9789241548151_eng.pdf. 9 Nov. 2017.
- 24.EPA. EPA, editor. 2015 https://safewater.zendesk.com/hc/en-us/articles/212077787-4-What-are-EPA-s-drinking-water-regulations-for-atrazine-
- 25.Giersch T. Journal of Agricultural Food and Chemistry. 1993;41:1006–1011. [Google Scholar]
- 26.PerkinElmer. 2018 Apr 24; http://www.perkinelmer.com/es/lab-products-and-services/application-support-knowledgebase/alphalisa-alphascreen-no-wash-assays/hook-effect.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


