Abstract
AphB is a LysR-type transcriptional regulator (LTTR) that cooperates with a second transcriptional activator, AphA, at the tcpPH promoter to initiate expression of the virulence cascade in Vibrio cholerae. Since it is not yet known whether AphB responds to a natural ligand in V. cholerae that influences its ability to activate transcription, we used a computational approach to identify small molecules that influence its activity. In silico docking was used to identify potential ligands for AphB, and saturation transfer difference NMR was subsequently employed to access the validity of promising targets. We identified a small molecule, BP-15, that specifically binds the C-terminal regulatory domain of AphB and increases its activity. Interestingly, molecular docking predicts that BP-15 does not bind in the putative primary effector-binding pocket located at the interface of RD-I and RD-II as in other LTTRs, but rather at the dimerization interface. The information gained in this study helps us to further understand the mechanism by which transcriptional activation by AphB is regulated by suggesting that AphB has a secondary ligand binding site, as observed in other LTTRs. This study also lays the groundwork for future design of inhibitory molecules to block the V. cholerae virulence cascade, thereby preventing the devastating symptoms of cholera infection.
Keywords: Transcription factor, Regulation, Compound Computational Screen, Saturation Transfer Difference NMR (STD-NMR), Regulatory Domain (RD)
INTRODUCTION
Cholera is a disease caused by the ingestion of food and water contaminated with pathogenic strains of the bacterium V. cholerae.1 The infection causes watery diarrhea that rapidly causes dehydration and, if treatment is not promptly given, can result in death.1–4 In order to be pathogenic, V. cholerae serogroup O1 must possess two genetic elements, the Vibrio pathogenicity island (VPI) which contains genes for the toxin coregulated pilus (TCP), and the CTX phage, a lysogenic bacteriophage that encodes the protein cholera toxin (CT).5–7 TCP is essential for V. cholerae colonization of host epithelial cells5 and CT is responsible for the symptoms associated with the disease.5, 8
A transcriptional cascade involving activator proteins encoded both within the VPI and the ancestral genome controls the expression of these two virulence factors. AphB, encoded in the ancestral genome, is a LysR-type transcriptional regulator (LTTR) that initiates the expression of the virulence cascade by activating the tcpPH promoter on the VPI.9, 10 In order to perform this function, AphB cooperates with a second transcriptional activator, AphA, also encoded in the ancestral genome.9, 11 TcpP is a membrane bound transcriptional activator that, once expressed, cooperates with a homologous transcriptional activator, ToxR, to activate the gene encoding ToxT.12 ToxT is the master virulence regulator in V. cholerae and directly activates the genes encoding TCP and CT, in addition to regulating its own expression.13, 14
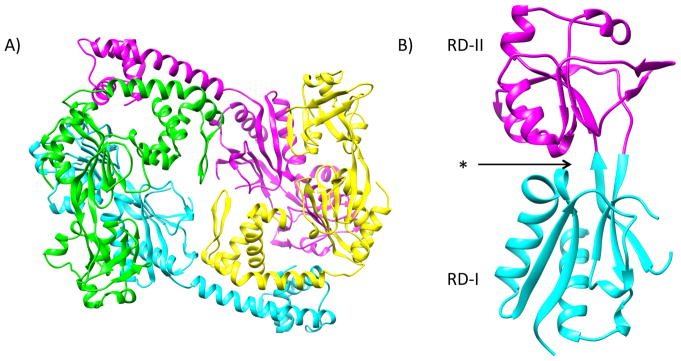
Proteins in the LTTR family are among the most abundant types of transcriptional regulators in prokaryotes and members are involved in regulating diverse sets of genes that influence a variety of biological processes.15 Homologs in this family possess a well-conserved winged helix-turn-helix DNA binding domain in the N-terminus and a poorly conserved regulatory domain in the C-terminus and are known to adopt different oligomeric states.15 The X-ray structure of tetrameric AphB has been solved to 2.2 Å (Figure 1)16 and shows two dimerization interfaces within the protein, one at the DNA binding domain and the other at the regulatory domain (Figure 1A). The regulatory domain comprises two distinct subdomains (RD-I and RD-II), at the interface of which lies the putative effector-binding pocket (Figure 1B).16 A tetramer of an LTTR consists of extended and compact subunits, which differ in the distance and orientation between the regulatory domain and the DNA binding domain.
Figure 1. Crystal Structure of AphB.
A) Ribbon structure of the AphB tetramer. Extended subunits are shown in cyan and magenta and compact subunits are shown in green and yellow (PDB ID: 3SZP). B) Monomer of the regulatory domain of AphB. The previously identified putative effector-binding pocket (denoted with an asterisk) is located between RD-I (cyan) and RD-II (magenta).16
For many LTTRs, the effector is a substrate or metabolite of the regulated genes, and effector binding into the pocket of the regulatory domain induces a conformational change in the protein that alters its ability to bind to DNA and activate transcription of specific target genes.15, 17, 18 The high-resolution crystal structure of TsaR bound to its effector molecule para-toluenesulfonate (TSA) is the only full-length effector-bound LTTR structure currently in the literature.18
In addition, only a few crystal structures of LysR regulatory domains have been solved with their effectors bound. These include BenM from Acinetobacter baylyi bound to both its effector molecules: cis, cis-muconate, in the pocket created between RD-I and RD-II, and benzoate in a second binding site located in RD-I.17 The LysR homolog, CatM, has been solved with its effector molecule cis, cis-muconate in its regulatory domain.17 The regulatory domain of DntR has been solved with salicylate bound at two distinct locations.19 Additionally, the LysR homologue CrgA has been shown to be activated by a specific protein-protein interaction with HPr, a phosphocarrier protein involved in PTS sugar metabolism, demonstrating that regulation of these proteins is not only mediated by small molecule ligands, but also effector proteins.20 In addition to its role in virulence, AphB regulates a diverse set of genes that are involved in the maintenance of pH homeostasis and management of oxidative stress in V. cholerae.21, 22 Although it is not known if AphB requires an effector for transcriptional activation, AphB appears to be influenced by both pH and environmental oxygen levels.16, 21. Mutations identified in the putative primary effector-binding pocket allow the protein to constitutively activate tcpPH expression at the non-permissive pH of 8.5 and in the presence of oxygen.16 These findings indicate that AphB is responsive to intracellular pH as well as to anaerobiosis and that the residues in the effector-binding pocket of the protein influence its ability to respond to both of these signals. The crystal structure of one of the effector-binding pocket mutants, N100E, showed a reorientation of the DNA binding domain of AphB, suggesting how effector binding could influence transcriptional activation.16
To gain insight into the mechanism of transcriptional activation of tcpPH by AphB, we set out to identify small molecules that influence its transcriptional activity. Using the computational screening tool AutoDock 4.0.1, we have identified an activator molecule for AphB. Utilizing saturation transfer difference NMR (STD-NMR), we have calculated the affinity of the ligand for AphB and show that it enhances its ability to activate transcription. The information gained from this study will allow for the design of potential inhibitor molecules to block the interaction of AphB and DNA, thus blocking the expression of the virulence cascade and preventing the progression of cholera.
MATERIALS AND METHODS
Virtual Screening
Molecular docking studies were performed using AutoDock 4.0.1, a molecular docking program obtained from the Scripps Research Institute.23 The receptor was obtained from the Protein Data Bank, ID: 3SZP. The Diversity Set library of 3000 molecules obtained from Life Chemicals Inc. in 2012 was used for screening. The ligands were flexible and the receptor rigid. AutoDock Tools 1.5.4 was used to add hydrogen atoms and atomic charges to the receptor and ligands. The grid box was centered on a putative binding pocket identified from X-ray crystallographic studies with dimensions of 30 x 30 x 30 points with grid point spacing of 0.375 Å. The lowest energy conformations from the screen as well as molecules with 50% homology were selected for experimental characterization by NMR and purchased from various vendors through ZINC.docking.org.24
A second grid box was centered on the EBD dimer at (12, −45.8, −27.544) with dimensions of 80 x 115 x 120 points with grid point spacing of 0.375 Å. A docking experiment with BP-15 was performed on the EBD per the AutoDock protocol.
STD-NMR spectroscopy of the full-length protein
All NMR spectroscopy experiments on full length AphB were performed at 25°C on a 700 Mhz Bruker Avance III NMR spectrometer equipped with a 5 mm TCI cryogenic probe. Water suppression was obtained with excitation sculpting. NMR samples were prepared using 5% 99.98% D2O and contained 300 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 8.0. Small molecules for screening were dissolved to 200 mM in D6DMSO and diluted to 200 μM in 700 μL. Samples tested for binding contained 10 μM AphB. The saturation transfer difference experiment was acquired with the on- and off- resonance data collected in alternating scans. For the on-resonance experiments, the protein was saturated at 200 Hz for 4 seconds. The total relaxation delay for each experiment was 8 seconds. Controls in the absence of AphB were collected in the same manner.
Equation 1 shows the calculation for the STD amplification factor (STD-AF). 25, 26
STD-NMR spectroscopy of N-terminal and C-terminal Constructs
All NMR spectroscopy experiments for the N- and C-terminal constructs were performed on a Bruker 600Mhz spectrometer equipped with a 1.7 mm TCI cryogenic probe. The N-terminal and C-terminal constructs of AphB (residues 1–91, and residues 78–291 respectively) were dialyzed into 300 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 8.0. Protein samples were prepared at 10 μM with 200 μM small molecule added to a total volume of 700 μL. Binding experiments were performed using a saturation at 200 Hz for 4 seconds. STD intensities were calculated using equation 1.
STD Build-up curve
All samples with small molecules contained 3–16% D6-DMSO and 10% D3-Acetonitryl to assist in solubility. Small molecule concentrations of 57 μM, 113 μM, 200 μM, 438 μM, 800 μM, 1100 μM were confirmed by a 50 μM trimethylsilylpropanoic acid standard, and added to the protein samples from a concentrated stock solution to minimize dilution effects. All AphB concentrations for the experiment were 10 μM. Data were obtained at 25°C on a 700 Mhz Bruker Avance III NMR spectrometer equipped with a 5mm TCI cryogenic probe. Saturation times used were 1, 2, 3, and 4 seconds. Data were acquired as described above. The data STD-AF calculated using equation 1 were plotted against saturation time for each ligand concentration.
Calculation of Dissociation Constant
The KD of the reaction was calculated using the STD build-up curve for concentrations of 57 μM, 113 μM, 200 μM, 438 μM, 800 μM, and 1100 μM. Data was plotted and analyzed using Kalediograph. Once the STD build-up curves for each concentration were plotted, they were fit to the monoexponential equation 2 seen below as reported by Angulo et al.27
Where STDmax is the max obtainable intensity when long saturation times are used, ksat is the saturation rate constant, and t is the saturation time. Once the fit was obtained, the STD-AF0, or the extrapolated amplification factor at zero was calculated using equation 3 below.
STD-AF0 was then plotted against ligand concentration to obtain a binding isotherm fit to equation 4 below.
Purification of AphB
AphB was purified using the IMPACT protein fusion and purification system (New England Biolabs) described by Kovacikova et al., 2004.11 To purify AphB, the plasmid pWEL218 was transformed into E. coli BL21-Star cells. Overnight cultures were grown from a single colony. These cultures were diluted 500-fold into ZYM-5052 auto inducing medium containing 100 μg/ml ampicillin as described by Studier.28 Cells were grown for 26 hours and resuspended in 1 M NaCl, 20 mM Tris, pH 8.0. Cells were then subjected to sonication and the spun supernatant applied to a chitin column at 4°C equilibrated with resuspension buffer. The column was washed with 10 column volumes of resuspension buffer before beginning overnight cleavage with 20 mM Tris-HCl pH 8.0, 500 mM NaCl and 100 mM DTT (16–20 hours, 4°C). Protein was eluted with cleavage buffer and concentrated before loading onto a Superdex 200 hL gel filtration column (GE).
Purification of N- and C-terminal domains of AphB
The region of AphB corresponding to the N-terminus (amino acids M1-G91) was cloned into the pTXB1 vector generating plasmid pBRT4 and the protein was expressed using the IMPACT protein fusion and purification as described above. The region corresponding to the C-terminus of AphB (amino acids M78-Q291) was cloned into the pTXB1 vector generating plasmid pRD1 and purified using the same method that was used for the N-terminus.
Reporter assay
All V. cholerae strains used in this study are listed in Supplementary Table 1. Strains were maintained at −80°C in LB medium in 30% glycerol. All cells used in the reporter assay were cultured in Luria-Bertani medium (LB) pH 6.5 at 30°C unless otherwise noted. β-galactosidase assays with V. cholerae tcpPH-lacZ fusions were performed after cells grew to mid-log phase (~3.5 hours).29 Experiments were performed in triplicate. Data was analyzed with Prism using a one sample t-test.
Western Blotting
Cell lysates from β-galactosidase assays were loaded into a 16% Tris-glycine gel and electrophoresis was performed in SDS running buffer. Samples were transferred to a PDVF membrane per the manufacturer’s protocol (Invitrogen iBlot) and incubated with 3% BSA in TBST (20 mM Tris, pH 7.5, 150 mM NaCl, 0.5% Tween 20) for 2 hours. The membrane was washed with TBST and incubated with an antibody against AphB (1:10,000) for 2 hours. Membranes were washed and incubated with a 1:5000 dilution of horseradish peroxidase-conjugated anti-mouse antibodies for 2 hours. Blots were washed in TBS and detected using a BioRad ECL kit. Intensities of bands were measured by Kodak Molecular Imaging software.
RESULTS AND DISCUSSION
Identification of small molecule ligands that bind to AphB
To shed light on whether a ligand is involved in AphB function, we attempted to computationally identify ligands that influence its activity. A set of 3000 compounds from the Diversity Set from Life Chemicals was screened against the putative pocket of AphB and results of the screen were sorted according to the AutoDock 4.0.1 algorithm.23 In order to test the binding of the compounds identified through the in silico screen, the top fourteen commercially available molecules were purchased through ZINC.docking.org for screening by STD-NMR.24
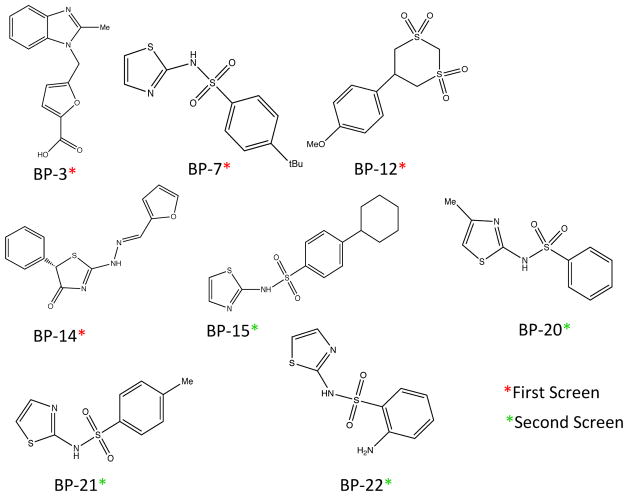
Compounds BP-1 through BP-14 were tested to determine if in vitro binding to AphB occurred. Analysis of the spectra showed a difference in the on- and off-resonance intensities of the peaks, demonstrating binding for 4 of the 14 small molecules identified from the computational screen (BP-3, BP-7, BP-12, BP-14, shown in Figure 2).
Figure 2. Structures of small molecules identified to bind AphB by STD-NMR.
Compounds identified by AutoDock screen are denoted with a red asterisk. Compounds identified through ZINC.docking.org homology search denoted with a green asterisk.
Once binding was identified through STD-NMR, a 50% homology search of these compounds was performed using the ZINC.docking.org database.24 This search resulted in ten more small molecules for screening, which had similar structural motifs that may promote binding. Most molecules in the second round contained either a sulfonamide or benzoxazole moiety. Figure 2 shows the structures of the small molecules that bound AphB from this second round of screening (BP-15, BP-20, BP-21, BP-22).
Small molecule activation of AphB at the tcpPH promoter
AphB is involved in the initiation of transcription of the tcpPH promoter in V. cholerae. Once produced, TcpP and TcpH are required for the expression of the virulence cascade.10 The level of expression of the tcpPH promoter is dependent on the activity of AphB, and changes in the tcpPH transcript can provide information about the activity of AphB in vivo.
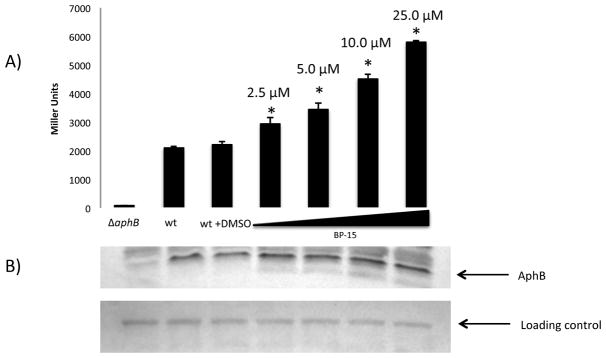
Of the small molecules identified in the screens, only BP-15 showed a biological effect in V. cholerae (Figure 3). To determine the effect of BP-15 on the transcriptional activity of AphB, a β-galactosidase assay was performed under inducing conditions in the classical biotype strain O395 containing a chromosomal fusion of the tcpPH promoter to lacZ. As shown in Figure 3A, addition of increasing amounts of BP-15 enhanced transcription from this promoter. Three-fold activation was observed in the presence of 25 μM BP-15, suggesting that the binding observed by STD-NMR has a biological effect on AphB. Additionally, the β-galactosidase assay was performed under non-inducing conditions (pH 6.5/37°C, 8.5/30°C, 8.5/37°C). We did not see a greater increase in the expression in the presence of the compound than we did under inducing conditions (data not shown).
Figure 3. Compound dependent activation of tcpPH-lacZ. A) From left to right.
GK121 (ΔaphB), KSK618 (wt tcpPH-lacZ), wt with 0.16% DMSO, wt with 2.5 μM BP-15 (P= 0.0022), wt with 5 μM BP-15 (P= 0.0007), wt with 10 μM BP-15 (P <0.0001), wt with 25 μM BP-15 (P <0.0001). B) Western blot of showing levels of AphB from β-galactosidase assay samples. Strains were grown aerobically at 30°C in LB medium pH 6.5. The GK121 ΔaphB strain represents the basal level of tcpPH-lacZ transcription.
Western blotting experiments show that the levels of AphB are similar in all samples except for the negative control (Figure 3B), implying that the increase in transcription is due to increased AphB activity, and not protein levels. The full blot and relative intensity of AphB in each lane are shown in Supplementary Figure 1. While the observed three-fold activation of tcpPH by AphB is relatively small, this is the first demonstration of small molecule based activation of AphB, and provides insight into the mechanism by which a natural activator of AphB could influence the virulence cascade in V. cholerae.
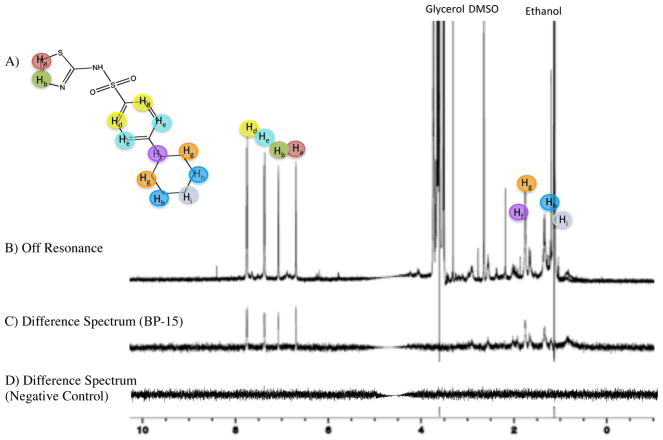
Data acquired from STD-NMR experiments show that BP-15 (Figure 4A) has a strong calculated STD. The difference spectrum indicates the largest effect was in the aromatic region of the spectra, suggesting that the thiazole moiety is interacting most closely with AphB (Figure 4C). All % STD values for BP-15 are summarized in Table 1.
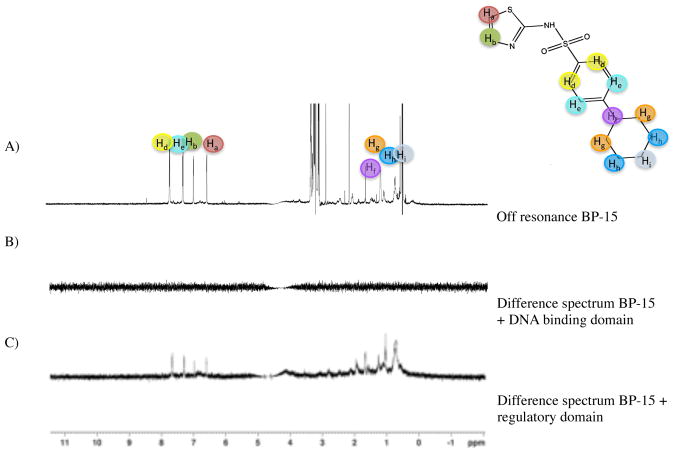
Figure 4. AphB binds to BP-15, identified through computational screening.
A) Structure of BP-15. B) 1D off-resonance STD-NMR spectrum of BP-15. The proton assignment is indicated by color. C) Difference spectrum between on- and off-resonance spectra in the presence of BP-15 and AphB. D) Difference spectrum of a compound that does not bind AphB.
Table 1.
% STD calculated for individual protons in BP-15.
| Proton | % STD | Normalized % STD |
|---|---|---|
| Ha | 23.8 | 100% |
| Hb | 21.5 | 90% |
| Hc | 19.5 | 81% |
| Hd | 19.2 | 80% |
| Hf | 19.9 | 83% |
| Hg | 19.7 | 82% |
The protons from the thiazole ring have a calculated STD of 23.8% and 21.5%. In contrast, STD observed for the unsaturated ring is lower, with an STD of 19.2–19.5%, and the aliphatic protons have a STD between 19.2–19.9%. These results suggest that the primary interaction of BP-15 with AphB occurs at the thiazole ring, possibly augmented by interactions between the negatively charged oxygen molecules attached to the sulfur group and the positively charged pocket of AphB (see below).
Small Molecule BP-15 Specifically Binds The C-terminal Regulatory Domain of AphB
To identify the region of AphB that the molecule BP-15 interacts with, and to determine if the molecule was binding at the previously identified pocket, STD-NMR experiments were performed separately on the two different domains of the protein. The N-terminal DNA binding domain (1–91) was screened for binding to BP-15, and showed no interaction with the DNA binding domain of AphB (Figure 5A and 5B). The C-terminal regulatory domain (78–291) containing the putative binding pocket was tested under the same conditions, and a difference spectrum was observed, as shown in Figure 5C. These data suggest that binding occurs at the regulatory domain, and not at the DNA binding domain.
Figure 5. The regulatory domain of AphB binds BP-15.
A) 1D NMR spectrum of BP-15. B) STD-NMR difference spectrum of BP-15 with the DNA binding domain of AphB (residues 1–91). C) STD-NMR difference spectrum of BP-15 with the regulatory domain of AphB (residues 78–291).
Small Molecule BP-15 Does Not Appear to Bind in the Putative Ligand Binding Pocket
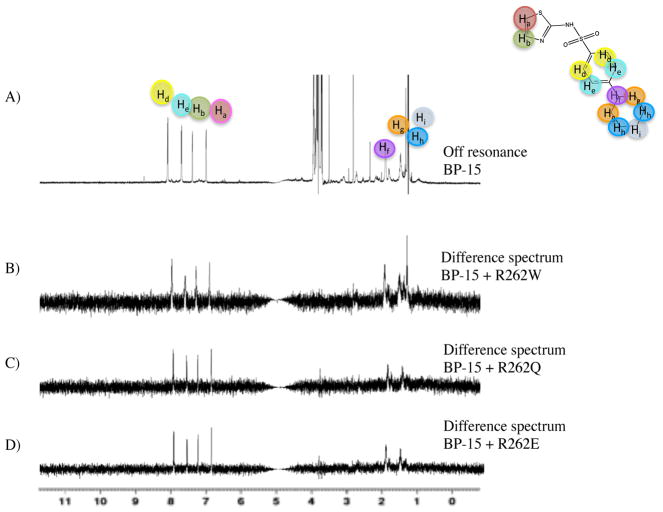
The X-ray structure of AphB revealed a putative effector-binding pocket containing a positively charged arginine residue (R262), which was hypothesized to be involved in effector binding.16 To probe the binding location of BP-15, STD-NMR was performed on mutants of AphB at R262 where either charge or size of the residue was altered to perturb binding. Figure 6A–D shows the STD-NMR spectra upon BP-15 binding to the R262W, R262Q and R262E mutants respectively, with all spectra indicating the binding interaction remains.
Figure 6. Mutations in the primary effector-binding pocket do not affect BP binding.
A) STD-NMR off-resonance spectrum of BP-15. B) Difference spectrum of BP-15 binding R262W mutant of AphB. C) Difference spectrum of BP-15 binding R262Q mutant. D) Difference spectrum of BP-15 binding R262E mutant.
These data suggest BP-15 is not binding at the putative effector-binding pocket that lies between RD-I and RD-II, and suggest AphB has a ligand binding site at a different location, as observed in the LTTRs BenM, DntR, and TsaR and PcpR.17–19, 30, 31
Ligand Binding Affinity Determined by STD-NMR
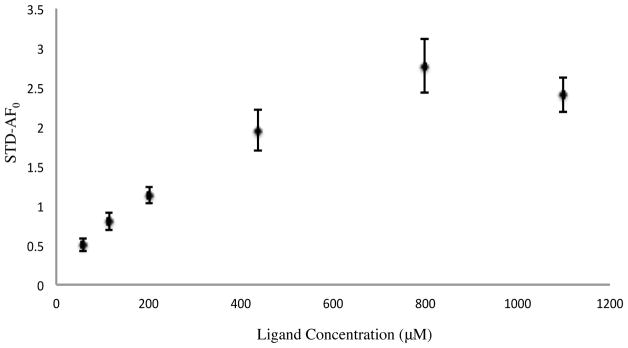
Using a method previously reported in the literature27, amplification of the STD effect as BP-15 was titrated into protein sample was measured, and these data were used to determine the binding affinity of the activator to AphB. Figure 7 shows the plot of STD-AF0 as a function of ligand concentration. Each point represents an average STD-AF0 for the 4 aromatic protons that interact with AphB (Ha, Hb, Hc, Hd) at each concentration measured. The dissociation constant of the AphB-activator complex was determined to be in the range of 324 μM ± 146 μM to 398 μM ± 114 μM as seen in Table 2.
Figure 7. STD-NMR increase used to determine the binding constant for BP-15.
Average STD-NMR build up curve for aromatic protons of BP-15 at concentrations of 57 μM, 113 μM, 200 μM, 438 μM, 800 μM, and 1100 μM with saturation times of 1, 2, 3, and 4 seconds. Concentrations were determined by comparing the intensity of the 1D NMR spectra of the ligand to an internal standard.
Table 2.
Summary of KD for aromatic protons calculated using a STD-NMR build up curve. The STD build-up curve for each proton was plotted and fit to equation 2. STD- AF0 was calculated using equation 3 at each saturation time and plotted vs. ligand concentration and fit to equation 4.
| Proton | KD μM |
|---|---|
| Ha | 324 +/− 146 |
| Hb | 398 +/− 142 |
| Hc | 372 +/− 118 |
| Hd | 398 +/− 114 |
This binding constant is consistent with the intensity of the difference spectra observed in Figure 3. The determined KD by STD-NMR titrations in the micromolar range for BP-15 is most likely weaker than the natural biological effector for AphB.
Small molecule binding model for BP-15 and AphB
To gain insight into the possible binding site of BP-15, we used AutoDock4 to model its interactions with AphB. A grid was created that encompassed the entire regulatory domain dimer of AphB, and BP-15 was docked using AphB as a rigid receptor. The resulting model predicts BP-15 does not bind in the putative pocket located at the interface of RD-I and RD-II, but rather at the dimerization interface (Figure 8).
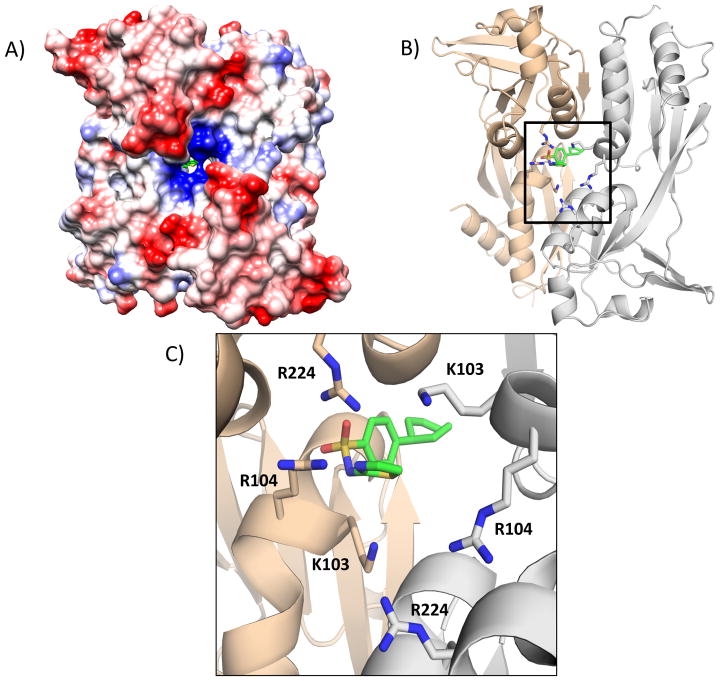
Figure 8. Putative Ligand Binding Pocket of AphB.
A) Electrostatic surface representation of the regulatory domain of AphB. A pocket is present at the dimerization interface containing positively charged residues. BP-15 shown in green. Red represents a negatively charged surface, blue represents a positively charged surface. B) Ribbon model of the dimerization interface of the regulator domain with BP-15 bound. The tan structure represents one protomer of the regulatory domain dimer, the gray structure represents the second protomer. C) Close-up view showing the cluster of positively charged residues surrounding the BP-15 ligand in the proposed secondary binding pocket. Figures were created with Pymol.30
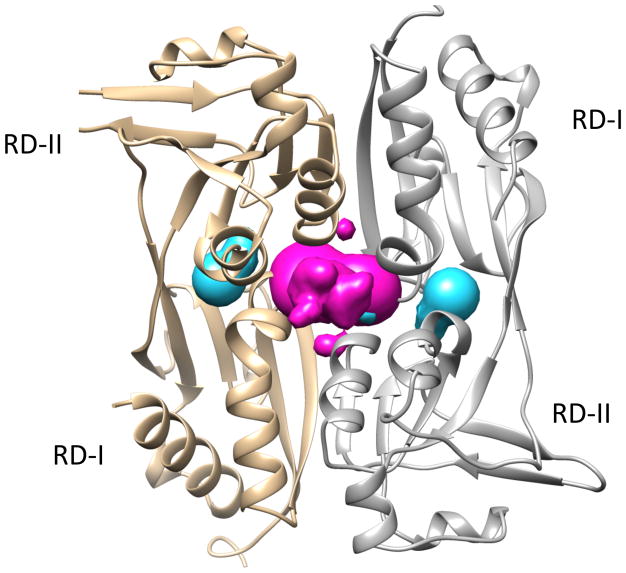
This result suggests that AphB has a ligand binding pocket that differs from the previously identified pocket, as has been described for other LTTRs.17–19 This putative pocket is located at the regulatory domain dimerization interface and contains K103, R104 and R224 from both chains of the dimer, creating a positively charged region on one side of the pocket as seen by the surface charge representation in Figure 8A. In the AutoDock model, the guanidino groups of these residues may interact with the negatively charged oxygen molecules from the sulfone moiety of BP-15. The proximity of R104 to the partially positive nitrogen of the thiazole ring may also support this interaction. These contacts are consistent with the STD-NMR data presented in Figure 3C. The hydrophobic cyclohexane ring of BP-15 extends through the interface of the regulatory domains to a less charged region of the pocket. The secondary pocket volume is 1622.7 Å3 as calculated using the CastP server.32 A comparison of the locations of the previously identified pocket between RD-I and RD-II and the newly identified pocket is shown in Figure 9.
Figure 9. CastP calculated pocket regions of AphB.
Previously identified putative binding pocket in cyan, newly identified binding pocket in magenta. Figure made with Chimera.33
Comparison with other LTTRs
Only a few LTTR structures with inducers bound have been described, and of those deposited in the PDB it appears that, unlike the primary effector binding sites, the location and characteristics of secondary effector binding sites are not conserved. For example, TsaR, which is involved in the degradation of para-toluenesulfonate (TSA) as the sole source of carbon for Comamonas testosteroni, (PDB: 3FXU) has an amino acid identity of 12.57% in the regulatory domain when compared to AphB. It is the only full-length LTTR structure bound to an effector molecule, and binds TSA at eleven different locations, including the primary effector-binding pocket between RD-I and RD-II, as well as at the HTH DNA binding domain and locations outside of the pocket in the regulatory domain.18
The X-ray structure of the regulatory domain of CatM from the soil bacterium Acinetobacter baylyi ADP1, (PDB ID: 2F7C) shows its effector, cis, cis-muconate, binds to the primary effector-binding pocket.17 Similarly, BenM from the same bacterium, (PDB ID: 2F78) binds the same molecule at the equivalent location; however, the structure also reveals a secondary binding pocket occupied by a benzoate molecule.17 Notably, this secondary effector-binding pocket is not in the same location as predicted for AphB. Interestingly, while BenM and CatM both act as transcriptional regulators involved in degradation of aromatic compounds, benzoate binding has not been observed for CatM. The complexity of the dual-inducer activation mechanism of some LTTRs is supported by biochemical work by Bundy et al., who have demonstrated that BenM responds positively to two different effector molecules.34
DntR (PDB ID: 2Y7K), an LTTR protein involved in the oxidative degradation of 2,4-dinitrotoluene by Burkholderia sp., binds its effector, salicylate, at the primary site as well as at a secondary site that differs from both BenM and AphB. Superposition of the apo and holo X-ray structures shows that a large conformational change occurs upon binding at the secondary effector site.17, 19 This finding demonstrates that effector binding to the secondary site can have an impact on both protein structure and function and supports the physiological relevance of the site in the function of this LTTR.
The X-ray structure of the regulatory domain PcpR (PDB ID: 4RPN), a protein involved in regulation of pentachlorophenol degradation in Sphingobium chlorophenolicum, has been solved to 2.2 Å bound to the effector pentachlorophenol (PCP).31 This structure shows PcpR contains a secondary effector binding pocket at the regulatory domain dimerization interface, a location similar to that observed for AphB, and both pockets appear to be important for function.31
In addition to being activated, LTTRs have also been shown to be inhibited by effector binding. The X-ray structure of the regulatory domain of AmpR from Citrobacter freundii was solved bound to its natural repressor ligand, MurNAc pentapeptide, revealing this inhibitor blocks access of the natural effector to the primary pocket.35 The X-ray structure of the regulatory domain of PqsR, from Pseudomonas aeruginosa, has also been solved bound to its inhibitor, 2-nonyl-4-hydroxyquinoline (NHQ), and the structure shows the inhibitory molecule inserts into a hydrophobic pocket near the primary effector-binding site located in RD-I.36
It has recently been demonstrated through molecular docking and biological studies that AphB can be inhibited by binding of the anti-viral drug ribavirin at its primary effector-binding pocket.37 This study also found that ribavirin inhibited the activity of the LTTR Hrg in Salmonella Typhi.37 Our discovery of a new binding pocket in AphB suggests there is likely an additional way to modulate protein function, through the design of an inhibitor that binds at this location.
CONCLUSIONS
AphB, involved in the regulation of the virulence cascade of V. cholerae, is an important drug target for the mitigation of the disease cholera. This study provides the first example of ligand-binding event increasing the activity of AphB at the tcpPH promoter and adds to the understanding of the mechanism of action of AphB at this promoter by demonstrating that AphB can be activated by a ligand, possibly in a manner similar to how it might respond to a natural effector. The data presented here suggest that, as has been described for other LTTRs, AphB has two binding sites. We propose the secondary site is located at the dimerization interface of the regulatory domain of AphB, as predicted by AutoDock4 and supported by mutational analysis of the primary effector-binding pocket. Future work will include mutational analysis of the amino acids at the dimerization interface, including K103 and R104, to observe the effect on both BP-15 binding and in vivo transcriptional activity. This demonstration of small molecule activation of AphB provides insight into the mechanism by which a natural activator of AphB could influence the virulence cascade in V. cholerae.
Supplementary Material
Acknowledgments
FUNDING INFORMATION
This work was funded by National Institutes of Health grants AI039654 to R.K.T, AI072661 to F.J.K and AI120068 to F.J.K and K.S. Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311)
Footnotes
This work is accompanied by Supplemental Online Material containing: Table S1-Strains and plasmids used in this study; Figure S1-Western blot of AphB.
References
- 1.World Health O. The Weekly Epidemiological Record (WER) 2015;90:517. [Google Scholar]
- 2.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology , Genetics , and Ecology of Toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandal S, Mandal MD, Pal NK. Cholera: a great global concern. Asian Pac J Trop Med. 2011;4:573. doi: 10.1016/S1995-7645(11)60149-1. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez J, Holmgren J. Cholera toxin structure, gene regulation and pathophysiological and immunological aspects. Cell Mol Life Sci. 2008;65:1347. doi: 10.1007/s00018-008-7496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Childers BM, Klose KE. Regulation of virulence in Vibrio cholerae: the ToxR regulon. Future Microbiol. 2007;2:335. doi: 10.2217/17460913.2.3.335. [DOI] [PubMed] [Google Scholar]
- 6.Davis BM, Moyer KE, Boyd EF, Waldor MK. CTX prophages in classical biotype Vibrio cholerae: functional phage genes but dysfunctional phage genomes. J Bacteriol. 2000;24:6992. doi: 10.1128/jb.182.24.6992-6998.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karaolis D, Johnson J, Bailey C, Boedeker E, Kaper J, Reeves P. A Vibrio cholerae pathogenicity island associated with epidemic. Proc Natl Acad Sci U S A. 1998;95:3134. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bharati K, Ganguly NK. Cholera toxin : A paradigm of a multifunctional protein Cholera toxin : an historical introduction. Indian J Med Res. 2011;133:179. [PMC free article] [PubMed] [Google Scholar]
- 9.Kovacikova G, Skorupski K. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol. 1999;181:4250. doi: 10.1128/jb.181.14.4250-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacikova G, Skorupski K. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol Microbiol. 2001;41:393. doi: 10.1046/j.1365-2958.2001.02518.x. [DOI] [PubMed] [Google Scholar]
- 11.Kovacikova G, Lin W, Skorupski K. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol Microbiol. 2004;53:129. doi: 10.1111/j.1365-2958.2004.04121.x. [DOI] [PubMed] [Google Scholar]
- 12.Krukonis E, Yu R, DiRita V. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol Microbiol. 2000;31:67–84. doi: 10.1046/j.1365-2958.2000.02111.x. [DOI] [PubMed] [Google Scholar]
- 13.Champion GA, Neely MN, Brennan MA, DiRita VJ. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 14.Yu RR, DiRita VJ. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J Bacteriol. 1999;181:2584–2592. doi: 10.1128/jb.181.8.2584-2592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 16.Taylor JL, De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ. The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH. Mol Microbiol. 2012;83:457. doi: 10.1111/j.1365-2958.2011.07919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezezika OC, Haddad S, Clark TJ, Neidle EL, Momany C. Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator. J Mol Biol. 2007;367:616. doi: 10.1016/j.jmb.2006.09.090. [DOI] [PubMed] [Google Scholar]
- 18.Monferrer D, Tralau T, Kertesz M, Dix I, Sola M, Uson I. Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold. Mol Microbiol. 2010;75:1199–1214. doi: 10.1111/j.1365-2958.2010.07043.x. [DOI] [PubMed] [Google Scholar]
- 19.Devesse L, Smirnova I, Lonneborg R, Kapp U, Brzezinski P, Leonard G, Dian C. Crystal structures of DntR inducer binding domains in complex with salicylate offer insights into the activation of LysR-type transcriptional regulators. Mol Microbiol. 2011;81:354–367. doi: 10.1111/j.1365-2958.2011.07673.x. [DOI] [PubMed] [Google Scholar]
- 20.Derkaoui M, Atunes A, Poncet S, Abdallah JN, Joyet P, Maze A, Henry C, Taha MK, Deutscher J, Deghmane AE. The phosphocarrier protein HPr of Neisseria meningitidis interacts with the transcription regulator. Mol Microbiol. 2016;5:788–807. doi: 10.1111/mmi.13349. [DOI] [PubMed] [Google Scholar]
- 21.Kovacikova G, Lin W, Skorupski K. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J Bacteriol. 2010;192:4181. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Wang H, Zhou Z, Sheng Y, Naseer N, Kan B, Zhu J. Thiol-based switch mechanism of virulence regulator AphB modulates oxidative stress response in Vibrio cholerae. Mol Microbiol. 2016;102:939–949. doi: 10.1111/mmi.13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olsen AJ. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J Comput Chem. 2009;30:2785. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irwin JJ, Sterling T, Mysinger M, Bolstad ES, Coleman RG. Zinc: A Free Tool to Discover Chemistry for Biology. J Chem Inf Model. 2012;52:1757. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer M, Meyer B. Group Epitope Mapping by Saturation Transfer Difference NMR To Identify Segments of a Ligand in Direct Contact with a Protein Receptor. J Am Chem Soc. 2001;123:6108. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 26.Bhunia A, Bhattacharjya S, Chatterjee S. Applications of saturation transfer difference NMR in biological systems. Drug Discovery Today. 2012;17:505–513. doi: 10.1016/j.drudis.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Angulo J, Enriquez-Navas PM, Nieto PM. Ligand – Receptor Binding Affinities from Saturation Transfer Difference (STD) NMR Spectroscopy: The Binding Isotherm of STD Initial Growth Rates. Chem - Eur J. 2010;16:7803. doi: 10.1002/chem.200903528. [DOI] [PubMed] [Google Scholar]
- 28.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expression Purif. 2005;41:207. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 30.The PyMOL Molecular Graphics System, Enhanced for Mac OS X, Version 1.6.0.0. Schrödinger, LLC; [Google Scholar]
- 31.Hayes R, Moural T, Lewis K, Onofrei D, Xun L, Kand C. Structures of the inducer-binding domain of pentachlorophenol-degrading gene regulator PcpR from Sphingobium chlorophenolicum. Int J Mol Sci. 2014;15:20736–20752. doi: 10.3390/ijms151120736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucl. Acids Res., 34:W116-W118. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Bundy B, Collier M, Hoover L, Neidle TLE. Synergistic transcriptional activation by one regulatory protein in response to two metabolites. Proc Natl Acad Sci U S A. 2001;99:7693–7698. doi: 10.1073/pnas.102605799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vadlamani G, Thomas MD, Patel TR, Donald LJ, Reeve TM, Stetefeld J, Standing KG, Vocadlo DJ, Mark BL. The β-Lactamase Gene Regulator AmpR Is a Tetramer That Recognizes and Binds the d-Ala-d-Ala Motif of Its Repressor UDP-N-acetylmuramic Acid (MurNAc)-pentapeptide. J Biol Chem. 2015;290:2630–2643. doi: 10.1074/jbc.M114.618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ilangovan A, Fletcher M, Rampioni G, Pustelny C, Rumbaugh K, Heeb S, Cámara M, Truman A, Chhabra S, Ram E, Jonas P. Structural Basis for Native Agonist and Synthetic Inhibitor Recognition by the Pseudomonas aeruginosa Quorum Sensing Regulator PqsR (MvfR) PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandal R, Ta A, Sinha R, Theeya N, Ghosh A, Tasneem M, Bhunia A, Koley H, Das S. Ribavirin supresses bacterial virulence by targeting LysR-type transcriptional regulators. Sci Rep. 2016;6:39454. doi: 10.1038/srep39454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.