Abstract
Dynamic maintenance of cell polarity is essential for development and physiology. Here we combine experiments and modeling to elucidate mechanisms that maintain cortical polarity in the C. elegans zygote. We show that polarity is dynamically stabilized by two coupled cross-inhibitory feedback loops: one involves the oligomeric scaffold PAR-3 and the kinase PAR-1; the other involves CDC-42 and its putative GAP CHIN-1. PAR-3 and CDC-42 are both required locally to recruit PAR-6/PKC-3, which inhibits PAR-1 (shown previously) and inhibits local growth/accumulation of CHIN-1 clusters. Conversely, PAR-1 inhibits local accumulation of PAR-3 oligomers, while CHIN-1 inhibits CDC-42 (shown previously), such that either PAR-1 or CHIN-1 can prevent recruitment of PAR-6/PKC-3, but loss of both causes complete loss of polarity. Ultrasensitive dependence of CHIN-1 cluster growth on PAR-6/PKC-3 endows this core circuit with bistable dynamics; while transport of CHIN-1 clusters by cortical flow can stabilize the AP boundary against diffusive spread of PAR-6/PKC-3.
Introduction
The ability to polarize is fundamental to cellular life. Cells typically establish polarity in response to transient, localized cues by creating asymmetric distributions of specific molecules or molecular activities (Li and Bowerman, 2010). But how they maintain these asymmetries after the cue is gone, despite dissipative processes such as diffusion and turnover, remains poorly understood.
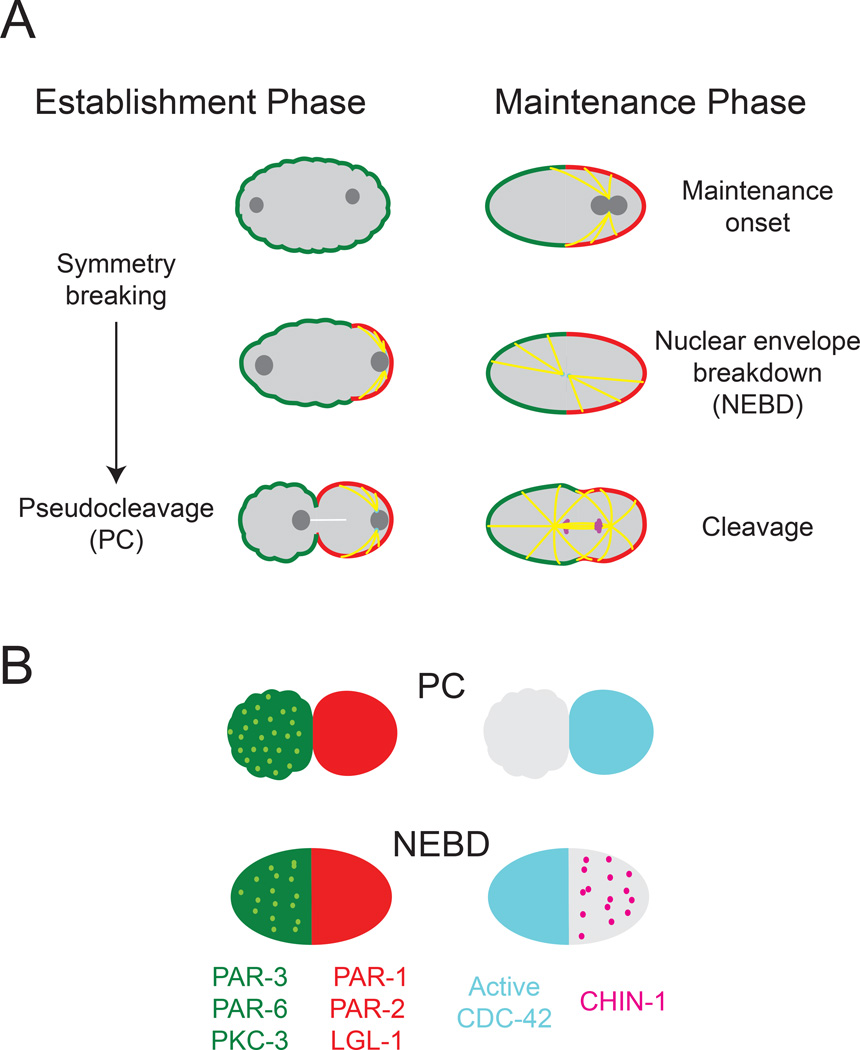
The C. elegans zygote polarizes during mitotic interphase in response to signal(s) from the sperm microtubule organizing center (the sperm MTOC) that forms near the site of sperm entry (Figure 1A; Munro and Bowerman, 2009). The response to these signals involves rapid redistribution of conserved polarity proteins known as PAR proteins into two complementary domains. Just before polarity is established, the oligomeric scaffold PAR-3, and a heterodimer of the adaptor PAR-6 and the atypical kinase PKC-3 (henceforth PAR-6/PKC-3), are enriched on the entire cell surface, where PKC-3 phosphorylates and inhibits cortical association of the kinase PAR-1, the ring-domain-containing protein PAR-2, and the tumor suppressor LGL-1 (Beatty et al., 2010; Boyd et al., 1996; Etemad-Moghadam et al., 1995; Guo and Kemphues, 1995; Hoege et al., 2010; Hung and Kemphues, 1999; Tabuse et al., 1998). Symmetry is broken when local inhibition of actomyosin contractility near the sperm MTOC triggers cortical flows that segregate PAR-3, PAR-6 and PKC-3 towards the anterior pole, allowing PAR-1, PAR-2, and LGL-1 to associate with a complementary posterior domain (Figure 1A) (Cheeks et al., 2004; Motegi and Sugimoto, 2006; Munro et al., 2004; Schonegg and Hyman, 2006). Recently, Motegi et al (2011) identified a second mode of flow-independent symmetry-breaking in which the sperm MTOC promotes local association of PAR-2 with the plasma membrane, where it recruits PAR-1, which phosphorylates and promotes dissociation of PAR-3 (Motegi et al., 2011).
Figure 1. Overview of polarization in the C. elegans zygote.
(A) Overview of key events that occur during polarity establishment and maintenance phases. (B) Distributions of the key polarity proteins at the end of establishment phase (PC) and during maintenance phase at nuclear envelope breakdown (NEBD). Light green dots represent clusters of PAR-3, PAR-6, and PKC-3 that co-localize at the cortex. Magenta dots represent clusters of CHIN-1 that appear during maintenance phase. Note the biphasic localization of active CDC-42 to the posterior during establishment phase and to the anterior during maintenance phase. In this and all subsequent figures, embryos are approximately 50µm in length, and posterior is to the right.
At the end of polarity establishment phase, the sperm MTOC vacates the posterior pole, but PAR asymmetries persist through the rest of the cell cycle. A currently favored idea is that these asymmetries are maintained through a mutual competition between anterior and posterior polarity proteins, which exchange dynamically with the cell surface, diffuse across the AP boundary, and act locally to promote one another’s dissociation (Munro and Bowerman, 2008, Hoege et al, 2010, Beatty et al, 2013, Goehring et al., 2011a). Simple mathematical models show that such a competition can give rise to bistable dynamics, leading to stable coexistence of complementary domains, if one or more exchange rates have ultrasensitive dependence on the local concentrations of participating factors (Dawes and Munro, 2011; Goehring et al., 2011b). Additional mechanisms, such as depletion of a limiting cytoplasmic factor, are required to stabilize the position of the boundary between these two domains (Dawes and Munro, 2011; Goehring et al., 2011b; Mori et al., 2008).
These simple models have been conceptually useful, but for deeper understanding it is essential to resolve a more detailed view of the molecular circuitry that mediates cross-inhibition and to identify key interactions that mediate ultrasensitivity, bistable dynamics and positional stability of the AP boundary. Current models propose a key role for PAR-2 in promoting local dissociation of PAR-6/PKC-3, which can be fulfilled in par-2 mutants by over-expressing LGL-1 (Beatty et al, 2010, Hoege et al, 2010). PAR-2 (and possibly LGL-1) is also required to prevent posterior directed flows that could redistribute PAR-6/PKC-3 during early maintenance (Munro et al., 2004, Beatty et al., 2010), but the relative importance of dissociation and flow for maintaining PAR-6/PKC-3 asymmetries has not been determined.
The roles of other factors beyond PAR-6/PKC-3, PAR-2 and LGL-1 in polarity maintenance remain poorly characterized. During polarity maintenance, the small GTPase CDC-42 becomes active at the anterior pole, while its putative GAP CHIN-1 accumulates at the posterior pole through unknown mechanisms (Kumfer et al, 2010). Both active CDC-42 and PAR-3 bind PAR-6/PKC-3 and both are required for cortical recruitment of PAR-6/PKC-3 (Gotta et al, 2001; Aceto et al., 2006; Li et al., 2010b, Beers and Kemphues, 2006). But the nature of this dual requirement, and how local recruitment (as opposed to local dissociation) shapes spatial distributions of PAR-6/PKC-3 has not been adequately explored. Depletion of CHIN-1 leads to uniform activation of CDC-42 during polarity maintenance, but with minimal effects on PAR asymmetries and embryo viability (Kumfer et al, 2010). Recent genetic studies suggest that roles for CHIN-1 may be masked by redundancies with other factors such as PAR-2 and LGL-1 (Beatty et al, 2013), but the nature of these redundancies has not been established. Similarly, PAR-1 can phosphorylate and displace PAR-3 under certain conditions (Motegi et al, 2011), but depletion of PAR-1 is reported to have no effect on PAR-3 asymmetries (Etemad-Moghadam et al., 1995) and minor effects on PAR-6/PKC-3 during polarity maintenance (Cuenca et al, 2003, Zonies et al, 2010), suggesting that it too may function redundantly with other factors. Finally, both PAR-3 and CHIN-1 form clusters at the cell membrane (Hung and Kemphues, 1999; Kumfer et al., 2010; Tabuse et al., 1998), and may therefore exhibit more complex dynamics of exchange and mobility than assumed in current models. However, the significance of clustering for polarity maintenance has not been explored.
Here, we combine quantitative imaging with genetic perturbations and mathematical modeling to identify core circuitry and dynamical mechanisms that stabilize cortical polarity in the zygote. We show that cortical asymmetries are stabilized by two dynamically coupled cross-inhibitory circuits in which (a) PAR-3 is required for local association of PAR-6/PKC-3 with CDC-42; (b) PAR-6/PKC-3 inhibit local accumulation of PAR-1 and CHIN-1 clusters; and (c) PAR-1 and CHIN-1 act redundantly to prevent local accumulation of PAR-6/PKC-3. We show that dynamic clustering of CHIN-1 and PAR-3 has two key consequences for circuit-level dynamics: Ultrasensitive dependence of CHIN-1 cluster growth on PAR-6/PKC-3 (and likely a similar dependence of PAR-3 clusters on PAR-1) endows the polarity circuit with bistable dynamics, while local coupling of CHIN-1 and PAR-3 clusters to cortical flow helps to stabilize AP boundary position. We propose that similar mechanisms may act in many other contexts where the highly conserved members of this polarity circuit form dynamically stable cellular asymmetries.
Results
PAR-2 and LGL-1 prevent loss of PAR asymmetry during maintenance by controlling spatial patterns of cortical flow
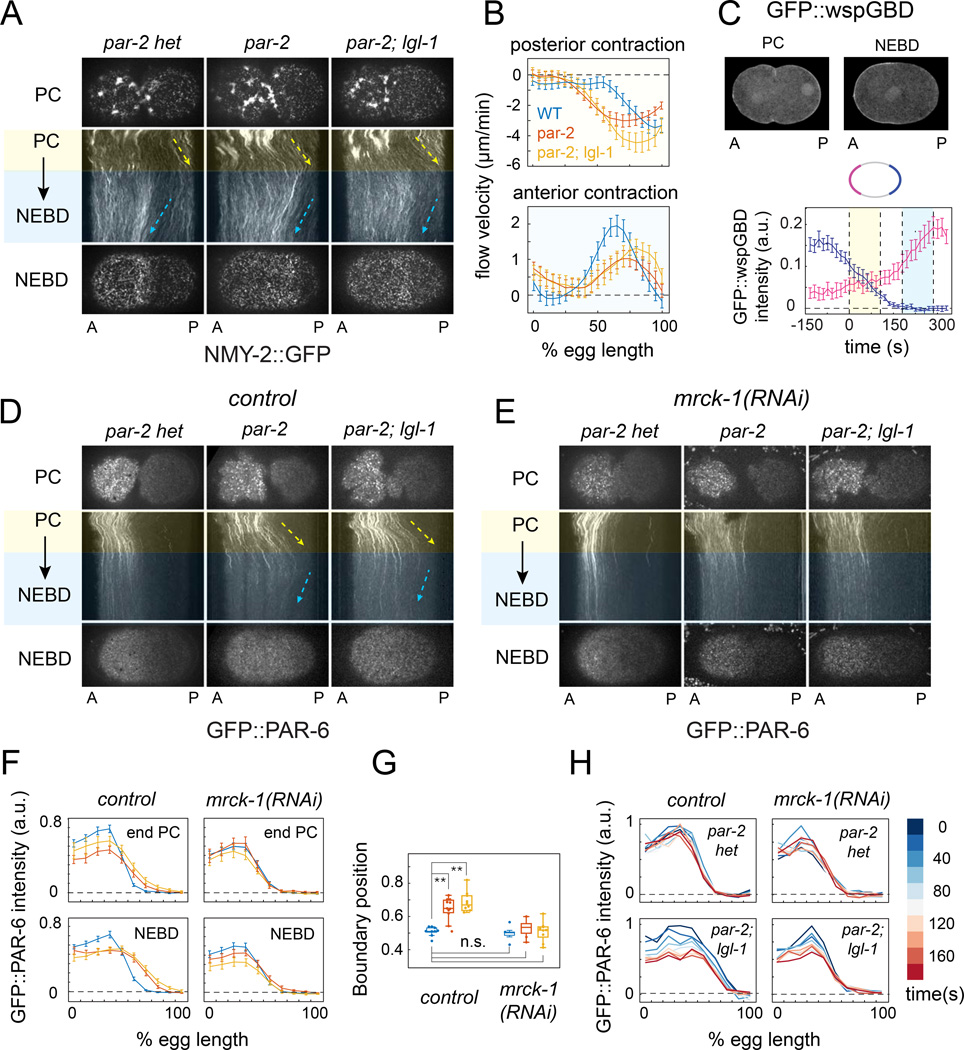
As a first step towards distinguishing flow-dependent and independent mechanisms for polarity maintenance, we quantified patterns of cortical flow in wild type, par-2 and par-2;lgl-1 embryos expressing non-muscle myosin II fused to GFP (NMY-2::GFP; Figure 2A & B). In wild type embryos, a previously uncharacterized period of posterior contraction and flow began with the onset of maintenance (end PC) and lasted ∼80 seconds, followed by anterior-directed flows that persist until metaphase (Munro et al, 2004; Figure 2A). Biphasic contraction and flow coincided with biphasic localization of active CDC-42, as previously revealed by a biosensor for active CDC-42 (GFP::GBDwsp-1; (Kumfer et al., 2010); Figure 2C), and both phases were completely abolished by depletion of the kinase MRCK-1, which is required for Myosin II activation downstream of CDC-42 (Kumfer et al., 2010 ; Figure S1A,B).
Figure 2. PAR-2 and LGL-1 prevent loss of PAR asymmetry during maintenance by patterning cortical flow.
(A) Dynamics of cortical myosin II (NMY-2::GFP) during maintenance phase in control (par-2(lw32) heterozygotes), par-2(lw32), and par-2(lw32);lgl-1(tm-2616) embryos. Top and bottom rows show surface views of cortical NMY-2::GFP at late PC and NEBD. Kymographs show patterns of cortical flow during maintenance phase. Dashed rectangle in top left of (A) indicates region from kymographs were constructed in (A), (D) and (E). Yellow and blue shading indicates posterior and anterior contraction phases respectively (see main text for details). Arrows indicate general direction and speed of cortical flow. (B) Cortical flow velocities during posterior (top) and anterior (bottom) contraction phases for wild type, par-2 and par-2;lgl-1 embryos. Dashed vertical line indicates approximate position of GFP:PAR-6 boundary (see F). Error bars indicate +/− 1 SEM for n = 10 embryos. (C) (top) Localization of a biosensor for active CDC-42 (GFP::wspGBD) at PC and NEBD in a wild-type embryo; (bottom) GFP::wspGBD fluorescence intensity vs. time for anterior (magenta) and posterior (blue) domains showing biphasic accumulation of the biosensor. Error bars indicate mean +/− 1 SEM for n = 12 embryos aligned to the onset of maintenance (t = 0). (D&E) Dynamics of cortical GFP::PAR-6 during maintenance phase in embryos of the indicated genotypes without (D), or with (E) mrck-1(RNAi). Surface views and kymographs are as in (A). (F) GFP::PAR-6 intensity vs. anterior/posterior position at the end of PC and NEBD in control (n = 10), par-2(lw32) (n = 11), par-2(lw32);lgl-1(tm2616) (n = 10), mrck(RNAi) (n = 10), par-2(lw32); mrck(RNAi) (n = 10), and par-2(lw32); lgl-1(tm2616); mrck(RNAi) (n = 10) embryos. Error bars indicate +/− 1 SEM. (G) Box and whisker plots showing the distributions of GFP::PAR-6 boundary position for the data in (F). ** p < 0.005 by Students T test. (H) GFP::PAR-6 intensity profiles measured for the same genotypes at 20 sec intervals during maintenance phase in single embryos. Heat map indicates time relative to the beginning of the anterior contraction phase. See also Figure S1 and Movies S1 & S2.
Posterior contraction and flow occurred with normal timing in par-2 and par-2;lgl-1 mutant embryos (Figure 2A). However, the domain of contraction was significantly broader than in wild type embryos (Figure 2B); it overlapped the PAR-6::GFP boundary (dashed vertical line in Figure 2B), and posterior contraction was accompanied by a rapid posterior expansion of GFP::PAR-6 (Figure 2D,F; Movie S1). In par-2;lgl-1 embryos, the magnitude of posterior flow, and posterior expansion of PAR-6::GFP, were slightly enhanced relative to par-2 alone (Figure 2B,F).
To test if redistribution by cortical flow is primarily responsible for loss of PAR-6 asymmetry in par-2 and par-2;lgl-1 embryos, we co-depleted these embryos of MRCK-1 to inactivate maintenance phase contractility. Indeed, posterior expansion of PAR-6::GFP during early maintenance was completely abolished in mrck-1(RNAi), par-2;mrck-1(RNAi) and par-2;lgl-1;mrck-1(RNAi) embryos (Figure 2E,F; Movie S2). The distributions of GFP::PAR-6 in these embryos were essentially wild type (Figure 2F), and remained stable through late maintenance phase (Figure 2G). Rapid posterior expansion of PAR-3::GFP also occurred during early maintenance in par-2 and par-2;lgl-1 embryos and was also rescued by co-depletion of MRCK-1 (Figure S1C,D and data not shown). We conclude: (a) that PAR-2 acts primarily during maintenance to restrict the domain of posterior contraction and prevent redistribution of anterior PAR proteins towards the posterior pole, (b) that LGL-1 makes a minor contribution in par-2 mutants by attenuating the magnitude of posterior flow, and (c) that other factors are completely sufficient to stabilize an anterior enrichment of PAR-6/PKC-3 in the absence of PAR-2 and LGL-1 when contractility is inhibited.
PAR-1 and CHIN-1 act redundantly to prevent loss of PAR-6 asymmetry during maintenance phase
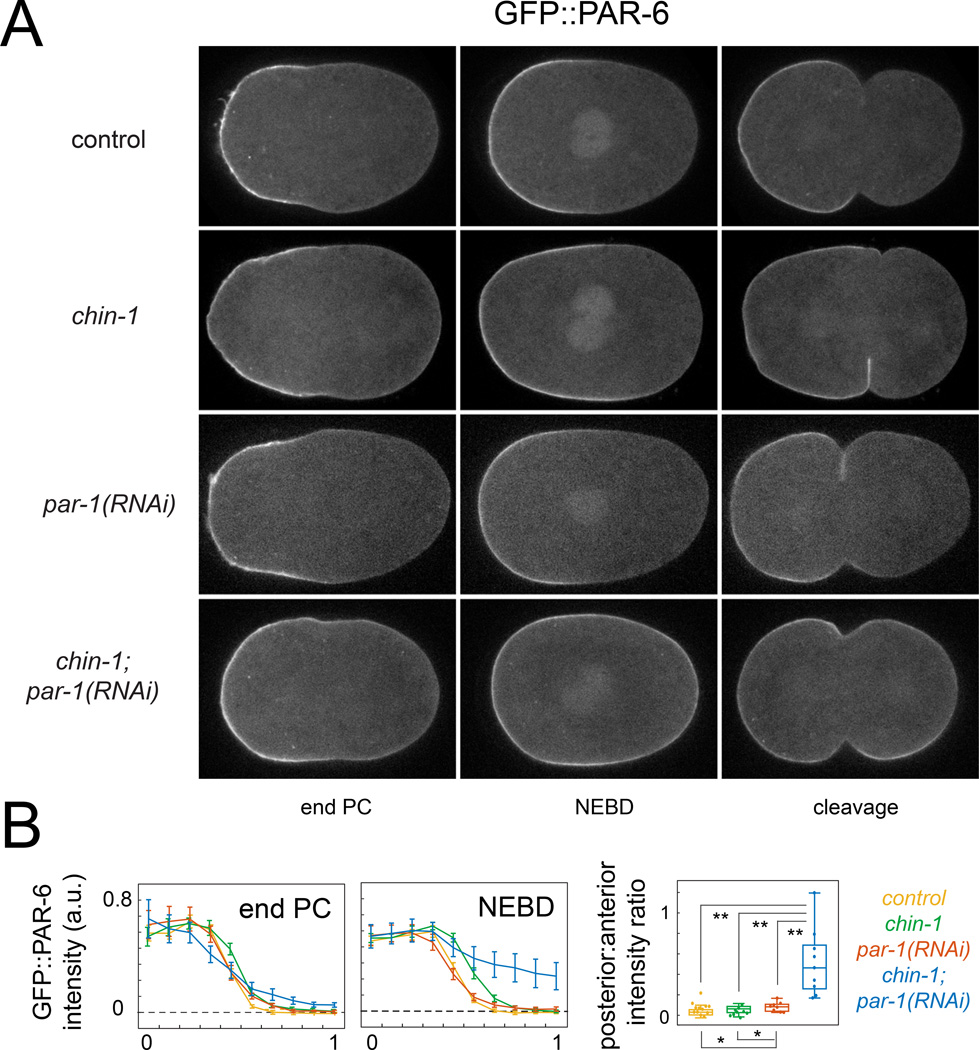
Two obvious candidates are the posterior factors PAR-1 and CHIN-1. Consistent with previous reports (Cuenca et al., 2003; Kumfer et al., 2010), depletion of CHIN-1 or PAR-1 alone had minor effects on PAR-6::GFP asymmetries. In chin-1(tm1909) mutant embryos, posterior contraction accompanied broadening of the GFP::PAR-6 domain during early maintenance, but this was completely rescued by co-depletion of MRCK-1 (Figure 3A,B; Figure S2A,B; Movie S4). In both chin-1(tm1909) and par-1(RNAi) embryos, the spatial profile of GFP::PAR-6 was stable from NEBD to late metaphase (Figure S2C).
Figure 3. PAR-1 and CHIN-1 act redundantly to prevent loss of PAR-6 asymmetry during maintenance phase.
(A) Equatorial views of GFP::PAR-6 at the end of PC, NEBD and cleavage and (B) GFP::PAR-6 intensity profiles in control (chin-1(tm1909) heterozygotes; n = 9), chin-1(tm1909) (n =12), par-1(RNAi) (n = 8), and chin-1(tm1909);par-1(RNAi) (n = 9) embryos at late PC and NEBD. Error bars indicate +/− 1 SEM. (C) Box and whisker plots showing distributions of Posterior: Anterior (P/A) intensity ratios for the data in (B). * p < 0.05; **p < 0.005 by Students T test. See also Figure S2 and Movies S3 & S4.
By contrast, in chin-1; par-1(RNAi) embryos, there was a rapid loss of GFP::PAR-6 asymmetry during early maintenance that was invariably followed by a symmetric cell division (Figure 3A,B; Movie S4). Unlike in par-2, par-2;lgl-1 and chin-1 mutant embryos, this loss of asymmetry was not rescued by co-depletion of MRCK-1 (Figure S2A,B; Movie S4). Thus, PAR-1 and CHIN-1 act redundantly during maintenance phase to prevent posterior accumulation of PAR-6 through mechanisms that do not involve local inhibition of contractility and flow.
PAR-3 acts locally to gate cortical association of PAR-6/PKC-3 with active CDC-42
Both PAR-3 and active CDC-42 bind PAR-6/PKC-3 and both are required for normal accumulation of PAR-6/PKC-3 during maintenance phase (Aceto et al., 2006; Beers and Kemphues, 2006; Hung and Kemphues, 1999; Tabuse et al., 1998). In principle, simultaneous posterior accumulation of active CDC-42 and PAR-3 in chin-1; par-1(RNAi) embryos could explain rapid posterior accumulation of PAR-6/PKC-3. A previous study reported normal distributions of PAR-3 in PAR-1-depleted embryos (Etemad-Moghadam et al., 1995). However, weak accumulation of PAR-3 might have escaped detection in these studies and might be sufficient to promote strong PAR-6/PKC-3 accumulation in the presence of active CDC-42. Alternatively, posterior activation of CDC-42 caused by loss of CHIN-1 might enhance posterior accumulation of PAR-3 (Aceto et al., 2006) in the absence of PAR-1.
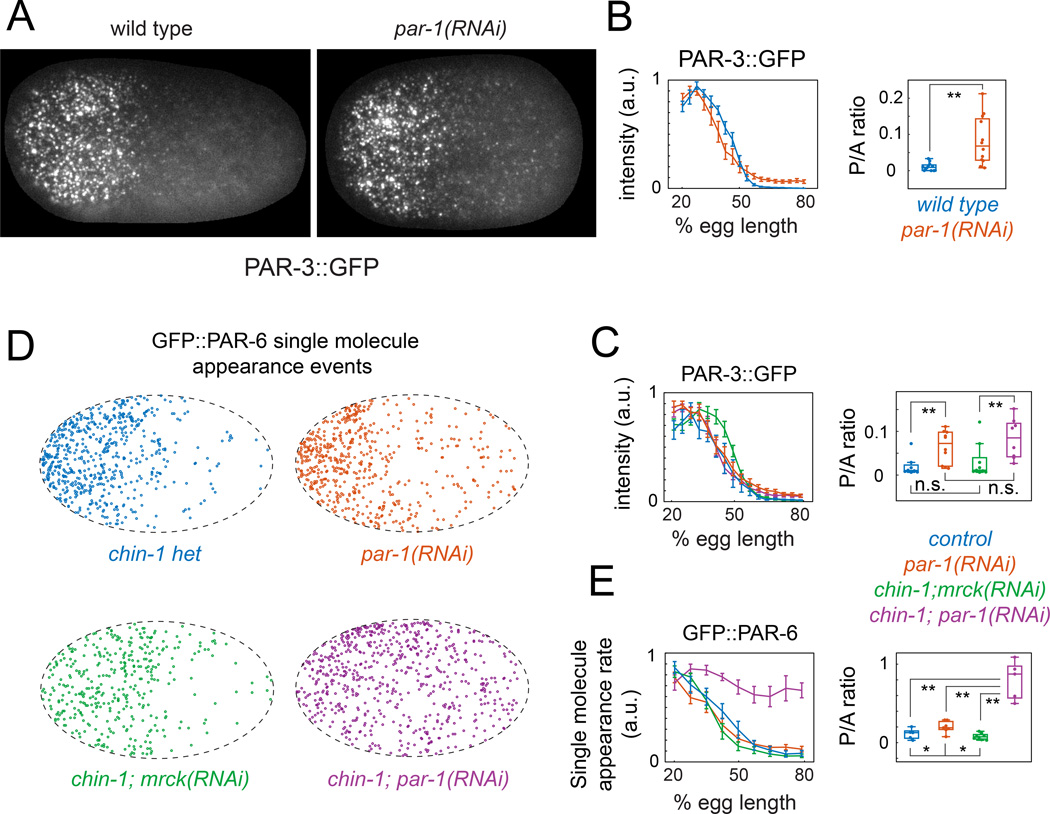
To look for weak accumulation of PAR-3 in par-1(RNAi) embryos, we used a strain expressing GFP::PAR-3 from the endogenous locus by CRISPR-mediated homologous recombination (Dickinson et al., 2013). We used imaging conditions that allow sensitive detection of single GFP molecules (Robin et al., 2014; see Supplementary Methods). In wild type embryos during maintenance phase, PAR-3::GFP was enriched on the anterior cortex in discrete clusters, with a broad range of sizes (Figure 4A; Figure S3A; Movie S5), consistent with previous reports (Hung and Kemphues, 1999; Tabuse et al., 1998) and with the known ability of PAR-3 to oligomerize (Benton and St Johnston, 2003a; Feng et al., 2007; Li et al., 2010a; Mizuno et al., 2003)). However, PAR-3 clusters were exceedingly sparse on the posterior cortex (∼ 0.05 clusters/µm2, Figure 4B, Figure S3B,C), with a mean intensity close to that measured for single molecules under the same imaging conditions (Figure S3A).
Figure 4. PAR-3 acts locally to gate cortical association of PAR-6/PKC-3 with active CDC-42.
(A) Surface views of cortical PAR-3::GFP at early maintenance in wild-type and par-1(RNAi) embryos. (B) PAR-3::GFP intensity vs. AP position measured during early maintenance phase in wild type (n = 12) and par-1(RNAi) (n = 12) embryos. (C) PAR-3::GFP intensity vs. AP position during early maintenance in wild type (n = 8), chin-1(tm1909);mrck- 1(RNAi) (n = 10), par-1(RNAi) (n = 8) and chin-1(tm1909);par-1(RNAi) (n = 8) embryos. (D) Spatial distributions of single molecule appearance events for GFP::PAR-6 during early maintenance in embryos with the indicated genotypes (E) Plots of single molecule appearance rates vs. AP position in wild-type (n = 6), chin-1(tm1909);mrck-1(RNAi) (n = 6), par-1(RNAi) (n = 6), and chin-1(tm1909);par-1(RNAi) (n = 6) embryos. Error bars in B,C,E indicate +/− 1 SEM. Box and whisker plots in B,C,E show distributions of Posterior::Anterior (P/A) ratios for the data shown in the graphs. *p < 0.05; **p < 0.005 by Students T test. See main text and experimental procedures for details. See also Figure S3 and Movies S5 & S6.
By contrast, during maintenance phase in par-1(RNAi) embryos, PAR-3::GFP accumulated on the posterior cortex to ∼10% of the anterior levels. (Figure 4A,B; Figure S3B; Movie S5). This reflected an increase in both the number and mean intensity of PAR-3::GFP clusters (Figure S3C & D), consistent with the possibility that PAR-1 inhibits oligomerization of PAR-3 (Benton and St Johnston, 2003b).
To ask if posterior activation of CDC-42 can enhance the weak posterior accumulation of PAR-3 caused by depletion of PAR-1, we compared embryos singly and doubly-depleted of PAR-1 and CHIN-1, using embryos that express transgenic PAR-3::GFP at levels similar to the CRISPR allele (Figure S3D). In chin-1(tm1909) embryos, PAR-3::GFP spread towards the posterior pole during early maintenance, but this was reversed by co-depletion of MRCK-1 (Figure S3E), and there was no significant difference in posterior PAR-3::GFP intensities between chin-1(tm1909);mrck-1(RNAi) and wild type embryos (Figure 4C). Likewise, we observed weak posterior accumulation of PAR-3::GFP to similar levels in par-1(RNAi) and chin-1(tm1909);par-1(RNAi) embryos (Figure 4C). Thus, posterior activation of CDC-42 does not enhance PAR-3 accumulation in either wild type or par-1(RNAi) embryos.
To ask if weak accumulation of PAR-3::GFP, combined with local activation of CDC-42, is sufficient for robust local recruitment of PAR-6/PKC-3, we used single molecule imaging to measure spatial differences in GFP::PAR-6 recruitment rates during maintenance phase in wild-type, chin-1(tm1909), par-1(RNAi), and chin-1(tm1909);par-1(RNAi) embryos. We used gfp(RNAi) to reduce levels of the GFP::PAR-6 transgene (Robin et al., 2014); then we imaged under conditions (low density and rapid photobleaching; see Supplementary Methods) that allow unambiguous detection of single molecule appearance events (Figure 4D, Movie S6). In wild-type embryos, the spatial distribution of average appearance rates was highly asymmetric (Figure 4D & E). The A to P ratio of appearance rates (9.2 +/− 4.1) matched the A to P ratio of single molecule densities (9.0 +/− 1.4) measured previously under low photobleaching conditions (Robin et al., 2014). Thus asymmetric recruitment, not asymmetric dissociation, determines asymmetric distributions of PAR-6 during maintenance phase in wild type embryos.
GFP::PAR-6 recruitment rates were also strongly asymmetric in chin-1(tm1909); mrck-1(RNAi) and par-1(RNAi) embryos (Figure 4E). However, in chin-1(tm1909);par-1(RNAi) embryos, GFP::PAR-6 recruitment rates were strikingly symmetrical (Figure 4E), consistent with the rapid loss of PAR-6 asymmetry observed in these embryos (Figure 3). We conclude that in the presence of active CDC-42, weak accumulation of PAR-3 is both necessary and sufficient to promote rapid PAR-6 recruitment and complete loss of PAR-6 asymmetry.
PAR-6/PKC-3 control CDC-42 activity by inhibiting local accumulation of CHIN-1 clusters
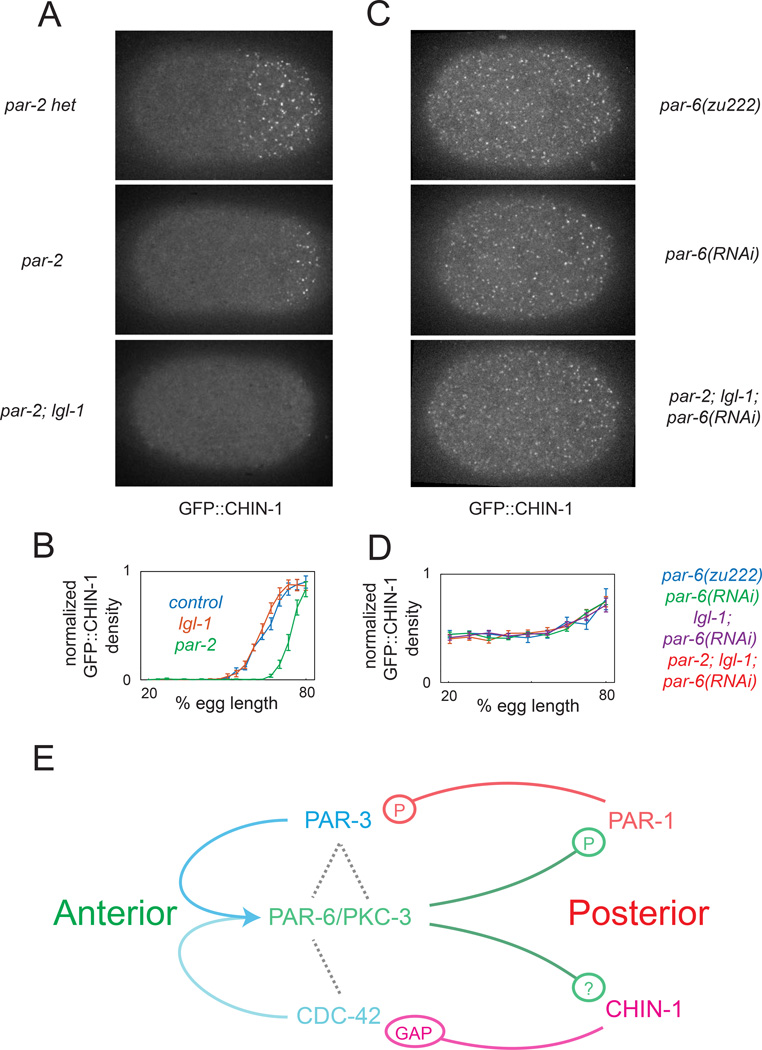
Our results show that posterior CHIN-1 acts redundantly with PAR-1 to prevent local accumulation of PAR-6/PKC-3, but what restricts CHIN-1 to the posterior pole? Kumfer et al (2010) suggested that posterior PAR-2 promotes CHIN-1 recruitment. However, in par-2(RNAi) embryos, CHIN-1 clusters accumulate with normal timing, but in a significantly smaller posterior domain, which is complementary to the expanded PAR-6 domain (Figure 5A,B; Figure S4A,B and data not shown). CHIN-1 clusters accumulated normally in lgl-1(tm2616) embryos, which show normal distributions of PAR-6 during maintenance (Figure 5C and data not shown). In par-2(lw32);lgl-1(tm2616) double mutants, they were either absent, or accumulated weakly in a small posterior domain (Figure 5A and data not shown).
Figure 5. PAR-6/PKC-3 inhibit local accumulation of CHIN-1 clusters.
(A) Surface views of CHIN-1::GFP at late maintenance phase in control (par-2(lw32) heterozygotes), par-2(lw32), and par-2(lw32);lgl-1(tm2616) embryos. (B) Plots of normalized CHIN-1::GFP intensity vs AP position for control (n = 8), lgl-1(tm2616) (n = 8), and par-2(lw32) (n =8) embryos. Boundary shift between control or lgl-1 and par-2 is significant at p < 0.0001 by Students t-test. (C) Surface views of CHIN-1::GFP at late maintenance phase in par-6(zu222), par-6(RNAi), and par-2(lw32);lgl-1(tm2616);par-6(RNAi) embryos. (D) Plots of normalized CHIN-1::GFP intensity vs. AP position at late maintenance in par-6(zu222) (n = 12), par-6(RNAi) (n = 12), lgl-1(tm2616);par-6(RNAi) (n = 12), and par-2(lw32);lgl-1(tm2616); par-6(RNAi) (n = 11). Error bars in (B) and (D) indicate +/− 1 SEM. (E) Schematic view of a core circuit for polarity maintenance based on our data and previous work. See text for details. See also Figure S4 and Movie S7.
An alternative possibility is that anterior PAR proteins inhibit local accumulation of CHIN-1 clusters and that PAR-2 and LGL-1 affect CHIN-1 accumulation indirectly by affecting the distribution of PAR-6/PKC-3. Indeed, in par-6(zu222) mutants or par-6(RNAi) embryos, CHIN-1 clusters accumulated with normal timing but throughout the cortex (Figure 5C; Figure S4A,B). Moreover, we observed similarly broad, normally timed, accumulation of CHIN-1::GFP clusters either in par-2;lgl-1 mutants subjected to par-6(RNAi) (Figure 5C,D), or in par-6(zu222) embryos subjected to par-2(RNAi) (Figure S4A). We obtained similar results using RNAi to deplete CDC-42, PAR-3, or PKC-3 instead of PAR-6 (Figure S4C). We conclude that PAR-6/PKC-3 act locally to inhibit CHIN-1 cluster growth independently of PAR-2 and LGL-1.
Dynamic analysis of CHIN-1 clusters reveals ultrasensitive dependence of cluster growth on PAR-6 levels
Based on our results and previous work, we propose that polarity is stabilized by two dynamically coupled feedback circuits (Figure 5E), one involving cross-inhibition between PAR-1 and PAR-3, the other cross-inhibition between CDC-42 and CHIN-1. These two circuits are coupled through a dual requirement for active CDC-42 and PAR-3 to recruit PAR-6.PKC-3, which inhibits local accumulation of both PAR-1 and CHIN-1. In particular, our results imply that PAR-3/PAR-1 feedback is sufficient to stabilize PAR-6/PKC-3 asymmetries in the absence of CHIN-1, when CDC-42 is uniformly active, while CDC-42/CHIN-1 feedback is sufficient to stabilize polarity in the absence of PAR-1, when PAR-3 is everywhere above the threshold for recruiting PAR-6/PKC-3.
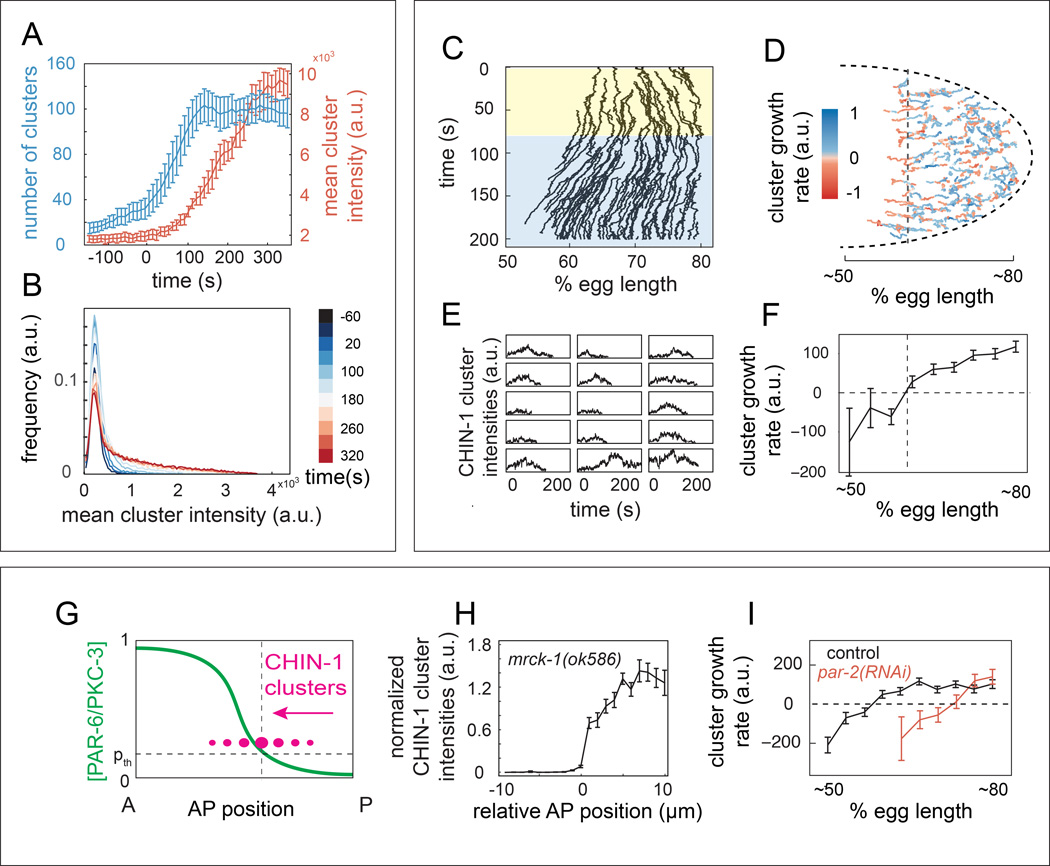
Both PAR-3 and CHIN-1 form clusters at the cell membrane, and we hypothesized that non-linear effects associated with cluster assembly might endow each sub-circuit with bistable dynamics. To test this possibility, we focused on the CDC-42/PAR-6/PKC-3/CHIN-1 sub-circuit and on CHIN-1 clusters, whose dynamics can be readily followed using high speed imaging and single particle tracking (SPT). CHIN-1 clusters first appeared at the onset of maintenance in a broad posterior domain. The number of clusters increased rapidly for the first ∼100 s and then remained ∼constant (Figure 6A; blue trace), while the mean cluster intensity increased steadily through late maintenance (Figure 6A; red traces; Figure 6B). Clusters often appeared to merge and split, however the majority of clusters could be readily tracked, through mergers and splits, over hundreds of seconds, implying stable association with the cell surface (data not shown).
Figure 6. Dynamic analysis of CHIN-1 clusters reveals ultrasensitive dependence of cluster growth on PAR-6/PKC-3 levels.
(A) Number (blue) and mean intensity (red) of CHIN-1 clusters vs. time relative to maintenance phase onset (t = 0s; error bars indicate mean +/− 1 SEM; n = 5 embryos). (B) Normalized distribution of CHIN-1 cluster intensities at successive time points during maintenance phase (averaged over n = 5 embryos). (C) Kymograph representation of cluster trajectories within the posterior half of a single embryo. Color overlays indicate posterior (yellow) and anterior (blue) contraction phases, as shown in Figure 2. (D) Cluster trajectories from the same embryo during maintenance. Blue = positive growth; red = negative growth. (E) Plots of intensity vs. time for individual clusters that cross the dashed line in (D). (F) Mean cluster growth rate vs. AP position for the embryo in (D). (G) Hypothesized transition of CHIN-1 clusters (magenta) from growth to decay as they cross a threshold level of PAR-6/PKC-3, indicated by the dashed horizontal line. (H) Plot of normalized CHIN-1::GFP intensity vs. AP position in mrck-1 (ok586) embryos. Error bars indicate mean +/− 1 SEM, n = 12. Data for individual embryos were aligned to the position at which CHIN-1::GFP intensity crossed 30% of its maximum value. (I) Plots of mean cluster growth rate vs. AP position for CHIN-1::GFP clusters during maintenance in wild type (n = 5) and par-2(RNAi) (n = 5) embryos. In F and I, error bars indicate mean +/− 1 SEM for n > 10 (F) and n > 50 (I) measurements of cluster growth rate per position. See also Figure S6 and Movie S7.
Significantly, we found that CHIN-1 clusters coupled strongly to cortical flow, moving towards the posterior during early maintenance, and towards the anterior during mid-late maintenance (Figure 6C; Movie S7). Strikingly, many of the clusters that moved anterior with flow near the edge of the CHIN-1 domain underwent a transition from growth to decay, leading ultimately to cluster disappearance (Figure 6D,E; Movie S7). These transitions were typically sharp (Figure 6E), and occurred within a narrow range of AP positions (vertical dashed lines in Figure 6D). Plotting mean growth rates vs. AP position for all CHIN-1 clusters in the same embryo revealed a general transition from cluster growth to decay at a similar AP position (Figure 6D,F). In wild-type embryos, cortical flow combined with decay tended to broaden the CHIN-1 boundary. However, in mrck-1(ok586) embryos, which lack maintenance phase flow, the fall-off in CHIN-1 cluster intensity became very sharp, increasing from ∼0 to 50% over a distance of < 1µm (Figure 6H), further confirming that a transition from growth to decay occurs at a sharply-defined AP position.
Since PAR-6/PKC-3 inhibits CHIN-1 cluster formation (Figure 5C,D), and the distribution of GFP:PAR-6 is stable during mid-late maintenance (Figure 2C,F), these data suggest that CHIN-1 clusters switch from growth to decay at a threshold density of PAR-6/PKC-3 (Figure 6G). To test this further, we examined CHIN-1 cluster dynamics in par-2(lw32) mutants in which the PAR-6 distribution is shifted posterior during early maintenance (Figure 2C,E). Indeed, the transition from mean cluster growth to decay still occurred in these embryos. However, there was a significant posterior shift in the transition point by ∼13% egg length relative to controls (p < 0.0001 by Students t-test; Figure 6I), which is similar in magnitude to the posterior shift in the GFP::PAR-6 boundary in par-2 mutants (compare Figure 6I and Figure 2E).
Ultrasensitive dependence of CHIN-1 cluster assembly on PAR-6/PKC-3 yields bistable dynamics
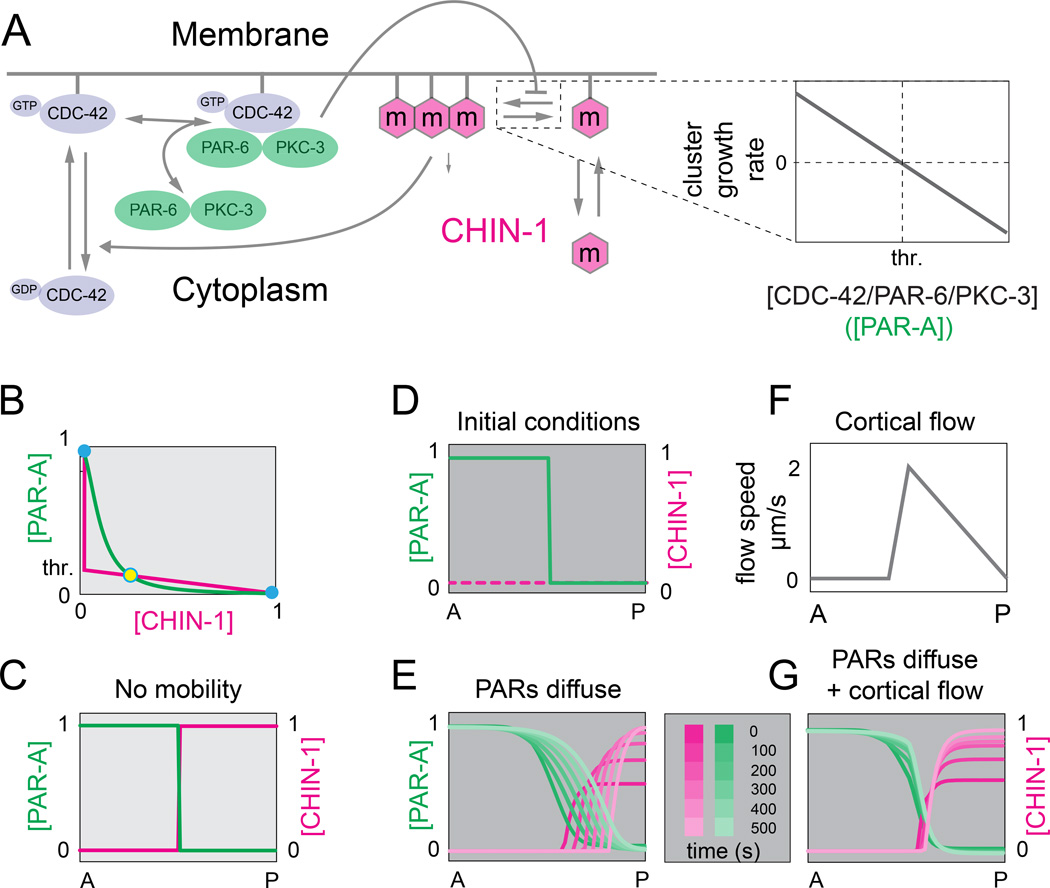
To ask whether ultrasensitive dependence of CHIN-1 cluster growth on local concentrations of PAR-6/PKC-3 could lead to bistable dynamics, we analyzed a mathematical model of the CDC-42/PAR-6/PKC-3/CHIN-1 sub-circuit. (Figure 7A; see Supplementary Methods for details). We assumed that PAR-1 is absent, and thus PAR-3 is not limiting for recruitment of PAR-6/PKC-3. We used mass action kinetics to model reversible activation of CDC-42, reversible binding of cytoplasmic PAR-6/PKC-3 heterodimers to active (membrane-bound) CDC-42, and reversible exchange of CHIN-1 monomers, which assemble at the membrane into CHIN-1 clusters. We assumed that CHIN-1 promotes local inactivation of CDC-42 at a rate proportional to local concentrations of CHIN-1 and CDC-42.
Figure 7. Ultrasensitive dependence of CHIN-1 cluster growth on PAR-6/PKC-3 levels yields bistable dynamics and a stable boundary position.
(A) Schematic view of model assumptions: CDC-42 cycles between inactive and active forms; PAR-6/PKC-3 dimers reversibly bind active CDC-42 from a cytoplasmic pool. CHIN-1 monomers cycle between the cytoplasm and plasma membrane, where they assemble into larger CHIN-1 clusters. CHIN-1 clusters act locally to promote inactivation and dissociation of CDC-42; CDC-42/PAR-6/PKC-3 trimers inhibit local clustering of CHIN-1, such that clusters grow below a threshold level of CDC-42/PAR-6/PKC-3 (expanded view at right; see Supplementary Modeling Procedures for details). (B) Steady state dependence of [CHIN-1] on [PAR-A] (magenta curve), and [PAR-A] on [CHIN-1] (green curve). PAR-A represents the CDC-42/PAR-6/PKC-3 trimer. Steady states occur where the two curves intersect. Solid blue circles indicate stable steady states; open blue circle indicates an unstable steady state. (C) Spatial coexistence of the two stable states in the absence of diffusion. (D) Initial conditions for the simulations shown in E and G. (E) Predicted PAR-A and CHIN-1 distributions vs. time for simulations that incorporate measured values for PAR-6/PKC-3 and CHIN-1 mobility and turnover. (F,G) Predicted PAR-A and CHIN-1 distributions vs. time (G) for simulations that incorporate same parameter values as in (E), and the flow field shown in (F). See Supplemental Modeling Procedures for details. See also Figure S5.
Because the molecular details are still unclear, we used a simple phenomenological description of CHIN-1 cluster growth that could be sharply constrained by experimental observations. We assumed that CHIN-1 clusters undergo net growth above a critical monomer concentration, and that CDC-42/PAR-6/PKC-3 modulates this critical concentration through λ = λ0 + η[PAR-6/PKC-3] where λ0 is the critical concentration in the absence of CDC-42/PAR-6/PKC-3 and η represents the steepness of the dependence. Together these assumptions imply a threshold concentration of PAR-6/PKC-3 (p) at which CHIN-1 clusters switch from growth to decay. Assuming that PAR-6/PKC-3 modulates monomer density relative to a fixed critical concentration leads to identical conclusions (see Supplementary Modeling Procedures for details).
Steady state analysis of the model equations shows that the CDC-42/PAR-6/PKC-3/CHIN-1 circuit could exhibit bistable dynamics, but whether it does so depends on the threshold p. and the strength with which CHIN-1 inhibits CDC-42. To estimate p, we compared steady state distributions of GFP::PAR-6 with the AP position at which CHIN-1 clusters switch from growth to decay (Figure S5A). To estimate the strength with which CHIN-1 inhibits CDC-42 activity, we plotted posterior levels of active CDC-42 against posterior levels of GFP:: CHIN-1 at successive time points during maintenance phase as CHIN-1 increases from minimal to maximal values (Figure S5B,C; Supplementary Modeling Procedures). For these measured values, the steady state analysis predicts two stable states, confirming that the CDC-42/PAR-6/PKC-3CHIN-1 sub-circuit is in fact tuned to exhibit bistable dynamics (Figure 7B,C; Figure S5D).
We then simulated spatiotemporal dynamics of the CDC-42/CHIN-1 circuit, using empirically measured values for PAR-6 diffusivity and turnover (Goehring et al, 201 1a; Robin et al, 2014) and CHIN-1 cluster mobility (Figure S6A), and initial conditions that mimic maintenance phase onset (Figure 7D, see Supplementary Modeling Procedures). Strikingly, the simulations predict that the position of the AP boundary is intrinsically unstable; diffusive spread of PAR-6/PKC-3 and a low threshold for inhibition of CHIN-1 cluster growth invariably lead to a posterior drift of the AP boundary (Figure 7E).
Consistent with this prediction, we found that the GFP::PAR-6 boundary drifts steadily towards the posterior pole in par-1(RNAi); mrck-1(RNAi) embryos which lack cortical flow, but not in either wild type or par-1(RNAi) embryos, which exhibit anterior directed flows during mid-late maintenance phase (Figure S2A,C, Figure S6C). We wondered if anterior transport of CHIN-1 clusters by cortical flow (Figure 6C) could counterbalance the tendency of PAR-6/PKC-3 to invade the posterior domain. Indeed, when we introduced the observed pattern of cortical flow into our simulations (Figure 2A; Figure 7F), and adjusted the turnover rates of clustered CHIN-1 to approximate the observed half-life of CHIN-1 clusters at the anterior edge of the CHIN-1 domain (∼30 s; see Figure 6E), our simulations now predicted a stably positioned AP boundary (Figure 7G). Thus the combination of diffusive spread of CDC-42/PAR-6/PKC-3, and a counterbalancing flow of CHIN-1 clusters, is sufficient to stabilize AP position.
Interestingly, PAR-3 clusters also exhibit slow sub-diffusive mobility (Figure S6B) and couple strongly to cortical flows near the edge of the anterior domain during polarity maintenance in both wild type and par-1(RNAi) embryos (Figure S6C,C’). The PAR-3 boundary position was stably maintained in both par-1(RNAi) embryos, but in mrck-1; par-1(RNAi) embryos, we observed a gradual spread of PAR-3::GFP during maintenance phase (Figure S6C,C’). Thus transport of PAR-3 clusters by cortical flow may also counteract an intrinsic tendency of the PAR-3 domain to spread in the absence of PAR-1.
Discussion
Zygotic polarity is stabilized by dynamic competition between anterior and posterior polarity proteins for occupancy of the cell surface. But a quantitative circuit level view of how this works has remained elusive. Here we identify a core circuit for polarity maintenance and show how dynamical clustering of CHIN-1 and PAR-3 can endow this circuit with bistable dynamics and a stably positioned boundary.
A core circuit for polarity maintenance
Previous models for polarity maintenance proposed a key role for PAR-2 in promoting local dissociation of anterior PAR proteins, and suggest that LGL-1 can serve this role when over-expressed in par-2 embryos. Here, we find that PAR-2 acts primarily during maintenance to prevent redistribution of anterior PAR proteins by posterior-directed cortical flows. PAR-2 restricts the domain of posterior contraction and flow to prevent overlap with the anterior PAR protein boundary. In par-2 mutants, LGL-1 plays a minor role in attenuating the magnitude of posterior flow and the posterior spread of PAR-6/PKC-3. However, absent maintenance phase contractility, neither PAR-2 nor LGL-1 are required to stabilize normal distributions of PAR-3 and PAR-6/PKC-3.
We have found that PAR-1 and CHIN-1 act redundantly, and independently of flow, to exclude PAR-6/PKC-3 from the posterior pole during maintenance phase. Using single molecule analysis, we find that PAR-6/PKC-3 asymmetries are not shaped by local dissociation, as previously proposed, but by asymmetrical recruitment (Figure 4D,E; see also Robin et al, 2014), which requires the local presence of both PAR-3 and active CDC-42. Neither high levels of active CDC-42 (in chin-1 embryos), nor the weak accumulation of PAR-3 (in par-1(RNAi) embryos) are sufficient to recruit PAR-6. However, in chin-1; par-1(RNAi) embryos, weak accumulation of PAR-3 is sufficient to promote rapid association of PAR-6/PKC-3 with CDC-42. One attractive possibility is that PAR-3 oligomers form a transient docking site for PAR-6/PKC-3 that enhances their probability to bind membrane-bound CDC-42, either through allosteric modulation or by inhibiting/displacing cytoplasmic factors that prevent PAR-6/PKC from binding to CDC-42 (Beers and Kemphues, 2006). Regardless of the details, this dual requirement allows either PAR-1 (by inhibiting PAR-3 (Motegi et al, 2011), or CHIN-1 (by inhibiting CDC-42 (Kumfer et al, 2010), to effectively inhibit local recruitment of PAR-6/PKC-3.
Synthesizing these results with previous work, we propose a core circuit for polarity maintenance (Figure 5E) in which (a) PAR-3 gates local association of PAR-6/PKC-3 with CDC-42, (b) PAR-6/PKC-3 inhibits local accumulation of PAR-1 (Motegi et al, 2011) and CHIN-1 clusters and (c) PAR-1 inhibits local accumulation of PAR-3 oligomers (Figure 4; Figure S3, Motegi et al, 2011), while CHIN-1 clusters locally inactivate CDC-42 (Kumfer et al, 2010), such that either are sufficient to prevent local recruitment of PAR-6/PKC-3. PAR-2 likely contributes by recruiting PAR-1 and by inhibiting the actions of PKC-3 towards itself and other targets (Griffin et al., 2011; Hao et al., 2006; Motegi et al., 2011) but these contributions are normally masked by redundant actions of CHIN-1. LGL-1 may also contribute by limiting overall levels of PAR-6/PKC-3 (Beatty et al, 2013), or their availability for recruitment by PAR-3 and CDC-42 (Wirtz-pietz and Knoblich, 2008). Whether the rescue of par-2 mutants by over-expressed LGL-1 relies on patterning cortical flow or these other contributions remains to be determined.
What are the advantages of this circuit design? Variants of this circuit polarize multiple cells using different cues during early C. elegans development (Anderson et al., 2008; Arata et al., 2010; Motegi et al., 2011; Munro and Bowerman, 2009; Munro et al., 2004). Using two redundant modes of cross-inhibition may provide a general way to preserve robust dynamic stabilization of cortical asymmetries as expression levels and/or activities of circuit elements vary with e.g. cell type, cell cycle progression and developmental stage. At the same time, a dual requirement for PAR-3 and CDC-42 to recruit PAR-6/PKC-3 allows a diversity of inputs to mediate symmetry breaking, such as local transport and/or inhibition of PAR-3 during zygotic symmetry breaking (Motegi et al, 2011) or cell contact-dependent inactivation of CDC-42 during radial symmetry breaking in later embryonic cells (Anderson et al., 2008).
Ultrasensitive dependence of CHIN-1 cluster growth on PAR-6/PKC-3 yields bistable dynamics
Our results provide strong quantitative evidence that ultrasensitive dependence of CHIN-1 cluster growth on PAR-6/PKC-3 endows the CDC-42/PAR-6/PKC-3/CHIN-1 sub-circuit, and thus the overall polarity circuit, with bistable dynamics. CHIN-1 clusters that move in cortical flow towards the anterior pole undergo a sharp transition from growth to decay at a particular AP position (Figure 6C–F), which corresponds to a particular steady state level of PAR-6/PKC-3 (Figure S5A) , and shifting the PAR-6/PKC-3 distribution towards the posterior pole produces a corresponding shift in the transition from growth to decay (Figure 2F, Figure 6I). Our mathematical analysis shows that ultrasensitive dependence of cluster growth on PAR-6/PKC-3, combined with an increase in membrane binding avidity with cluster size (Lemmon, 2008), and strong inhibition of CDC-42 by CHIN-1 (Figure S5B,C), is sufficient to endow the CDC-42/CHIN-1 sub-circuit with bistable dynamics (Figure 7B).
Dynamic clustering of membrane-associated proteins has been widely observed (Dodgson et al., 2013; McGill et al., 2009, Yap et al., 1997, Douglass and Vale, 2005; Himanen et al., 2007, Greenfield et al., 2009, Wu, 2013). A likely basis for clustering in many of these contexts is weak multivalent protein-protein interactions (reviewed in Banjade and Rosen, 2014; Wu, 2013), which can lead naturally to sharp transitions in cluster nucleation and growth above critical concentrations and/or binding affinities of participant molecules (Flory, 1953). Although the molecular basis for CHIN-1 cluster assembly is currently unknown, the kinetics of CHIN-1 cluster nucleation and growth (Figure 6A,B), and the sharp transition from growth to decay for individual clusters (Figure 6E), are consistent with a scenario in which CHIN-1 clusters form through multivalent association of membrane-bound subunits, with a critical concentration for cluster nucleation and growth. Ultrasensitive dependence on PAR-6/PKC-3 would arise naturally if PAR-6/PKC-3 modulated the critical concentration, or the concentration of subunits, or both. Importantly, however, the prediction of bistability in our mathematical model does not depend on these unknown details.
Although we have focused on CHIN-1, local inhibition of PAR-3 oligomerization by PAR-1 (Benton and St Johnston, 2003a), combined with increased avidity of PAR-3 binding with oligomer size, could also endow the PAR-3/PAR-1 sub-circuit with bistable dynamics in the presence of uniformly active CDC-42 (Supplementary Modeling procedures; see also Dawes and Munro, 2011). This possibility is consistent with the sharpness of the PAR-3 boundary in wild type and chin-1 embryos (Figure 4A and data not shown), the increase in mean PAR-3 cluster size on the posterior cortex of par-1 (RNAi) embryos (Figure S3D), and the failure of PAR-3 variants that lack a functional oligomerization domain to associate with the cortex (Li et al., 2010a). We suggest that ultrasensitive dependence of cluster growth and membrane binding avidity on opposing regulators may be a general mechanism to produce sharply delimited and stably polarized cortical domains from cross-inhibition of dynamically exchanging proteins.
Clustered polarity proteins couple PAR asymmetries to the actomyosin cortex
Our results suggest that dynamic coupling of polarity proteins to cortical flow plays a key role in shaping boundary position during maintenance phase. Changes in AP boundary position correlate strongly with flow in both wild type and various mutant (e.g. par-2, par-2; lgl-1, chin-1) embryos, while boundary positions are stable and essentially identical in the same backgrounds when MRCK-1 is also depleted (Figure 2A,D-G; Figure 3; Figure S2C, Figure S3F). Previous studies have proposed that physical advection of freely diffusing, and slowly exchanging, PAR proteins (e.g. PAR-6 and PAR-2) could explain their redistribution by cortical flow (Goehring et al., 2011b). However, this assumes a bulk flow of the membrane in which the PAR proteins are diffusing, which has not been observed in C. elegans or in other contexts where movements of cell-surface proteins couple to cortical flows (Kucik et al., 1990; Lee et al., 1990).
Based on our results, we favor an alternative model in which dynamically clustered polarity proteins PAR-3 and CHIN-1 provide the essential coupling between the zygotic polarity circuit and the actomyosin cortex. For both PAR-3 and CHIN-1, we observe sharply reduced mobility of clusters relative to single molecules and strong local coupling to cortical flows (Figure S6, Figure 6C,D). Because PAR-3 acts locally to gate recruitment of PAR-6/PKC-3, the redistribution of PAR-3 will necessarily force redistribution of PAR-6/PKC-3. Likewise, redistribution of CHIN-1 clusters will necessarily shape distributions of active CDC-42 and thus PAR-6/PKC-3 recruitment. Thus tight coupling of PAR-3 and CHIN-1 clusters to cortical flow could be sufficient to mediate redistribution of all members of the cross-inhibitory circuit, without a need for bulk membrane flow.
Quantitative support for this idea comes from our analysis of CDC-42/CHIN-1 sub-circuit dynamics in PAR-1-depleted embryos. Using empirically constrained values for protein mobility, exchange kinetics and strengths of cross-inhibition, our simulations predict an intrinsic tendency for PAR-6/PKC-3 to spread towards the posterior pole, which we observe experimentally in embryos doubly depleted of PAR-1 and MRCK-1, but not in embryos depleted of PAR-1 alone, which still exhibit anterior-directed cortical flows (Figure S2A,C). Our simulations suggest that cortical transport of CHIN-1 clusters at experimentally observed rates could be sufficient to counterbalance diffusive spread of PAR-6/PKC-3 and thus stabilize the AP boundary. Interestingly, actomyosin contractility is also required to prevent posterior spread of PAR-3::GFP in PAR-1-depleted embryos, which will also contribute to limiting the posterior spread of PAR-6/PKC-3 (Figure S6C).
Because core polarity circuit includes factors such as CDC-42 and PAR-2 that control the distribution of Myosin II, the movements of PAR-3 and CHIN-1 will not just depend on cortical flows; they will also feedback to pattern those flows. Thus ultimately, it will be necessary to understand how a unique and stable boundary position emerges from the interplay of reaction-diffusion, cortical transport, and the mechanical balance of cortical forces.
Experimental Procedures
C. elegans culture and strains
We cultured C. elegans strains under standard conditions (Brenner, 1974). See Supplementary Experimental Procedures, strains, for a list of mutations and transgenes used in this study. Unless otherwise specified, strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources.
RNA interference
We performed RNAi using the feeding method (Timmons et al., 2001). Unless otherwise specified, bacteria targeting specific genes were obtained from the library of (Kamath et al., 2003). See Supplementary Experimental Procedures for details.
Live Imaging
We mounted embryos for live imaging as previously described (Munro et al., 2004). For single molecule imaging experiments, we mounted embryos in egg salts containing ∼100 uniformly sized polystyrene beads (18.7 ± 0.03 µm diameter, Bangs labs, #NT29N) to achieve uniform compression of the embryo surface across experiments (Robin et al., 2014).
We collected spinning disc confocal images using a Nikon Ti-E inverted microscope equipped with solid state 50mW 481nm and 561nm Sapphire lasers (Coherent Inc), a Yokogawa CSU-X1 spinning disk scan head, and Rolera em-c2 and Andor iXon3 897 EM-CCD cameras. We collected near-TIRF images using an Olympus IX71 inverted microscope equipped with an Olympus OMAC TIRF illumination system, a 50-mW 481nm Sapphire laser (Coherent) and an Andor iXon3 897 EMCCD camera. See Supplementary Experimental Procedures for details of image acquisition.
Image analysis
We performed all image analysis in Matlab (www.mathworks.com).
Measuring cortical intensities of GFP::PAR-6 or GFP::wspGBD
We measured cortical intensities of GFP::PAR-6 and GFP::wspGBD from single equatorial confocal sections. We smoothed each image with a 4×4 mean filter and subtracted the mean background pixel intensity measured in a region outside the embryo. For each of 200 equally spaced boundary positions, we measured pixel intensities along a ray perpendicular to the boundary, directed inwards. We determined the position along the ray at which the smoothed and background-subtracted intensity first exceeded 90% of the value measured in posterior cytoplasm. We measured the mean intensity in the original image for 300nm along the ray beyond this position and then divided by the mean intensity measured over an additional 2µm along the same ray. We then binned these data to obtain average measurements at ten equally spaced positions along the AP axis. To control for variability of transgene expression in different genetic backgrounds, we normalized the intensity data for each embryo by the mean intensity within a region of posterior cytoplasm. See Supplementary Experimental Procedures for details.
Visualization and analysis of cortical flow
We produced kymographs using ImageJ (imagej.nih.gov/ij/). We measured cortical flow velocities for cortical NMY-2::GFP by Particle Image Velocimetry (PIV) using freely available Matlab software (www.oceanwave.jp/softwares/mpiv). See Supplementary Experimental Procedures for details.
Single particle detection and tracking
We performed all single particle detection and localization using a Matlab implementation (http://people.umass.edu/kilfoil/downloads.html) of the Crocker and Grier method (Crocker, 1996). This method identifies regions below a characteristic feature size in which the maximum pixel intensity exceeds a user-defined threshold and estimates their centroid positions to sub-pixel accuracy. We chose thresholds and feature sizes to optimize detection for different types of particles (single molecules and CHIN-1 or PAR-3 clusters; see Supplementary Experimental Procedures for details).
We performed particle-tracking analysis using freely available µTrack software (Jaqaman et al., 2008). µTrack first links particles frame-to-frame, then links these short segments into longer sequences. Both linking steps use statistical models for particle motion to compute costs for different possible linkage assignments (particle appearance, disappearance, displacement, fusion and fission), and then identify the assignments that globally minimize these costs. For all analyses reported here, we used a motion model provided with µTrack that represents a mixture of Brownian and directed motion. See Supplementary Experimental Procedures for details.
MSD analysis
To measure mean-square-displacement (MSD) for each lag time τ, we partitioned all particle trajectories into contiguous segments of length τ; then we averaged the MSD over all such segments. We only report data for values of τ with > 100 independent measurements. See Supplementary Experimental Procedures for details.
Quantitative analysis of PAR-3 and CHIN-1 clusters
To measure background-subtracted intensities for individual clusters, we measured the total pixel intensity (Itot) in a circular mask with radius equal to the feature size used for detection. We measured the background intensity (Ibg) as the average pixel intensity within an annular region, two pixels in width, surrounding this mask. Then we computed the background-subtracted intensity as:
where Amask is the area of the circular mask. To measure total intensity in detected clusters vs. AP position, we used ImageJ to trace the outlines of the region in which clusters were in focus at the embryo surface. We fit an ellipse to this outline and took its major axis to be the AP axis. We partitioned the embryo surface into 20 bins of equal width along this axis, assigned each detected particle into one of those bins on the basis of its position, then measured the mean intensity in each bin as the sum of all background-subtracted cluster intensities divided by the bin’s area.
To measure instantaneous cluster growth rates, we smoothed the intensity data for each cluster trajectory using a 5 frame moving average; then we measured the frame-to-frame differences in these smoothed intensities. We then measured mean growth rate vs. AP position as described above for mean intensities. See Supplementary Experimental Procedures for details.
Measuring GFP::PAR-6 single molecule recruitment rates
We tuned GFP::PAR-6 expression to single molecule levels and performed single molecule imaging, detection and tracking as described previously (Robin et al, 2014). We used imaging conditions (high signal: noise and low density of single molecules) that allowed accurate reconstruction of single molecule trajectories (Jaqaman et al, 2008, Robin et al, 2014; see Supplementary Experimental Procedures for details). We assumed that the beginning of each trajectory represents a single recruitment event. For each embryo, we collected recruitment events for 100 sec following the onset of maintenance phase, binned the data with respect to AP position as described above, then divided the total numbers in each bin by bin area and elapsed time to obtain plots of appearance rate vs. AP position. See Supplementary Experimental Procedures for details.
Mathematical and numerical analysis of the CDC-42/PAR-6/PKC-3/CHIN-1 circuit
We converted the network diagram in Figure 7a into a system of ordinary differential equations following basic assumptions that are outlined in the main text and further discussed in the Supplementary Modeling procedures. We scaled the model equations to facilitate direct comparison to experimental data. We used standard methods to perform steady state analysis and assess conditions for bistability in the absence of diffusion or cortical transport. To analyze spatiotemporal dynamics, we introduced terms representing diffusion (of CDC-42 and CDC-42/PAR-6/PKC-3) and transport (of CHIN-1). Then we invoked rotational symmetry of the ellipsoidal zygote to solve the resulting equations numerically on a 1D domain, representing a section of the cell surface, running posterior to anterior. See Supplementary Modeling Procedures for details.
Supplementary Material
Highlights.
Zygotic polarity is stabilized by two redundant cross-inhibitory feedback loops
PAR-1 opposes PAR-3 clusters; PAR-6/PKC-3 oppose CHIN-1 clusters
Threshold dependence of CHIN-1 cluster growth on PAR-6/PKC-3 yields bistable dynamics
Cortical transport of CHIN-1 and PAR-3 clusters stabilizes AP boundary position
Acknowledgements
We thank Michael Glotzer, Ken Kemphues, Kraig Kumfer and Geraldine Seydoux for strains, Ken Kemphues, Michael Glotzer and members of the Munro Lab for valuable discussions. This work was supported in part by NIGMS 1RO1GM098441. This paper is dedicated to Ken Kemphues for his many seminal contributions to this field and for his generous spirit.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
A.S., A.A. and Y.L. designed and performed experiments, analyzed data and helped to prepare figures. S.L. performed simulations. E.M. conceived, funded and supervised the study, conducted experiments, simulations and data analysis, and prepared figures and wrote the paper with help from A.S., A.A. and S.L.
References
- Aceto D, Beers M, Kemphues KJ. Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. 2006;299:386–397. doi: 10.1016/j.ydbio.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DC, Gill JS, Cinalli RM, Nance J. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science. 2008;320:1771–1774. doi: 10.1126/science.1156063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arata Y, Lee J-Y, Goldstein B, Sawa H. Extracellular control of PAR protein localization during asymmetric cell division in the C. elegans embryo. Development. 2010;137:3337–3345. doi: 10.1242/dev.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjade S, Rosen MK. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife. 2014:3. doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty A, Morton DG, Kemphues K. PAR-2, LGL-1 and the CDC-42 GAP CHIN-1 act in distinct pathways to maintain polarity in the C. elegans embryo. Development. 2013;140:2005–2014. doi: 10.1242/dev.088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty A, Morton D, Kemphues K. The C. elegans homolog of Drosophila Lethal giant larvae functions redundantly with PAR-2 to maintain polarity in the early embryo. 2010;137:3995–4004. doi: 10.1242/dev.056028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers M, Kemphues K. Depletion of the co-chaperone CDC-37 reveals two modes of PAR-6 cortical association in C. elegans embryos. 2006;133:3745–3754. doi: 10.1242/dev.02544. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol. 2003a;13:1330. doi: 10.1016/s0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003b;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Boyd L, Guo S, Levitan D, Stinchcomb DT. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos…. 1996;122:3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeks RJ, Canman JC, Gabriel WN, Meyer N, Strome S, Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Current Biology. 2004;14:851–862. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Crocker J. Methods of Digital Video Microscopy for Colloidal Studies. Journal of Colloid and Interface Science. 1996;179:298–310. [Google Scholar]
- Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes AT, Munro EM. PAR-3 Oligomerization May Provide an Actin-Independent Mechanism to Maintain Distinct Par Protein Domains in the Early Caenorhabditis elegans Embryo. Biophys. J. 2011;101:1412–1422. doi: 10.1016/j.bpj.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Meth. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson J, Chessel A, Yamamoto M, Vaggi F, Cox S, Rosten E, Albrecht D, Geymonat M, Csikasz-Nagy A, Sato M, et al. Spatial segregation of polarity factors into distinct cortical clusters is required for cell polarity control. Nat Commun. 2013;4:1834. doi: 10.1038/ncomms2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan L-N, Zhang M. The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. Embo J. 2007;26:2786–2796. doi: 10.1038/sj.emboj.7601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory PJ. Principles of Polymer Chemistry. Ithaca, NY: Cornell University Press; 1953. [Google Scholar]
- Goehring NW, Hoege C, Grill SW, Hyman AA. PAR proteins diffuse freely across the anterior-posterior boundary in polarized C. elegans embryos. 2011a;193:583–594. doi: 10.1083/jcb.201011094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Trong PK, Bois JS, Chowdhury D, Nicola EM, Hyman AA, Grill SW. Polarization of PAR proteins by advective triggering of a pattern-forming system. Science. 2011b;334:1137–1141. doi: 10.1126/science.1208619. [DOI] [PubMed] [Google Scholar]
- Gotta M, Abraham MC, Ahringer J. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr Biol. 2001;11:482–488. doi: 10.1016/s0960-9822(01)00142-7. [DOI] [PubMed] [Google Scholar]
- Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Odde DJ, Seydoux G. Regulation of the MEX-5 gradient by a spatially segregated kinase/phosphatase cycle. Cell. 2011;146:955–968. doi: 10.1016/j.cell.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Hao Y, Boyd L, Seydoux G. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev Cell. 2006;10:199–208. doi: 10.1016/j.devcel.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen J-P, Saha N, Nikolov DB. Cell-cell signaling via Eph receptors and ephrins. Current Opinion in Cell Biology. 2007;19:534–542. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Constantinescu A-T, Schwager A, Goehring NW, Kumar P, Hyman AA. LGL can partition the cortex of one-cell Caenorhabditis elegans embryos into two domains. Curr Biol. 2010;20:1296–1303. doi: 10.1016/j.cub.2010.05.061. [DOI] [PubMed] [Google Scholar]
- Hung TJ, Kemphues KJ. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development. 1999;126:127–135. doi: 10.1242/dev.126.1.127. [DOI] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Meth. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kay AJ, Hunter CP. CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Current Biology. 2001;11:474–481. doi: 10.1016/s0960-9822(01)00141-5. [DOI] [PubMed] [Google Scholar]
- Kucik DF, Elson EL, Sheetz MP. Cell migration does not produce membrane flow. The Journal of Cell Biology. 1990;111:1617–1622. doi: 10.1083/jcb.111.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumfer KT, Cook SJ, Squirrell JM, Eliceiri KW, Peel N, O’Connell KF, White JG. CGEF-1 and CHIN-1 regulate CDC-42 activity during asymmetric division in the Caenorhabditis elegans embryo. Mol Biol Cell. 2010;21:266–277. doi: 10.1091/mbc.E09-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Gustafsson M, Magnusson KE, Jacobson K. The direction of membrane lipid flow in locomoting polymorphonuclear leukocytes. Science. 1990;247:1229–1233. doi: 10.1126/science.2315695. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Li B, Kim H, Beers M, Kemphues K. Different domains of C. elegans PAR-3 are required at different times in development. 2010a;344:745–757. doi: 10.1016/j.ydbio.2010.05.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kim H, Aceto DG, Hung J, Aono S, Kemphues KJ. Binding to PKC-3, but not to PAR-3 or to a conventional PDZ domain ligand, is required for PAR-6 function in C. elegans. 2010b;340:88–98. doi: 10.1016/j.ydbio.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Bowerman B. Symmetry breaking in biology. Cold Spring Harbor Perspectives in Biology. 2010;2:a003475–a003475. doi: 10.1101/cshperspect.a003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, McKinley RFA, Harris TJC. Independent cadherin-catenin and Bazooka clusters interact to assemble adherens junctions. The Journal of Cell Biology. 2009;185:787–796. doi: 10.1083/jcb.200812146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Suzuki A, Hirose T, Kitamura K, Kutsuzawa K, Futaki M, Amano Y, Ohno S. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J Biol Chem. 2003;278:31240–31250. doi: 10.1074/jbc.M303593200. [DOI] [PubMed] [Google Scholar]
- Mori Y, Jilkine A, Edelstein-Keshet L. Wave-pinning and cell polarity from a bistable reaction-diffusion system. Biophys. J. 2008;94:3684–3697. doi: 10.1529/biophysj.107.120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F, Seydoux G. The PAR network: redundancy and robustness in a symmetry-breaking system. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2013;368:20130010–20130010. doi: 10.1098/rstb.2013.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F, Sugimoto A. Sequential functioning of the ECT-2 RhoGEF, RHO-1 and CDC-42 establishes cell polarity in Caenorhabditis elegans embryos. 2006;8:978–985. doi: 10.1038/ncb1459. [DOI] [PubMed] [Google Scholar]
- Motegi F, Zonies S, Hao Y, Cuenca AA, Griffin E, Seydoux G. Microtubules induce self-organization of polarized PAR domains in Caenorhabditis elegans zygotes. Nat. Cell Biol. 2011;13:1361–1367. doi: 10.1038/ncb2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Bowerman B. Cellular symmetry breaking during Caenorhabditis elegans development. Cold Spring Harbor Perspectives in Biology. 2009;1:a003400–a003400. doi: 10.1101/cshperspect.a003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Robin FB, McFadden WM, Yao B, Munro EM. Single-molecule analysis of cell surface dynamics in Caenorhabditis elegans embryos. Nat Meth. 2014;11:677–682. doi: 10.1038/nmeth.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L, Gönczy P. Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos. WormBook. 2014:1–43. doi: 10.1895/wormbook.1.30.2. [DOI] [PubMed] [Google Scholar]
- Schonegg S, Hyman AA. CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. 2006;133:3507–3516. doi: 10.1242/dev.02527. [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 1998;125:3607–3614. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Wirtz-Pietz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric cell division: Aurora A phosphorylates the PAR complex to regulate numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS, Brieher WM, Gumbiner BM. Molecular and functional analysis of cadherin-based adherens junctions. Annu Rev Cell Dev Biol. 1997;13:119–146. doi: 10.1146/annurev.cellbio.13.1.119. [DOI] [PubMed] [Google Scholar]
- Zonies S, Motegi F, Hao Y, Seydoux G. Symmetry breaking and polarization of the C. elegans zygote by the polarity protein PAR-2. Development. 2010;137:1669–1677. doi: 10.1242/dev.045823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.