ABSTRACT
Background: Non-human primates appear to represent the most faithful model of human disease, but to date the oral microbiome in macaques has not been fully characterized using next-generation sequencing.
Objective: In the present study, we characterized the clinical and microbiological features of naturally occurring periodontitis in non-human primates (Macaca mulatta).
Design: Clinical parameters of periodontitis including probing pocket depth (PD) and bleeding on probing (BOP) were measured in 40 adult macaques (7–22 yrs), at six sites per tooth. Subgingival plaque was collected from diseased and healthy sites, and subjected to 16S rDNA sequencing and identification at the species or higher taxon level.
Results: All macaques had mild periodontitis at minimum, with numerous sites of PD ≥ 4 mm and BOP. A subset (14/40) had moderate-severe disease, with >2 sites with PD ≥ 5mm, deeper mean PD, and more BOP. Animals with mild vs moderate-severe disease were identical in age, suggesting genetic heterogeneity. 16S rDNA sequencing revealed that all macaques had species that were identical to those in humans or closely related to human counterparts, including Porphyromonas gingivalis which was present in all animals. Diseased and healthy sites harboured distinct microbiomes; however there were no significant differences in the microbiomes in moderate-severe vs. mild periodontitis.
Conclusions: Naturally occurring periodontitis in older macaques closely resembles human adult periodontitis, thus validating a useful model to evaluate novel anti-microbial therapies.
KEYWORDS: oral microbiome, macaques, naturally occurring periodontitis, next-generation sequencing, 16S rRNA
Introduction
Periodontitis is a microbiome-driven disease that mainly affects older individuals and results in chronic immune activation, inflammation, soft tissue and bone destruction, and ultimately tooth loss. Accumulating evidence indicates that periodontitis contributes to the risk and severity of systemic conditions in which inflammation plays a key role, including type 2 diabetes, atherosclerosis and cardiovascular disease, pre-term birth, arthritis, and certain cancers [1–3]. The current preventive regimens and treatments for periodontitis are focused on removal of bacterial biofilms through oral hygiene measures and surgical procedures. Such approaches are sometimes ineffective, and are labour intensive, costly, and difficult to implement from a public health perspective.
Given the microbial aetiology of periodontitis, new anti-microbial agents and vaccines represent a logical strategy for disease prevention and to augment current treatment regimens. Appropriate pre-clinical models are imperative for developing these interventions. Although much information has been gained from studies of periodontitis in various animals including mice, rats, and dogs, the most faithful model of human disease remains the non-human primate [4–11]. Previous studies have demonstrated that non-human primates are closely similar to humans in terms of clinical presentation of disease [12], periodontal structures, host immune responses and to a limited extent the composition of the oral microbiome [4,13,14]. The microbiology of periodontitis in non-human primates has mainly been evaluated using closed end technologies that targeted putative pathogens, but did not comprehensively characterize the entire oral microbiome [14,15]. Furthermore, most studies have been limited to younger animals with ligature-induced versus naturally occurring disease.
In the present study, we characterized naturally occurring periodontitis in a population of older non-human primates (Macaca mulatta), from the clinical and microbiological perspectives. Microbiological analyses employed next-generation 16S rRNA sequencing in order to provide a systematic analysis of the oral microbiome in this model system.
Materials and methods
Animals and ethics statement
Rhesus monkeys (Macaca mulatta) in a breeding colony at the FIOCRUZ Primate Research Center in Manguinhos (Rio de Janeiro, Brazil) were used for all studies. Their care and maintenance have been described previously [16]. This research complied with all relevant national (CONCEA: Brazilian Government Council for Control of Animal Experimentation) and international guidelines for care and use of animals in research. All the procedures involving monkeys conformed to the recommendations of the Weatherall report for the use of non-human primates in research (http://www.acmedsci.ac.uk/images/project/nhpdownl.pdf). Animal experimentation was approved by the Institutional Ethical Review Board (CEUA-FIOCRUZ, resolution #P-55/12–6). To minimize suffering before interventions, including sampling or clinical procedures, animals were anaesthetized with ketamine hydrochloride 10 mg kg−1 (Cetamin, Synthec Vet, São Paulo, Brazil) (62.5 mg/kg)/xylazine (12.5 mg/kg), and midazolam 0.10 mg kg−1 (Dormonid, Farma-Roche, São Paulo, Brazil), injected intramuscularly.
Clinical characteristics of the M. mulatta population
The animals were outbred older adults of both sexes that did not receive regular oral hygiene interventions, all of which had at least mild periodontitis. To characterize the status of animals with respect to naturally occurring periodontitis, 40 animals were assessed by clinical examination. Periodontal measurements were recorded, including: pocket depth (PD) and bleeding on probing (BOP) at six sites per tooth (mesial/buccal, buccal, distal/buccal, distal/lingual, lingual, mesial/lingual). Sixteen posterior teeth per animal were evaluated including the first and second premolars and first and second molars. PD measurements (mm) were made by a trained periodontist using a North Carolina periodontal probe (UNC-15, Hu-Friedy, Chicago, IL); BOP was recorded as a dichotomous measure. A total of 3,720 sites were evaluated.
Clinical and demographic data (Table 1) entry was error-proofed by an investigator (A.P.V.C.), and transferred to an Excel spreadsheet. PPD and percentage of sites with BOP were averaged within each animal and then across groups. Animals were categorized into mild and moderate-severe periodontitis groups, based on the presence of more than two sites with PPD ≥ 5mm (Table 2).
Table 1.
Clinical characteristics of the M. mulatta colony.
| Mean ± SD | Range/animal | Percentage of animals | |
|---|---|---|---|
| Age (yr) | 14.1 ± 4.3 | 7 – 22 | |
| Bleeding on probing (% sites) | 51.2 ± 15.6 | 9.4 – 74.2 | 100 |
| Pocket depth (mm) | 2.48 ± 0.41 | 1 – 10 | |
| Sites (%) 1–3 mm | 89.3 ± 9.7 | 5.2 – 40.5 | 100 |
| 4 mm | 7.1 ± 4.2 | 2.1 – 18.8 | 100 |
| 5 mm | 1.8 ± 3.2 | 0 – 14.6 | 37.5 |
| 6 mm | 0.8 ± 1.7 | 0 – 7.3 | 27.5 |
| 7–10 mm | 1.0 ± 0.7 | 0 – 3.3 | 32.5 |
Table 2.
Clinical parameters of mild vs moderate-severe periodontitis.
| Group | n | Mean PD (mm) | BOP (%) | Age |
|---|---|---|---|---|
| Mild | 26 | 2.30 ± 0.15 | 44.5 ± 13.8 | 14.2 ± 3.6 |
| Moderate-severe | 14 | 2.93 ± 0.31a | 63.6 ± 9.9a | 13.9 ± 5.5 |
a p < 0.0001 vs mild (t test).
Sample collection
After periodontal screening, all sites presenting PD ≥ 4 mm, and four to six clinically healthy sites (PD ≤ 2 mm) in each animal were selected for microbiome sampling. Supragingival plaque was removed with sterile gauze and discarded, and subgingival plaque was taken from the selected periodontal sites with Gracey curettes (Hu-Friedy), pooled and suspended in TE buffer (50 mM Tris HCL, 1 mM EDTA pH 7.6), placed on ice, and stored at −80°C until further analysis. Whole genomic bacterial DNA was extracted and purified from the pooled subgingival microbiome samples using the MasterPure™ DNA purification kit (Epicentre®, Madison, WI [17].
16S rRNA cloning and sequencing
DNA isolated from plaque from all animals was PCR-amplified for the 16S rRNA gene using a universally conserved primer set and a primer set specific for Bacteriodetes taxa. Plaque from PCR positive animals was subjected to definitive taxonomic identification, by cloning, sequencing, and informatics analysis. Four libraries were constructed, two with broad range primers and two with Bacteroidetes-specific primers. 16S rRNA PCR amplification, cloning and sequencing were performed as previously described [17]. More than 80 clones were picked from each library, amplified, and sequenced, yielding 330 partial sequences. Partial sequences of approximately 800 bp each were edited, aligned, and checked for chimeric inserts. Identifications were made using the BLAST tool on the Human Oral Microbiome Database (HOMD, http://www.homd.org/) and confirmed using BLAST on NCBI, the Ribosomal Database Project (RDP), and an in-house database containing 16S rDNA sequences from the oral cavity of younger macaques [18].
Next-generation sequencing analysis
HOMINGS (Human Oral Microbial Identification by Next-Generation Sequencing) employs a ProbeSeq program for species detection with modifications as previously described [19]. Briefly, 50 ng of genomic DNA was used for each initial single round of PCR. Amplification of the 16S rRNA gene (V3–V4 region) was followed by purification and processing using a modified next-generation sequencing method as described [18] (MiSeq, Illumina, Inc., San Diego, CA). Species-specific 16S rRNA-based oligonucleotide probes were used in the BLAST program ProbeSeq for HOMINGS, to identify the frequency of oral bacterial species/taxa and frequency [19]. Partial matches were not considered as a match. Sequences that were not detected by a single species probe were subsequently processed against genus level probes (two or more species within the genus). All hits were accumulated by species/genus by animal. Chimeric sequences were not removed from analyses for this study. In a separate analysis, chimeric sequences ranged from 10 to 15% of the total reads. The relative proportions of detected taxa did not vary significantly (data not shown).
Statistical analyses
Differences in the clinical parameters between groups were evaluated by Student’s t test. Antibody levels were analysed by Spearman rank correlation tests. HOMINGS hits data (0–300,000 range) were converted into relative proportions, and species/genera total abundance per animal and site (diseased vs healthy) was determined. The Wilcoxon signed-rank test was used to determine the statistical significance of differences found between healthy and diseased sites in all 40 animals, and overall in the mild vs moderate-severe periodontitis subgroups. For this analysis, p < 0.05 in combination with an FDR value of 0 was considered statistically significant. To determine how bacterial community composition varied across samples, we compared microbial sequencing profiles for each sample using correspondence analysis (CoA), providing a statistical visualization for picturing the associations between the levels of a two-way contingency table [20]. MeV 4_8_1 [21] was used for statistical software and CoA analysis. Statistical analyses including outlier determinations were performed using XLSTAT-Pro (version 2014.4.06) and/or SAS Enterprise Guide® (version 6.1).
Results
Clinical characteristics of naturally occurring periodontitis in adult macaques
Forty adult M. mulatta monkeys were screened for clinical signs of naturally occurring periodontitis. Animals ranged in age from 7 to 22 years; 68% were females. Most animals (32/40) had a complete posterior dentition, with 96 sites measured per animal (six sites/tooth). Eight of 40 had one or more missing posterior teeth owing to undetermined causes.
As summarized in Table 1, all animals had at least some periodontitis, with numerous sites with probing PDs of 4 mm or greater, considered to represent significant periodontal pathology in this model [22]. A subset of macaques (14/40) had more extensive pathology, with more than two sites with PD ≥ 5 mm (mean ± SD: 9.0 ± 6.3 deep sites/animal), significantly greater mean pocket depth and sites with BOP, consistent with moderate to severe periodontitis (Table 2). Of note, the mild and moderate-severe disease groups were nearly identical in age, suggesting that naturally occurring periodontitis disproportionately affects a subset of susceptible macaques, similar to its clinical prevalence in humans [23].
Comparisons between M. mulatta and human microbiomes
We first evaluated the overall similarity of the macaque oral microbiome to that of humans. Based on sequence analysis of 330 16S rRNA clones from subgingival plaque from seven macaques, 56% of 108 bacterial taxa detected were identical or closely similar to human oral species, including those that are health-associated as well as putative periodontal pathogens (Figure 1). Health-associated species included many Streptococcus, Lactobacillus, and Gemella spp. Putative periodontal pathogens included Porphyromonas gingivalis, Tannerella forsythia, Filifactor alocis, Parvimonas micra, several Treponema phylotypes, Fusobacterium spp., Dialister invisus, a Desulfobulbus phylotype, and Aggregatibacter actinomycetemcomitans. Caries-associated species, namely Streptococcus mutans and Scardovia wiggsiae, were also present. Forty-eight species were unique to the macaque; however, all were clearly closely related to human counterparts (Tables 3 and 4).
Figure 1.

Cluster analysis of macaque and human oral microbiomes in health and disease.
Table 3.
Macaque-specific speciesa.
| Aggregatibacter actinomycetemcomitans |
| Aggregatibacter aphrophilus HOT_545 |
| Aggregatibacter segnis HOT_762 |
| Alloprevotella sp. HOT_473 |
| Bacteroidetes [G-3] sp. HOT_280 |
| Bacteroidetes [G-3] sp. HOT_503 |
| Bacteroidetes [G-5] sp. HOT_505 |
| Cardiobacterium valvulum |
| Catonella morbi HOT_165 |
| Catonella sp. HOT_451 |
| Centipeda periodontii HOT_726 |
| Granulicatella elegans |
| Haemophilus parainfluenzae HOT_718 |
| Lachnoanaerobaculum sp. HOT_083 |
| Lachnospiraceae [G-3] sp. HOT_100 |
| Leptotrichia buccalis |
| Neisseria meningitidis HOT_669 |
| Paenibacillus sp. HOT_786 |
| Parvimonas sp. HOT_393 |
| Peptococcus sp. HOT_167 |
| Peptostreptococcaceae [XI][G-4] sp. HOT_369 |
| Peptostreptococcaceae [XI][G-7] sp. HOT_106 |
| Peptostreptococcaceae [XIII][G-2] sp. HOT_790 |
| Porphyromonas endodontalis |
| Porphyromonas sp. HOT_277 |
| Porphyromonas sp. HOT_279 |
| Porphyromonas sp. HOT_284 |
| Prevotella intermedia |
| Prevotella sp. HOT_293 |
| Prevotella sp. HOT_820 |
| Sanguibacter keddieii |
| Selenomonas dianae |
| Selenomonas sp. HOT_126 |
| Selenomonas sp. HOT_892 |
| Selenomonas sputigena HOT_151 |
| Streptococcus anginosus HOT_543 |
| Streptococcus constellatus |
| Streptococcus gordonii HOT_622 |
| Streptococcus intermedius HOT_644 |
| Streptococcus peroris HOT_728 |
| Streptococcus sp. HOT_056 |
| Streptococcus sp. HOT_068 |
| TM7 [G-3] sp. HOT_351 |
| Treponema sp. HOT_237 |
| Treponema sp. HOT_518 |
| Veillonella denticariosi HOT_887 |
| Veillonella parvula HOT_161 |
| Veillonella sp. HOT_780 |
a16S rRNA sequences 95–99% identical to the human counterpart.
bred: putative periodontal pathogen.
Table 4.
Human species found in macaques.
| Atopobium sp. HOT_810 |
| Bacteroides heparinolyticus |
| Campylobacter gracilis HOT_623 |
| Capnocytophaga sp. HOT_324 |
| Clostridiales [F-1][G-2] sp. HOT_402 |
| Clostridiales [F-2][G-1] sp. HOT_075 |
| Clostridiales [F-2][G-2] sp. HOT_085 |
| Desulfobulbus sp. HOT_041 |
| Dialister invisus HOT_118 |
| Erysipelotrichaceae [G-1] sp. HOT_904 |
| Eubacterium [XI][G-1] infirmum |
| Eubacterium [XI][G-6] nodatum |
| Filifactor alocis |
| Fusobacterium nucleatum ss. polymorphum |
| Fusobacterium nucleatum ss. vincentii |
| Fusobacterium nucleatum ss. animalis |
| Fusobacterium nucleatum ss. nucleatum |
| Fusobacterium periodonticum |
| Fusobacterium sp. HOT_203 |
| Gemella haemolysans |
| Gemella morbillorum HOT_046 |
| Kingella oralis |
| Lachnoanaerobaculum orale |
| Lachnoanaerobaculum umeaense |
| Lachnospiraceae [G-2] sp. HOT_096 |
| Lachnospiraceae [G-5] sp. HOT_080 |
| Lactobacillus [XVII] catenaformis |
| Mitsuokella sp. HOT_521 |
| Mogibacterium timidum |
| Mycoplasma faucium HOT_606 |
| Oribacterium sp. HOT_102 |
| Oribacterium sp. HOT_372 |
| Parvimonas micra HOT_111 |
| Parvimonas sp. HOT_110 |
| Peptostreptococcaceae [XI][G-1] sp. HOT_383 |
| Peptostreptococcaceae [XI][G-2] sp. HOT_091 |
| Peptostreptococcus stomatis |
| Porphyromonas gingivalis HOT_619 |
| Porphyromonas sp. HOT_279 |
| Prevotella buccae HOT_560 |
| Prevotella denticola |
| Scardovia wiggsiae |
| Selenomonas infelix HOT_639 |
| Selenomonas sp. HOT_442 |
| Solobacterium moorei |
| SR1 [G-1] sp. HOT_345 |
| SR1 [G-1] sp. HOT_875 |
| Streptococcus cristatus |
| Streptococcus downei HOT_594 |
| Streptococcus mitis |
| Streptococcus mitis bv 2 HOT_398 |
| Streptococcus mutans |
| Streptococcus sp. HOT_071 |
| Syntrophomonadaceae [VIII][G-1] sp. HOT_435 |
| TM7 [G-1] sp. HOT_346 |
| Treponema lecithinolyticum HOT_653 |
| Treponema sp. HOT_238 |
| Treponema sp. HOT_250 |
| Veillonellaceae [G-1] sp. HOT_129 |
| Veillonellaceae [G-1] sp. HOT_132 |
a16S rRNA sequences 100% identical to the human counterpart.
bRed: putative periodontal pathogen.
Since P. gingivalis is of particular interest as a potential ‘keystone pathogen’ in periodontitis, we amplified DNA from subgingival sites using P. gingivalis-specific primers and found that all monkeys tested in a screen (50/50) were positive for this species. Several of these PCR products were subjected to more extensive sequence analysis, which showed that macaque P. gingivalis species were 100% identical to human species, based on sequencing of up to 300 bp of the 16S rRNA gene amplicon (data not shown).
Relationship of the macaque oral microbiome at diseased and healthy sites
Subgingival plaque sample pools were subjected to next-generation sequencing for the 16S rRNA gene. Samples from 24/40 monkeys gave amplified products by PCR sufficient for comprehensive sequence analysis. A minimum of 50,000 sequences were obtained from each sample, from both diseased and clinically healthy sites in the two disease severity groups (mild and moderate-severe). Sequences were identified in silico by reference to the HOMD using probeSeq [19]. Approximately 45% of the sequences were identified at the species level and 21% at the genus level in HOMD. The balance (34%) was not identified by reference to either HOMD or RDP databases.
The results revealed that 125/767 species were significantly different in diseased vs healthy sites in this adult macaque population, representing 16% of the total species/taxa identified. Seventy-eight were more prevalent in diseased sites and 47 were found at higher levels in healthy sites (see Table S1 for the complete list). Notable species higher in diseased sites included: Atopobium sp., Bacteroidetes sp. oral taxons 280, 281, 365, D. invisus, P. micra, numerous Prevotella spp., Streptococcus intermedius, TM7, and several species of Treponema. Species higher in health included: Actinomyces, Capnocytophaga, Campylobacter, Corynebacterium, Fusobacterium, Leptotrichia, and Propionibacterium spp. Of note, P. gingivalis was present in all subgingival samples, but relative proportions were not different in diseased vs healthy sites (mean levels: 0.32% of total). A CoA of the beta diversity of the microbiome based on profiles at diseased vs healthy sites shows a clear and highly significant separation (Figure 2).
Figure 2.
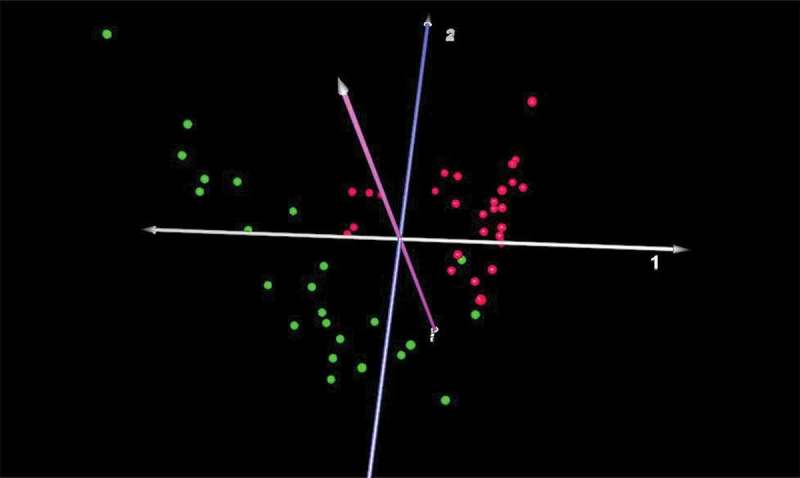
Correspondence analysis of microbiomes from healthy and diseased sites in macaques. Microbiomes present in healthy (green) vs. diseased (red) sites were clearly distinct (n = 24).
Animals were further sub-grouped on the basis of periodontitis severity as detailed above (mild vs moderate-severe), and the microbial profiles at healthy versus diseased sites in these animals analysed. As shown in Figure 3, four distinct clusters were present, albeit with considerable overlap between the microbiomes at both diseased and healthy sites. Comparisons of diseased or healthy sites in the mild vs moderate-severe disease groups did not reveal any statistically significant differences, indicating a high degree of concordance in microbiome composition that was independent of overall disease severity.
Figure 3.
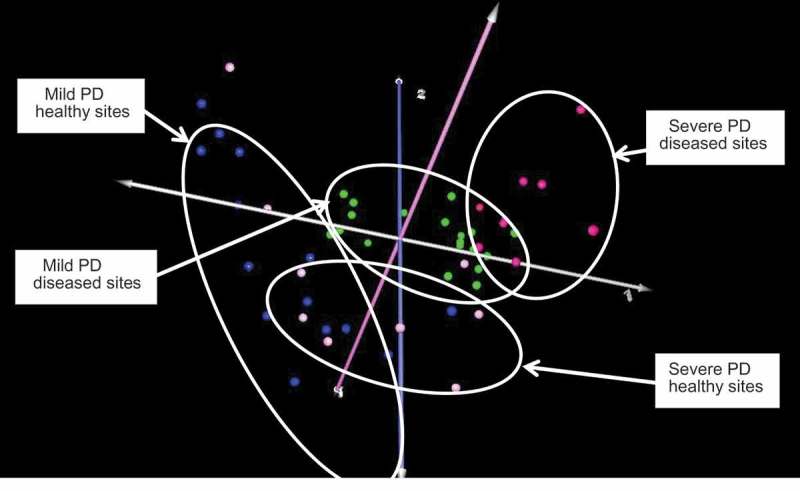
Correspondence analysis of healthy and diseased sites in mild or moderate-severe periodontitis. Key: mild periodontal disease (PD), healthy sites: blue; mild PD, diseased sites: green; moderate-severe PD, healthy sites: pink; moderate-severe PD, diseased sites: red. There was considerable overlap between healthy or diseased sites regardless of overall severity of periodontitis.
Discussion
Many animal models of periodontitis have been developed over the past several decades, which have been employed in studies to predict clinical outcomes in humans. For a model to be useful, it must closely resemble the clinical, structural, immunologic, and microbiologic features of the human disease. Whereas there is general agreement that non-human primates meet most of these criteria, there remain gaps in knowledge, in particular regarding the composition of the oral microbiome at sites of disease vs health. In the present study, we characterized naturally occurring chronic periodontitis in a colony of M. mulatta with respect to these parameters, with particular emphasis on the composition of the oral microbiome.
Our findings demonstrated that M. mulatta exhibits essentially all of the clinical features of chronic adult periodontitis, with a range of PDs and widespread inflammation as evidenced by BOP. The oral microbiome at subgingival sites in these animals was closely similar to that found in humans, with 56% of 108 bacterial taxa found to be identical or closely similar to human oral species. The balance of the microbiome consisted of unique macaque species that, nevertheless, were related to human counterparts. Disease-associated species included P. gingivalis, T. forsythia, F. alocis, P. micra, Treponema phylotypes, Fusobacterium spp., D. invisus, and A. actinomycetemcomitans. Of note, sequence analysis of macaque P. gingivalis revealed 100% identity to the human species P. gingivalis HOT-609. The macaque oral microbiome in periodontitis is thus very comparable with that of humans, possibly reflective of the co-evolution of the microbiome in genetically related hosts.
There was a clear segregation of species associated with diseased vs healthy sites (Figure 2) that was even more definitive than is typically seen in humans [24]. Prominent in disease were: Atopobium spp., Bacteroidetes spp. oral taxa 280, 281, 365, D. invisus, P. micra, Prevotella spp., S. intermedius, TM7, and several Treponema spp. Although several of the consensus ‘red complex’ periodontal pathogens were present in higher numbers at diseased compared with healthy sites, most of the periodontitis-associated organisms we have identified in macaques have recently been associated with human disease [25]. Taxa present in higher numbers in health included: Actinomyces, Capnocytophaga, Campylobacter, Corynebacterium, Fusobacterium, Leptotrichia, and Propionibacterium. Taken together, we conclude that the same range of periodontitis- and health-associated micro-organisms were present in the macaque as has been reported in humans.
Only one previous study employed next-generation sequencing to characterize the oral microbiome in macaques, albeit in Simian Immunodeficiency Virus (SIV) infection [18]. Prominent taxa in dental plaque in that study included: Streptococcus spp., Lachnospiraceae, G. morbillorum, Selenomonas, Filifactor, Peptostreptococcus, Clostridiales, Capnocytophaga spp., and Bacteriodetes spp. The periodontal status of animals was not assessed; however, Porphyromonas and Treponema were not found in plaque, and Tannerella was rare. The tongue microbiome was considerably altered in SIV infection, with high levels of G. morbillorum and reductions in Streptococcus. This microbial profile is distinct from that observed in the present study, likely owing to the modifying effects of SIV infection, age of the animals, and lack of significant periodontitis in that population.
We identified P. gingivalis in all animals at levels that were not significantly higher in diseased vs. healthy sites. This finding suggests that P. gingivalis might not be critical for periodontal pathogenesis in macaques. Indeed, the list of species that have been associated with periodontitis in humans has grown dramatically with the widespread adoption of next-generation sequencing [25]. However, alternative explanations may shed light on this observation. Although ‘healthy’ sites did not exhibit clinical measures of PD > 3 mm or BOP, the amount of plaque was elevated in virtually all sites, since animals did not receive mechanical plaque control, suggesting some degree of incipient inflammation and subclinical disease in most sites. It is also known from human studies that clinically healthy sites in diseased individuals usually harbour higher mean counts of pathogens than healthy sites of non-periodontitis subjects [26]. Another factor may relate to the metabolic activity of the microbiome. Recent studies of the metatranscriptome in progressing human periodontitis have revealed a dramatic lack of concordance between microbial profiles and their transcriptional activity, i.e. which species were metabolically active within the community [27–30]. Signature pathways associated with tissue destruction were identified, including cell motility, lipid A and peptidoglycan biosynthesis, and transport of iron, potassium, and amino acids [30]. Consensus periodontal pathogens T. forsythia and P. gingivalis upregulated various TonB-dependent receptors, peptidases, proteases, aerotolerance genes, iron transport genes, hemolysins, and CRISPR-associated genes. Such results indicate that gene-expression patterns may better define the disease state, compared with the simple composition of the microbiome. Community-wide transcriptomic studies are now needed to identify the most transcriptionally active species in macaques with periodontitis, as well as the pathways they express in disease vs health.
There were distinct subgroups within the macaque population with respect to disease severity, with about 35% of animals exhibiting moderate-severe periodontitis. Given that the mean ages of animals in these two subgroups were nearly identical, this suggests that there is heterogeneity among macaques with respect to susceptibility (Table 2), a picture previously observed in a retrospective study of this animal model [31]. The microbial challenge in the moderate-severe vs mild disease groups was not significantly different. Hence, the macaque disease can be viewed as representative of the ‘natural history’ of periodontitis in this species, with greater susceptibility likely due mainly to genetic factors. In humans, the summary effect of genetics on periodontitis, based on family and twins studies, has been estimated at approximately 50%, independent of smoking and other behavioural variables [32–34]. Of interest, these susceptibility findings are not dissimilar to studies in adult humans age 30 and older, conducted as part of the National Health and Nutrition Examination Survey (NHANES) [23,35]. As in the present study, NHANES also carried out full mouth examinations of six sites per tooth, to provide a comprehensive disease assessment; the prevalence of moderate and severe disease was found to be 8.9% and 30.0% respectively. The relationship of monkey to human age is approximately 0.3; thus, the population we have evaluated roughly corresponds to a mean age of 42 years in humans.
In summary, the present findings indicate a strong parallel between all of these factors, in naturally occurring periodontitis in M. mulatta, and chronic adult periodontitis in humans. We conclude that M. mulatta therefore represents an excellent model for human periodontitis, and for evaluating therapeutic regimens and agents for human disease.
Supplementary Material
Acknowledgements
This study was funded by Sanofi-Pasteur Inc., Cambridge, MA. The authors also acknowledge a capital infrastructure award from the Massachusetts Life Sciences Center, which supported the purchase of next-generation sequencing equipment and software for the Forsyth Salivary Diagnostics Center. The authors thank Dr Lina Faller for advice on informatics analyses.
Biographies
A. P. V. Colombo is Associate Professor, Institute of Microbiology at the Federal University of Rio de Janeiro (UFRJ), Head of the Oral Microbiology Lab and Head of the Medical Microbiology Department at the same Institution. She is a Professor and researcher in the postgraduate programs in Science (Microbiology) and Dentistry at UFRJ. Dr Colombo received a DMD (1990) and a certificate in Periodontology (1991) from the School of Dentistry at UFRJ. In 1997, she earned a DMSc in Oral Biology from Harvard School of Dental Medicine and a certificate in Microbiology from The Forsyth Institute. In 2007, she was a Visiting Scholar with Dr Bruce Paster at Forsyth. Her research focuses on the aetiology and pathogenesis of periodontal diseases and endodontic infections, particularly on characterizing the microbiome associated with the different forms of these infections, as well as testing the efficacy of alternative therapeutic approaches for treating these diseases. She also works on isolating and identifying pathogens associated with human infections at other body sites from the oral cavity.
B. J. Paster is presently Senior Member of the Staff, previously the director of the HOMINGS Oral Bacterial Identification Core, and director of the Sequencing Core Facility at The Forsyth Institute in Cambridge, Massachusetts. He is also Professor in the Department of Oral Medicine, Infection & Immunity, Harvard School of Dental Medicine and Professor II in the Dental Faculty in the University of Oslo. In 1975, he received a BS in Microbiology at the University of Rhode Island, in 1981, a PhD in Microbiology at the University of Massachusetts, Amherst, and, in 2014, an honorary PhD at the University of Oslo. He was a post-doc for Dr Carl Woese at the University of Illinois from 1981–1983 and later in 1983–1986 for Dr Ronald Gibbons at The Forsyth Institute.
G. Grimaldi is presently Emeritus Research Scientist at the Gonçalo Moniz Institute, FIOCRUZ Bahia (Salvador, Brazil). In 1972, he received an MD in Pathology at the Federal University of Bahia, and, in 1987, a PhD in Microbiology at the Federal University of Rio de Janeiro (Brazil). He was a post-doctoral fellow with Dr John David at the Harvard Medical School from 1981 to 1983 and later from 1986 to 1991 with Dr Diane McMahon-Pratt at the Yale University School of Medicine. At that time, his research was focused on studies employing monoclonal antibodies for the characterization and classification of leishmanial parasites from humans, wild mammals, and sand flies in the neotropical region. Dr Grimaldi’s current research is focused on the use of non-human primates as models for the study of human diseases, including immunological studies and drug and vaccine development against infectious diseases.
T. G. B. Lourenço is a postgraduate student in the Doctoral Programme in Science (Microbiology) at the Federal University of Rio de Janeiro (UFRJ). In 2011, she obtained a BS in Microbiology and Immunology, and in 2014 an MS in Science (Microbiology) from the Institute of Microbiology at UFRJ. She previously worked on determining the anti-microbial susceptibility of subgingival biofilms in periodontitis patients treated with antimicrobials, and on characterizing the microbiota of different forms of periodontal diseases by the Human Oral Microbial Identification Microarray. Currently, her research is focused on the association between oral and intestinal microbiomes in patients with periodontal diseases. In 2016, she was a visiting scholar at MIT, under the supervision of Dr Eric Alm.
A. Teva is currently Adjunct Professor at the National School of Public Health, FIOCRUZ Rio de Janeiro, and Medical School of Goytacazes, RJ (Brazil). Dr Teva received his Doctor-in-Science Degree (PhD) from the Oswaldo Cruz Institute/FIOCRUZ. His research is in the area of the immunology of infectious disease with special emphasis in vaccine and diagnostic development for leishmaniasis.
A. Campos-Neto is currently Director of DetectoGen Inc., Westborough, MA, and Adjunct Professor in the Department of Infectious Disease and Global Health at Cummings School of Veterinary Medicine, Tufts University. Dr Campos-Neto received his MD from the Medical School of Triangulo Mineiro, MG, Brazil, and his Doctor-in-Science Degree (PhD) from the Federal University of Rio de Janeiro, RJ, Brazil. He was a post-doc trainee at Harvard Medical School under the mentorship of Prof. Stuart Schlossman. His research is in the areas of immunology of infectious disease with special emphasis on vaccine and diagnostics development for leishmaniasis, tuberculosis, and Lyme disease.
J. McCluskey PhD, is Director, Bacteriology, US Research, Sanofi-Pasteur. Dr McCluskey has over 12 years’ industry research experience at Sanofi-Pasteur with a focus on developing vaccines and alternative therapeutic modalities against important human bacterial pathogens. She joined Sanofi-Pasteur in 2004 as an industrial postdoc at the French research site in Marcy l’Etoile before moving to the Toronto, Canada R&D site as a Senior Scientist, Bacteriology in 2006. In 2010, she relocated to Cambridge, USA to continue her career as a bacteriologist within the Cambridge R&D group. Here, she continued to focus on vaccine development against key bacterial pathogens. Between 2010 and the present day, she has significantly increased her scientific, managerial, and project leadership responsibilities and now holds a Director, Bacteriology position within the company’s R&D group. Her research interests have been in the field of vaccine development against a wide-ranging number of bacterial pathogens including, Streptococcus pneumonia, Neisseria meningitidis, Bordetella pertussis, Staphylococcus aureus, and Porphyromonas gingivalis. Prior to joining industry, Dr McCluskey completed two postdoctoral fellowships at the Universities of Edinburgh and Glasgow in the UK, both focusing on bacterial pathogens and their mechanisms of causing disease. She obtained her MSc and PhDs in Bacteriology from Universities of Glasgow and Leicester, respectively.
H. Kleanthous PhD, is Associate Vice President, U.S. Head of Research, Sanofi-Pasteur, Cambridge, MA. Dr Kleanthous has over 22 years’ industry experience in the research & development of recombinant live attenuated and subunit-based vaccines against viral and bacterial pathogens. He joined Sanofi-Pasteur as the North American Head of Discovery Research in 2008 with responsibility for evaluating and developing novel viral vaccine platforms and delivering novel targets to the Development pipeline. Previously, Dr Kleanthous was Vice President of Research at Acambis Inc., with responsibility for developing a new exploratory portfolio. His research interests have been in the field of replication-defective viral vaccine platforms, targeting Flaviviruses, Papilloma, and Herpes viruses, as well as their use for foreign antigen delivery. In recent years Dr Kleanthous has focused efforts on the research and development of Broadly Protective Influenza Vaccines. Prior to joining industry, Dr Kleanthous was a scientific investigator at academic teaching hospitals and the Central Public Health Laboratory Service in the UK, where he developed his expertise in the area of infectious diseases. He obtained his PhD in the field of Molecular Microbiology from the University of London.
T. E. Van Dyke is VP and Senior Member of Staff at the Forsyth Institute and Professor of Oral Medicine Infection and Immunity, Faculty of Medicine, Harvard University. He received a DDS (1973) from Case Western Reserve University, an MS (1979) and Periodontics Certificate (1980) from SUNY at Buffalo, PhD (1982) from SUNY at Buffalo, and an honorary Doctor Medicinae Dentaire degree from Justus Liebig University Giessen. He received the Balint Orban Memorial Prize in 1981, became a Diplomate of the American Board of Periodontology in 1989, received the IADR Award for Basic Research in Periodontology in 2001, the Norton Ross Award for Excellence in Clinical Research in 2002, and the William J. Gies Periodontology Award in 2008. His research focuses on the role of inflammation in oral and associated systemic diseases, and on the development of novel agonists that resolve inflammation.
P. Stashenko is Senior Member of Staff, Director of the Forsyth Host-Microbiome Center, and Associate Professor of Oral Medicine, Infection and Immunity at the Harvard School of Dental Medicine (HSDM). He served as Vice President for Research & Development, and subsequently as President and CEO of Forsyth from 2008 to 2016. Dr Stashenko received his BA in Biology from New York University, a DMD and certificate in endodontics from HSDM, and a PhD in immunology from Harvard University. His research has focused on: monoclonal antibodies against human CD20 and other B cell antigens; the osteoimmunology of inflammatory bone resorption in oral infections; and cross-talk between the oral microbiome and host immune response in infections and oral cancer. He was a member of several NIH study sections and the NIDCR/NIH National Advisory Council, and has been recognized as a top 5% recipient of NIH grants from 1985 to 2010.
Funding Statement
This work was funded by the Massachusetts Life Sciences Center Capital Infrastructure Program, and by Sanofi-Pasteur Inc., Cambridge, MA.
Disclosure statement
Drs J. McCluskey and H. Kleanthous are employees of Sanofi-Pasteur. No other conflicts of interest were reported by the authors.
Supplemental data
Supplemental data for this article can be accessed here.
References
- [1]. Han YW, Houcken W, Loos BG, et al. Periodontal disease, atherosclerosis, adverse pregnancy outcomes, and head-and-neck cancer. Adv Dent Res. 2014;26(1):47–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Kholy KE, Genco RJ, Van Dyke TE.. Oral infections and cardiovascular disease. Trends Endocrinol Metab. 2015;26(6):315–321. [DOI] [PubMed] [Google Scholar]
- [3]. Scannapieco FA, Cantos A. Oral inflammation and infection, and chronic medical diseases: implications for the elderly. Periodontol 2000. 2016;72(1):153–175. [DOI] [PubMed] [Google Scholar]
- [4]. Eke PI, Braswell L, Arnold R, et al. Sub-gingival microflora in Macaca mulatta species of rhesus monkey. J Periodontal Res. 1993;28:72–80. [DOI] [PubMed] [Google Scholar]
- [5]. Schou S, Holmstrup P, Kornman KS. Non-human primates used in studies of periodontal disease pathogenesis: a review of the literature. J Periodontol. 1993;64(6):497–508. [DOI] [PubMed] [Google Scholar]
- [6]. Madden TE, Caton JG. Animal models for periodontal disease. Methods Enzymol. 1994;235:106–119. Review. [DOI] [PubMed] [Google Scholar]
- [7]. Miller DR, Aufdemorte TB, Fox WC, et al. Periodontitis in the baboon: a potential model for human disease. J Periodontal Res. 1995;30:404–409. [DOI] [PubMed] [Google Scholar]
- [8]. Eke PI, Braswell LD, Fritz ME. Microbiota associated with experimental peri-implantitis and periodontitis in adult Macaca mulatta monkeys. J Periodontol. 1998;69:190–194. [DOI] [PubMed] [Google Scholar]
- [9]. Albuquerque C, Morinha F, Requicha J, et al. Canine periodontitis: the dog as an important model for periodontal studies. Vet J. 2012;191(3):299–305. [DOI] [PubMed] [Google Scholar]
- [10]. Abe T, Hajishengallis G. Optimization of the ligature-induced periodontitis model in mice. J Immunol Methods. 2013;394(1–2):49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Struillou X, Boutigny H, Soueidan A, et al. Experimental animal models in periodontology: a review. Open Dent J. 2010;29(4):37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. Review. [DOI] [PubMed] [Google Scholar]
- [13]. Oz HS, Puleo DA. Animal models for periodontal disease. J Biomed Biotechnol. 2011;2011:754857 Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Sela MN, Kornman KS, Ebersole JL, et al. Characterization of treponemes isolated from human and non-human primate periodontal pockets. Oral Microbiol Immunol. 1987;2(1):21–29. [DOI] [PubMed] [Google Scholar]
- [15]. Persson GR. Immune responses and vaccination against periodontal infections. J Clin Periodontol. 2005;32 Suppl 6:39–53. [DOI] [PubMed] [Google Scholar]
- [16]. Amaral VF, Ransatto VAO, Conceição-Silva F, et al. The Asian rhesus macaques (Macaca mulatta) as an experimental model for study of cutaneous leishmaniasis. Exp Parasitol. 1996;82(1):34–44. [DOI] [PubMed] [Google Scholar]
- [17]. Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183(12):3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Ocon S, Murphy C, Dang AT, et al. Transcription profiling reveals potential mechanisms of dysbiosis in the oral microbiome of rhesus macaques with chronic untreated SIV infection. PLoS One. 2013;8(11):e80863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Gomes BP, Berber VB, Kokaras AS, et al. Microbiomes of endodontic–periodontal lesions before and after chemomechanical preparation. J Endod. 2015;41(12):1975–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Belstrøm D, Holmstrup P, Bardow A, et al. Comparative analysis of bacterial profiles in unstimulated and stimulated saliva samples. J Oral Microbiol. 2016;16:30112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Saeed AI, Bhagabati NK, Braisted JC, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. [DOI] [PubMed] [Google Scholar]
- [22]. Sun HT, Zhang J, Hou N, et al. Spontaneous periodontitis is associated with metabolic syndrome in rhesus monkeys. Arch Oral Biol. 2014;59:386–392. [DOI] [PubMed] [Google Scholar]
- [23]. Eke PI, Dye BA, Wei L, et al. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J Periodontol. 2015;86(5):611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Belstrøm D, Paster BJ, Fiehn NE, et al. Salivary bacterial fingerprints of established oral disease revealed by the human oral microbe identification using next generation sequencing (HOMINGS) technique. J Oral Microbiol. 2016;8:30170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Pérez-Chaparro PJ, Gonçalves C, Figueiredo LC, et al. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res. 2014;93(9):846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Teles R, Sakellari D, Teles F, et al. Relationships among gingival crevicular fluid biomarkers, clinical parameters of periodontal disease, and the subgingival microbiota. J Periodontol. 2010;81(1):89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Benítez-Páez A, Belda-Ferre P, Simón-Soro A, et al. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genomics. 2014;15:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Jorth P, Turner KH, Gumus P, et al. Metatranscriptomics of the human oral microbiome during health and disease. mBio. 2014;5(2):e01012–e01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. Duran-Pinedo AE, Chen T, Teles R, et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 2014;8:1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Yost S, Duran-Pinedo AE, Teles R, et al. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 2015;7(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Gonzalez OA, Orraca L, Kensler TB, et al. Familial periodontal disease in the Cayo Santiago rhesus macaques. Am J Primatol. 2016;78:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Laine ML, Crielaard W, Loos BG. Genetic susceptibility to periodontitis. Periodontol 2000. 2012;58(1):37–68. [DOI] [PubMed] [Google Scholar]
- [33]. De Graaff J, Timmerman MF, Van der Weijden GA, et al. The effect of sibling relationship on the periodontal condition. J Clin Periodontol. 1993;9:683–690. [DOI] [PubMed] [Google Scholar]
- [34]. Michalowicz BS, Diehl SR, Gunsolley JC, et al. Evidence of a substantial genetic basis for risk of adult periodontitis. J Periodontol. 2000;71(11):1699–1707. [DOI] [PubMed] [Google Scholar]
- [35]. Eke PI, Dye BA, Wei L, et al. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91(10):914–920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


